Abstract
Identifying genomic alterations driving breast cancer is complicated by tumor diversity and genetic heterogeneity. Relevant mouse models are powerful for untangling this problem because such heterogeneity can be controlled. Inbred Chaos3 mice exhibit high levels of genomic instability leading to mammary tumors that have tumor gene expression profiles closely resembling mature human mammary luminal cell signatures. We genomically characterized mammary adenocarcinomas from these mice to identify cancer-causing genomic events that overlap common alterations in human breast cancer. Chaos3 tumors underwent recurrent copy number alterations (CNAs), particularly deletion of the RAS inhibitor Neurofibromin 1 (Nf1) in nearly all cases. These overlap with human CNAs including NF1, which is deleted or mutated in 27.7% of all breast carcinomas. Chaos3 mammary tumor cells exhibit RAS hyperactivation and increased sensitivity to RAS pathway inhibitors. These results indicate that spontaneous NF1 loss can drive breast cancer. This should be informative for treatment of the significant fraction of patients whose tumors bear NF1 mutations.
Keywords: breast cancer, mouse genetics, neurofibromin, NF1, genomic instability
TWIN and family studies indicate that only ∼25% of breast cancer cases have a heritable basis, and thus the majority (∼75%) appear to be “sporadic” (Lichtenstein et al. 2000). Hence, much effort is now being placed on genomic analysis of breast and other cancers, focusing on cancer genome alterations in addition to inherited genetic variation. Comprehensive large-scale studies have been, and are being conducted in an attempt to identify genes and pathways that are commonly altered in various cancers, and which may thus represent cancer “drivers” with causative roles. However, the prevalence of passenger mutations, genetic heterogeneity, and the diversity of tumor etiologies and subtypes complicates unequivocal identification of drivers. Therefore, validation of cancer drivers—especially novel ones—requires orthogonal lines of evidence and experimental confirmation. In this regard, mouse models can play an important role.
One putative cancer driver that has emerged is NF1 (Neurofibromin 1). Best known for causing the autosomal dominant genetic disorder neurofibromatosis type 1, cancer genome resequencing studies are finding evidence that NF1 is mutated (either by deletion or intragenic mutation) at significant rates in different cancers. NF1 is a negative regulator of the RAS oncogene. It stimulates the GTPase activity of RAS (and thus is a “RasGAP”), pushing it to the inactive GDP-bound state. NF1 is the third most prevalently mutated or deleted gene in glioblastoma multiforme (GBM) (The Cancer Genome Atlas Research Network 2008), one of the most significantly mutated genes in lung adenocarcinoma (Ding et al. 2008), and the fourth most (intragenically) mutated gene in ovarian carcinoma (The Cancer Genome Atlas Research Network 2011). Although NF1 alteration has not yet been implicated as a significant breast cancer driver, loss of heterozygosity (LOH) of NF1 has been noted in occasional cases (Guran and Safali 2005; Lee et al. 2010), as have intragenic mutations (Stephens et al. 2012). Women with neurofibromatosis type 1 (NF-1; a result of inheriting a mutant NF1 allele), have an increased risk of, or association with, breast cancer (Sharif et al. 2007; Salemis et al. 2010). Additionally, siRNA-mediated NF1 knockdown in epithelial-like breast cancer cells induced the expression of epithelial-to-mesenchymal transition-related transcription factors (Arima et al. 2009). However, it remains uncertain whether the NF1 mutations in these cases actually contribute to cancer etiology or maintenance.
Here, we took a comparative oncogenomic approach for breast cancer driver identification, exploiting the highly relevant mouse model C3H-Mcm4Chaos3/Chaos3. These mice bear a point mutation in the minichromosome maintenance 4 (Mcm4) gene that destabilizes the essential MCM2-7 replicative helicase. The resulting genomic instability (GIN) causes >80% of nulliparous females to develop mammary adenocarcinomas exclusively (Shima et al. 2007). The controlled genetic background and singular tumor etiology allows identification of recurrent mutational events likely to be involved in driving tumorigenesis. Strikingly, nearly all mammary tumors contained Nf1 deletions. Furthermore, examination of The Cancer Genome Atlas (TCGA) data revealed that over a quarter of all human breast carcinomas are missing at least one copy of NF1. These findings indicate a potentially prominent role for spontaneous NF1 aberrations in spontaneous breast cancers.
Materials and Methods
Mice
Chaos3 mammary tumors originated in mice congenic in C3HeB/FeJ except 16898. Chaos3 tumor 16898 and other Chaos3 tumors arose in a mixed C57BL/6J and C3HeB/FeJ background. MMTV-neu and PyVT mammary tumors occurred in the FvB background.
Microarray expression profiling
RNA was hybridized to custom murine Agilent microarrays and normalized as described (Herschkowitz et al. 2007, 2012). Data were deposited into the Gene Expression Omnibus (accession no. GSE36240). Chaos3 tumors were clustered in relation to other mouse models using an unsupervised analysis, and differentiation score was calculated as described (Herschkowitz et al. 2007; Prat et al. 2010). Significance analysis of microarray (SAM) results were used to define a Chaos3 gene signature (upregulated, FDR 0%), which was analyzed using the UNC337 human tumor data set (Prat et al. 2010). Genes significantly differentially expressed between Chaos3 tumors and those of other mouse models are presented in Supporting Information, File S1.
Partial exome resequencing
A custom mouse 5-Mb Sequence Capture array (NimbleGen) was used to enrich DNA corresponding to ∼1200 breast cancer candidate gene exons (File S2), followed by Illumina GAIIx sequencing. Candidate genes were selected and ranked based on breast cancer specificity and frequency in primary literature, existing cancer arrays, and cancer databases.
The sequence capture was performed as follows. Genomic DNA libraries of ∼200-bp fragment size were constructed for four Chaos3 mammary tumors and one inbred C3H WT spleen following the standard protocol of Illumina (San Diego). One microgram of tumor and control library DNA was hybridized to the 385 K or 720 K capture array using an X1 mixer on the NimbleGen Hybridization system (Roche-NimbleGen) at 42° for 3 days. Arrays were washed; then the captured molecules were eluted from the slides using a NimbleGen Elution Station. Eluted molecules were vacuum dried and amplified by ligation-mediated (LM)-PCR. Real-time PCR of eight control amplicons was performed in the precapture and postcapture library to estimate the target fold enrichment, which varied from 30- to 744-fold.
The read data from each sample were aligned to the mouse C57BL/6, NCBI Build 37 (mm9) reference sequence using Novoalign (http://novocraft.com, v 2.05, academic version). Default alignment settings were used, but nonuniquely mapped reads or reads failing on alignment quality were discarded (-r NONE -Q 9). The percentage of on-target reads for mutant samples ranged from 34.5 to 62.9%, reflecting a 230-fold average enrichment for the target breast cancer candidate genes. Genome Analysis Toolkit (GATK) version 1.04413 was used sequentially for base quality recalibration, depth of coverage estimation, variant calling, and variant evaluation (DePristo et al. 2011). Substitution variants discovery and genotyping were performed with the GATK Unified Genotyper across all samples simultaneously. Single sample SNP calling was used to complement joint-sample SNP calling. The raw SNP calls were filtered per GATK recommendations with standard hard filtering parameters or variant quality score recalibration (DePristo et al. 2011). Criterion required SNP loci to have ≥5× coverage, variant frequency in ≥25% of reads, missing bases <30%, no significant strand bias, and not overlapping indels. Indels were called with GATK IndelGenotyperV2 under both single sample and paired sample modes using C3H as the “normal” tissue to identify novel indels against C3H. No novel indels were identified in targeted coding regions. Known SNPs between C3H and C57BL/6J were mined from the Mouse Genome Database (http://www.informatics.jax.org/mgihome/projects/overview.shtml#snp), dbSNP (Sherry et al. 2001), and Sanger Mouse Genome Project (Keane et al. 2011) (http://www.sanger.ac.uk/resources/mouse/genomes/). There were 3,097 known C3H SNPs in seqcap target regions from traditional Sanger sequencing. GATK joint estimation from in-house data identified 2990 filtered SNPs, representing a 96.6% sensitivity. Known C3H SNPs were filtered out, and novel SNPs were identified for annotation and validation. Variation consequence was annotated with Ensembl Variation API (http://www.ensembl.org/info/docs/api/variation/index.html) and custom perl scripts. Binary Sequence Alignment/Map (BAM), Blue Elephant Definition (BED), and Variant Call Format (VCF) files were generated to visualize alignments and variations using the Integrative Genomics Viewer (IGV) software (Robinson et al. 2011). Variants were manually examined in IGV before proceeding to Sanger sequence validation as follows. Variant positions were amplified in corresponding tumor samples and inbred C3H control genomic DNA. Following Fast AP and Exo1 (Fermentas) treatment, PCR products were Sanger sequenced and analyzed using SeqMan. GeneCard, Ingenuity Pathway Tool, Biocarta, and KEGG databases were used to annotate genes.
Array comparative genomic hybridization
Five micrograms of genomic DNA from tumor and reference samples were labeled and hybridized to 3 × 720 K mouse Nimblegen CGH whole genome tiling arrays. The arrays consist of 50–75mer probes and a median spacing of 3.5 kb, with a subset of probes concentrated on exons. Two reference samples were used independently to ensure recurring CNAs were not artifacts caused by the reference sample. The first reference sample was collected from a C3H WT inbred mouse and run with tumor samples 2044b, 12351, and 12353. The second reference sample selected originated from a C3H congenic Chaos3+/+ mouse and run as the reference for the remaining samples. DNA labeling, hybridization, and posthybridization processing were performed according to the manufacturer’s protocol. Nimblegen software was used to normalize test/reference ratios and perform background correction. Copy number changes were identified and segmented with Nimblegen CGH-segMNT algorithm using unaveraged and 10× averaging windows. The significance threshold was set at ± 0.15 log2 ratio and required a minimum of two consecutive probes to exhibit a change in order to call a segment. Amplifications and deletions were visualized using Nimblegen software and confirmed by manually examining log2 ratios for regions of interest. In addition to using Nimblegen software, the normalized log2 ratio data were also analyzed using KCSmart software (Klijn et al. 2008) to identify significantly recurrent CNAs. The kernel width was 1 Mb, and the resolution of the sample point matrix was 5 Kb. Simple Bonferroni multiple testing correction P < = 0.025 was used as threshold for declaring significant regions. Select genes within CNAs were validated via qPCR. See File S3 for the primer list.
Human breast cancer data and CNA calls for comparison with Chaos3 CNAs were taken from the publicly available Cancer Genome Atlas portal (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp) 2010 update. The regions considered to have undergone segmental deletions by the publicly available Cancer Genome Atlas analysis (“level 4” data set) are those indicated in Figure 2B. The Memorial Sloan-Kettering Cancer Center cBio portal provides a breakdown by mammary tumor subtype for individual genes (http://www.cbioportal.org/public-portal/index.do). According to the cBioPortal data available as of May 2012, the genes between the NOS2 and NF1 interval (which were not classified as significantly segmentally deleted in the limited level 4 data set mentioned above), are hemizygously deleted at rates similar to NF1 itself. Critical regions within each Chaos3 CNA were identified as the region with the greatest overlap across multiple Chaos3 tumors.
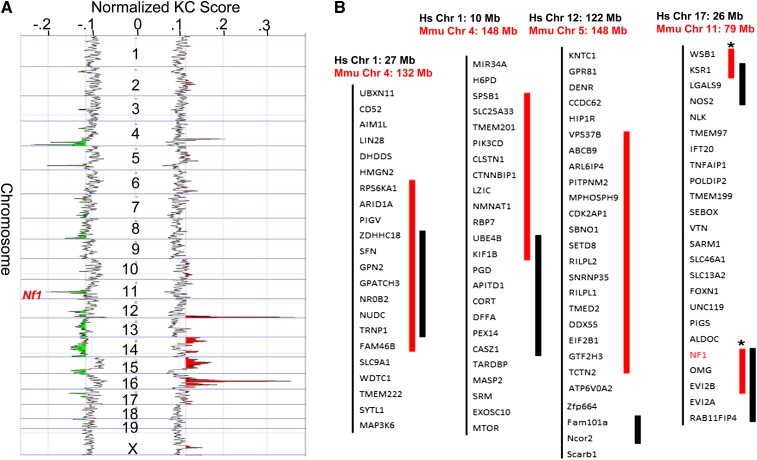
Figure 2 .
Recurrent CNAs in Chaos3 mammary tumors overlap with those in human breast cancer, including Nf1 deletion. (A) KCSmart analysis of combined aCGH data from 12 Chaos3 tumors, (nine mammary and three nonmammary). The most significant amplification peaks (red) lie on Chrs 12 and 16, and deletions (green) on Chrs 4, 5, and 11. (B) Overlap of mouse (Mmu) Chaos3 recurrent deletions with human (Hs) breast tumor CNAs. Human gene orders are shown. Thick red bars indicate the critical regions of mouse deletions, the subregions that, among all alterations in those regions, are common across all or most Chaos3 tumors. Of Chaos3 tumors with CNAs in these regions, the percentage of those containing the critical region is as follows: Chr 4 132 M = 86%; Chr 4 148 M = 71%; Chr 5 = 86%; Chr 11 = 86% (100% for Nf1, Ksr1, and Wsb1). The thick black bars to the right of the gene symbols indicate corresponding recurrent segmental CNAs in human breast cancers (limited level 4 data set from TCGA). Note that the Chr 11 deletions are single events in mice (asterisks on red part indicate these sequences are juxtaposed and contiguous in the mouse genome), and it is possible that the interval between NOS2 and NF1 may also be deleted as single events in human breast cancers, since the intervening genes are present in the hemizygous state in a high percentage of tumors according to extended TCGA data sets.
Cell culture experiments
Primary Chaos3 tumor biopsies were homogenized, cultured, treated with colcemid, and metaphase spreads were made (Shima et al. 2007). Imaged chromosomes were counted using ImageJ. Tumor cell lines were treated with the MEK1 inhibitor PD98059 or MTOR inhibitor rapamycin (Chang et al. 2007; Leong et al. 2010). Cell proliferation was assessed via 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma) and values read on a 96-well ELISA plate reader.
Active RAS pull-down and Western blotting
Levels of activated RAS were obtained using an active RAS pull-down kit (Thermo Scientific). Rabbit anti-NF1 (Novus Biologicals) was used at 2 μg/ml for Western analyses.
Results
Gene expression profiling reveals that the Mcm4Chaos3/Chaos3 mouse is a highly relevant luminal breast cancer model
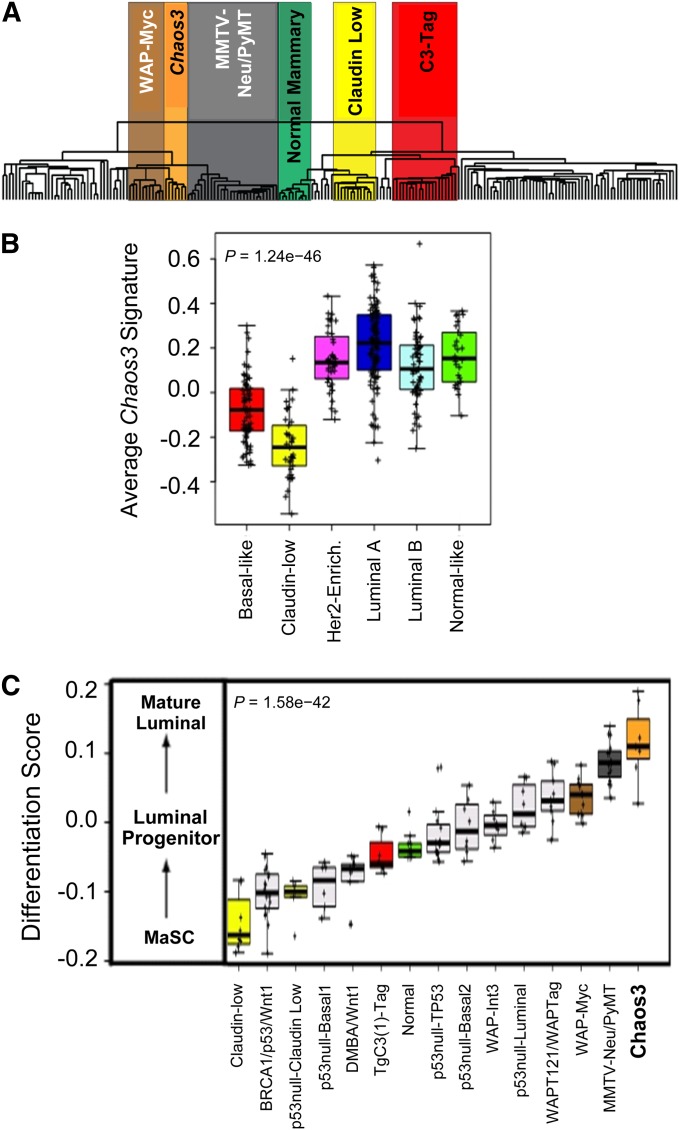
Human breast tumors can be classified into subtypes using gene expression signatures that are also conserved within mouse models of mammary cancers (Perou et al. 2000; Herschkowitz et al. 2007). Mcm4Chaos3/Chaos3 mammary tumors (referred to as “Chaos3”) are histologically classified as adenocarcinomas (Shima et al. 2007), and comparison with the 185 mouse mammary tumor data set of Herschkowitz et al. (2012) shows clustering near three luminal adenocarcinoma mouse models (Figure 1A). Consistent with this, the Chaos3 gene signature was most highly expressed in the human luminal A subtype, and was also high in HER2-enriched and luminal B tumors (Figure 1B). Luminal breast tumors are the most prevalent type in humans (Carey et al. 2006). SAM revealed that Chaos3 tumors have a distinct gene expression pattern from all other mouse models, including dramatic upregulation of Mucl1, a diagnostic marker in human breast cancer (Table S1) (Hube et al. 2004). Tumor differentiation score (D score) analysis showed that Chaos3 tumors more closely resemble mature human luminal cells than any mouse model analyzed to date (Figure 1C). Together, these results show that Chaos3 mice are an excellent human breast cancer model.
Figure 1 .
Chaos3 tumors model key human features. (A) Expression microarray dendrogram of Chaos3 mammary tumors and 185 other mouse mammary carcinomas and normal mammary tissue samples. The Chaos3 tumors cluster together as a distinct group near luminal murine models: MYC, PyMT, and Her2/Neu. Genes significantly differentially expressed between Chaos3 tumors and those of other mouse models are presented in File S1. (B) Boxplot of the Chaos3 gene signature in the UNC337 human breast tumor data set. Chaos3 tumors have higher signature expression in human luminal, HER2-enriched, and normal-like intrinsic subtypes. (C) Chaos3 differentiation score (D score) in relationship with other genetically engineered mouse models (GEMMs). The high D score shows that Chaos3 tumors more closely resemble the expression signature of mature human luminal cells relative to all other mouse models analyzed. (B and C) P-values reflect statistical significance of ANOVAs. MaSC, mammary stem cell.
Evidence that Chaos3 mammary tumors are driven by recurrent CNAs overlapping those common to human breast cancers
Primary Chaos3 cells have increased stalled replication forks that persist through metaphase, leading to chromosome breaks and improper chromosomal segregation (Shima et al. 2007; Kawabata et al. 2011a,b). Similar to human breast tumors (Hanahan and Weinberg 2011), Chaos3 tumors had high levels of aneuploidy and drastic variation in chromosome number, even within the cells of a single tumor (Figure S1). With such intratumor variation, we expect that only early and/or highly selected mutations would be readily detectable and highly recurrent across multiple cases. To uncover mutations potentially driving carcinogenesis in Chaos3 mice, we first performed partial exome resequencing of mammary tumors. Surprisingly, we discovered few somatic point mutations in the targeted exonic regions and calculated the mutation rate at 1.1 × 10−7, or 0.25 mutations/Mb, which is not above the background rate in other genomic studies of breast cancer (Greenman et al. 2007; Kan et al. 2010). The mutated genes are involved in diverse functions or are simply large (i.e., Titin), and together they do not implicate a commonly affected pathway underlying carcinogenesis (Table S1). These results indicate that elevated intragenic mutagenesis is not the primary mechanism driving Chaos3 carcinogenesis, and that other initiators such as CNAs may be responsible.
Saccharomyces cerevisiae containing the Chaos3 mutation have elevated translocations as well as segmental amplifications and deletions (Li et al. 2009). To examine genomic copy number changes, we performed array comparative genomic hybridization (aCGH) on 12 Chaos3 tumors (nine Chaos3 mammary and three nonmammary), and two MMTV-Neu mammary tumors. Chaos3 nonmammary tumors can be obtained by genetic perturbations or altering the strain background (Chuang et al. 2010; Kawabata et al. 2011b). Strikingly, the Chaos3 tumors exhibited highly recurrent CNAs in a small number of regions. Nearly all tumors had amplifications on chromosomes (Chr) 12 and 16 regardless of tumor type (Figure 2A; Table 1). Deletions on Chrs 5 and 11 were found in Chaos3 mammary tumors specifically, and there was a small commonly deleted region on Chr 4 that was shared by Chaos3 and MMTV-neu mammary tumors (Table 1). We screened TCGA human breast cancer genomic data and found commonly deleted regions overlapping the syntenic regions of recurrent Chaos3 deletions (Figure 2B, Table S2, and Table S3). Interestingly, the Chr 12 amplifications had precise breakpoints (Table 1) that flank a region enriched in immunoglobulin (Ig) gene fragments. While none of the six recurrently amplified regions were unique to Chaos3 mammary tumors, genes in these regions have roles in metastasis, pluripotency, signal transduction, or are upregulated in cancer (Table 2). The recurrently deleted regions contain several genes that overlap with human breast cancer CNAs and/or have potential or suggested roles in cancer: Ube4b (an ubiquitin ligase that negatively regulates Trp53) (Wu et al. 2011), Kif1b (a potential haploinsufficient tumor suppressor) (Munirajan et al. 2008), and Rad9 (deleted in three of the nine tumors, it is involved in the Ataxia telangiectasia and Rad3 related (ATR) DNA damage response pathway; mutation or misregulation of this gene is associated with various cancers, including breast) (Lieberman et al. 2011) (Table 2, Table S2, and Table S3).
Table 1 . Recurrent copy number alterations in Chaos3 tumors.
| Tumor/sample type | Sample name | Boundaries (Mb) |
Boundaries (Mb) |
Boundaries (Mb) |
Boundaries (Mb) |
Boundaries (Mb) |
Boundaries (Mb) |
Boundaries (Mb) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amplifications | Chr 16 45–54 | Chr 16 38–39 | Chr 12 114–117 | Chr 17 13–14 | Chr 17 77–78 | Chr 16 27M | |||||||||
| Chaos3 MT | 15259 | 45.8 | 50.4 | 38.8 | 39.6 | 114.5 | 117.1 | 13.4 | 13.7 | — | — | — | — | ||
| Chaos3 MT | 12351L | 45.8 | 50.5 | 38.8 | 39.0 | 114.7 | 117.1 | — | — | — | — | — | — | ||
| Chaos3 MT | 12353A | 45.0 | 50.5 | 38.8 | 39.0 | 114.7 | 117.1 | — | — | — | — | — | — | ||
| Chaos3 MT | 12115B | 44.9 | 53.9 | 38.8 | 39.5 | qPCR | qPCR | — | — | — | — | — | — | ||
| Chaos3 MT | 12352 | 45.1 | 49.6 | 38.8 | 39.0 | 114.7 | 117.1 | — | — | 77.5 | 78.0 | 27.6 | 27.7 | ||
| Chaos3 MT CL | 2044B | 45.8 | 50.5 | 38.8 | 39.5 | 114.6 | 117.1 | 13.3 | 13.7 | — | — | — | — | ||
| Chaos3 MT | 11929A | 45.8 | 49.7 | 38.8 | 39.6 | 114.7 | 117.1 | 13.2 | 13.7 | — | — | — | — | ||
| Chaos3 MT | 16168 | 47.3 | 47.3 | — | — | 114.7 | 117.1 | — | — | 78.1 | 78.1 | 27.6 | 27.7 | ||
| Chaos3 MT | 16898 | 46.3 | 49.3 | — | — | 114.7 | 117.1 | 13.2 | 13.6 | 76.3 | 78.5 | — | — | ||
| Chaos3 Mdl | 17883 | 45.1 | 49.6 | 38.8 | 39.6 | 114.7 | 117.1 | 13.3 | 13.6 | 77.2 | 78.4 | 27.6 | 27.7 | ||
| Chaos3 HS Uterus | 16862 | 45.8 | 50.4 | 38.8 | 39.6 | 114.7 | 115.1 | — | — | — | — | 27.6 | 28.0 | ||
| Chaos3 BT | 10658 | 45.8 | 48.6 | 38.8 | 39.6 | 114.7 | 117.1 | 3.5 | 27.5 | 67.2 | 95.3 | 27.6 | 27.7 | ||
| MMTV-Neu MT | 3750 | 45.8 | 49.3 | — | — | 115.2 | 117.1 | — | — | 76.4 | 78.9 | — | — | ||
| MMTV-Neu MT | 3744 | 46.3 | 49.3 | 38.8 | 39.6 | 114.8 | 117.2 | 13.2 | 13.7 | 77.6 | 78.5 | — | — | ||
| Deletions | Chr11 78–79 | Chr5 124–125 | Chr4 148–149 | Chr4 132–133 | Chr10 79–80 | Chr19 3–7 | Chr19 32–33 | ||||||||
| Chaos3 MT | 15259 | 78.9 | 79.6 | 124.3 | 125.1 | 148.6 | 149.6 | 132.7 | 133.3 | 79.9 | 80.3 | 4.0 | 4.5 | — | — |
| Chaos3 MT | 12351L | 78.5 | 79.7 | — | — | 149.1 | 149.3 | — | — | — | — | 3.9 | 4.6 | — | — |
| Chaos3 MT | 12353A | 78.1 | 79.6 | 123.5 | 125.1 | 147.6 | 150.1 | 133.0 | 134.5 | 78.1 | 78.9 | — | — | — | — |
| Chaos3 MT | 12115B | 78.9 | 79.4 | 122.7 | 125.3 | 148.6 | 149.4 | 132.9 | 133.8 | — | — | — | — | — | — |
| Chaos3 MT | 12352 | qPCR | qPCR | 124.5 | 125.1 | 148.1 | 149.1 | 131.9 | 132.2 | 80.6 | 80.7 | — | — | — | — |
| Chaos3 MT CL | 2044B | 78.6 | 79.6 | 123.8 | 125.1 | qPCR | qPCR | 133.0 | 134.5 | 79.8 | 80.3 | 4.2 | 4.8 | 33.1 | 33.5 |
| Chaos3 MT | 11929A | 78.9 | 79.3 | 124.3 | 125.3 | — | — | 133.2 | 133.6 | 79.0 | 81.0 | — | — | — | — |
| Chaos3 MT | 16168 | qPCR | qPCR | — | — | 148.1 | 149.5 | — | — | — | — | — | — | 32.2 | 32.6 |
| Chaos3 F1 MT | 16898 | 78.2 | 79.2 | 124.9 | 125.8 | 148.4 | 149.3 | 132.6 | 133.5 | 79.0 | 80.4 | 3.0 | 7.5 | — | — |
| Chaos3 Mdl | 17883 | — | — | — | — | — | — | — | — | — | — | — | — | 32.6 | 33.1 |
| Chaos3 HS Uterus | 16862 | — | — | — | — | — | — | 131.2 | 133.6 | 79.4 | 79.6 | — | — | — | — |
| Chaos3 BT | 10658 | — | — | — | — | — | — | 131.3 | 132.6 | — | — | — | — | — | — |
| MMTV-Neu MT | 3750 | — | — | — | — | 147.2 | 155.6 | 122.2 | 140.2 | 79.0 | 81.2 | 3.0 | 9.2 | 26.0 | 61.3 |
| MMTV-Neu MT | 3744 | — | — | — | — | 122.2 | 155.6 | 122.2 | 155.6 | 79.0 | 81.1 | — | — | — | — |
Recurring CNAs among 12 Chaos3 tumors and two MMTV-Neu mammary tumors identified by aCGH. The values are physical locations of the deleted or amplified regions according in the mouse genome sequence. In some cases, map locations were refined by qPCR as indicated. Samples analyzed are primary tumors except where indicated. MT, mammary tumor; CL, cell line; HS, histiocytic sarcoma; Mdl, mediastinal tumor; BT, bone tumor.
Table 2 . Cancer and immunity-related genes in Chaos3 mammary tumor CNAs.
| Amplified |
Deleted |
|||||||
|---|---|---|---|---|---|---|---|---|
| Function | Chr 16 | Chr 12 | Function | Chr 4 | Chr 5 | Chr 11 | Chr 10 | Chr 19 |
| Pluripotency | Dppa4, Dppa2 | Tumor suppressor | Cdk2ap1 | Nf1* | ||||
| Signal transduction | Adam6* | DNA checkpoint/repair | Kntc1, Gtf2h3, Setd8 | Rad9 | ||||
| Immunity/inflammation | Pvrl3, Retnlb, Retnla | Ig/abparts* | Apoptosis/necrosis | Dffa, Ube4b*Kif1b* | Oaz1* | |||
| Upregulated in cancer | Igsf11 | Signal transduction | Pik3cd | Ksr1* | Csnk1g2* Mknk2* | |||
| Immunity/inflammation | Lingo3* | |||||||
| Cancer related | Arid1a, Sfn* | Sbno1 | Minpp1* | |||||
The genes commonly altered specifically in mammary tumors are underlined, and those that additionally have CNAs in human breast cancers are marked with an asterisk. Ig/abparts, Ig locus and antibody parts gene feature conserved between mice and humans.
Nf1 is deleted in nearly all Chaos3 mammary tumors and a high proportion of human breast cancers
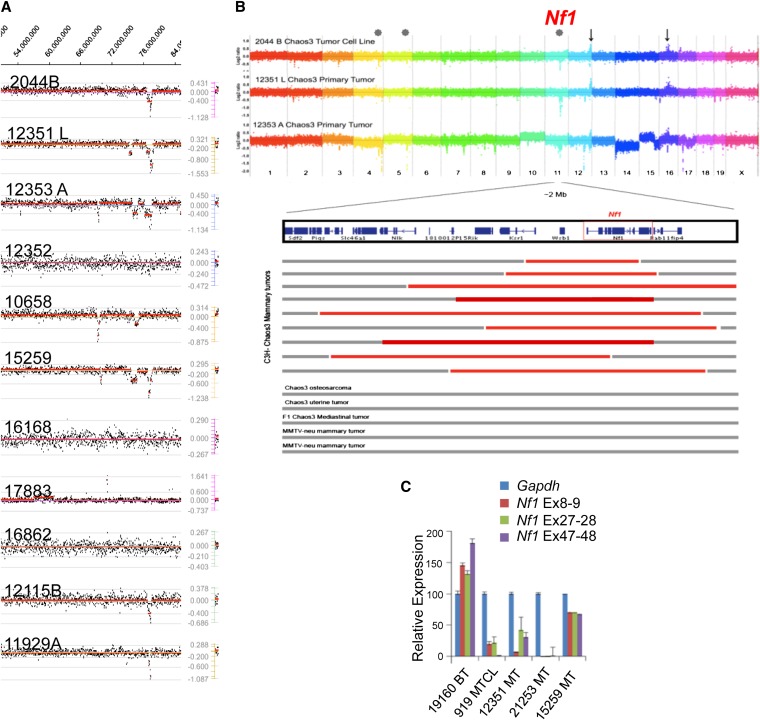
Particularly striking to us is the set of Chaos3 deletions on Chr 11 that overlaps with a recurring cluster of CNAs on human Chr 17. All Chaos3 mammary tumors examined by aCGH but none of the MMTV-Neu driven mammary tumors or Chaos3 nonmammary tumors contained Chr 11 deletions (Figure 3, A and B, Table 1, Table S2, and Table S3). The small deletions have nested breakpoints that define a commonly deleted region, and all of which contained the tumor suppressor Nf1 (Neurofibromin 1), Omg (oligodendrocyte myelin glycoprotein; this gene lies within an intron of Nf1), Wsb1 (WD repeat and SOCS box-containing protein 1), and Ksr1 (kinase suppressor of RAS) (Figure 3B). Except for Nf1, none have a plausible role as a mammary tumor suppressor. Omg is expressed primarily in neural tissues and is required for myelination in the central nervous system. Ksr1 promotes oncogenic RAS and MAPK signaling in mice and cells, so its deletion would be expected to actually inhibit tumor growth (Lozano et al. 2003; Goettel et al. 2011). WSB1 appears to participate in an E3 ubiquitin ligase complex not known to be associated with cancers. The gene Rab11fip4, which is deleted in many but not all of the Chaos3 mamary tumors, acts as a regulator of endocytic traffic but is expressed predominantly in retina and neural tissues, with little or no detectable expression in mouse or human mammary tissue (microarray data viewable at BioGPS.org and GeneCards.org).
Figure 3 .
Nf1 is deleted in Chaos3 mammary tumors. (A) Recurrent deletions detected by aCGH on Chr 11 at ∼79 Mb, specific to Chaos3 mammary tumors. The broken red line indicates significant log2 ratios. Tumor 17883 is a mediastinal lymphoma/leukemic tumor, 16862 is a histiocytic sarcoma in the uterus, 10658 is a bone tumor, and the other tumors are mammary. Mammary tumors 16168 and 12352 did not have significant detectable deletion by aCGH, but did by qPCR (Table S4). (B, Top) aCGH results of two primary Chaos3 mammary tumors and one Chaos3 mammary tumor cell line. Dots substantially above the log2 ratio line correspond to loci amplified in the tumor, and dots below are underrepresented. Arrows mark loci commonly amplified in Chaos3 tumors regardless of tumor type, and asterisks mark commonly deleted loci segregating specifically with mammary tumors. (Bottom) Expanded view of Chr 11 deletion. Red bars indicate aCGH or qPCR confirmed deletion in all nine Chaos3 mammary tumors overlapping Nf1. (C) qRT–PCR analysis of Nf1 mRNA levels across the transcript in Chaos3 tumors. Percentage of expression is relative to an MMTV-PyVT tumor as control, which does not have loss of Nf1. Error bars show standard error of the mean. Mammary tumor 15259 is heterozygously deleted for Nf1, and the others are homozygously deleted. Residual signal may reflect biopsy contamination or tumor heterogeneity.
These data and information led us further to explore the potential role of Nf1 as a driver of Chaos3 mammary tumors. We analyzed the DNA of the aCGH samples and additional Chaos3 mammary tumors by qPCR. Overall, 59/60 contained Nf1 deletions, with 51.6% appearing homozygous and 46.6% heterozygous (Table 3 and Table S4). Nf1-deleted tumors showed absence or severe reduction of mRNA and protein (Figures 3C and 4A). NF1 negatively regulates RAS, which controls proliferation, differentiation, cell adhesion, apoptosis, and cell migration through the MAPK and PI3K signal transduction pathways (Figure 4B). RAS proteins are often deregulated in cancers, leading to increased invasion and metastasis and decreased apoptosis (Pylayeva-Gupta et al. 2011).
Table 3 . qPCR analysis of deletions in Chaos3 mammary tumors.
| DNAs |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 15259 | 12351L | 12353A | 12352 | 2044B | 11929A | 16168 | 16898 | 12115B | WT | ||
| Chr 4 | Kif1b | 53.0 | 141.1 | 77.8 | 25.9 | 61.3 | 96.5 | ||||
| Pik3cd | 55.8 | 134.6 | 89.9 | 29.2 | 55.4 | 103.3 | |||||
| Eno1 | 58.7 | 87.3 | 105.3 | 39.3 | 51.5 | 108.4 | |||||
| Rere | 51.7 | 108.9 | 104.4 | 32.4 | 48.9 | 100.6 | |||||
| Chr 11 | Slc46a1 | 82.2 | 36.7 | ||||||||
| Tnfaip1 | 92.9 | 71.4 | 79.3 | 65.0 | 83.4 | 59.8 | |||||
| Nlk | 103.6 | 63.5 | 105.4 | 67.5 | 116.0 | 50.5 | |||||
| Nf1 (5′) | 17.4 | 35.5 | 25.5 | 12.7 | 63.7 | 16.7 | 9.2 | 9.6 | 31.2 | 107.3 | |
| Omg (Nf1 3′) | 52.9 | 12.0 | 16.7 | 0.7 | 48.0 | 16.3 | 75.1 | 38.7 | 109.4 | ||
| Rab11fip4 | 49.8 | 65.9 | |||||||||
qPCR values are presented in percentage of genomic DNA compared to C3H wild type. Boldfaced data indicate heterozygous or homozygous deletion (<80% control signal). Cancer-related genes, deleted at high frequency in mammary tumors specifically, are underlined. Nf1 deletion was validated at the 5′ and 3′ ends of the gene (Omg lies within an intron near the 3′ end of Nf1). Note that copy number differences between Nf1 5′ and 3′ are observed in some tumors, indicating a breakpoint within the Nf1 gene. Deletion calls were made as follows: Heterozygous = 15–80%; homozygous = <15%; if either Nf1 or Omg were <15%, the tumor sample was called as homozygously deleted (see text for overall summary; data for tumors not examined by aCGH are not shown here) because full Nf1 transcripts cannot be made. Nucleotide positions are from the mm9 mouse assembly. Sample 2044B is a tumor-derived cell line. WT, wild type.
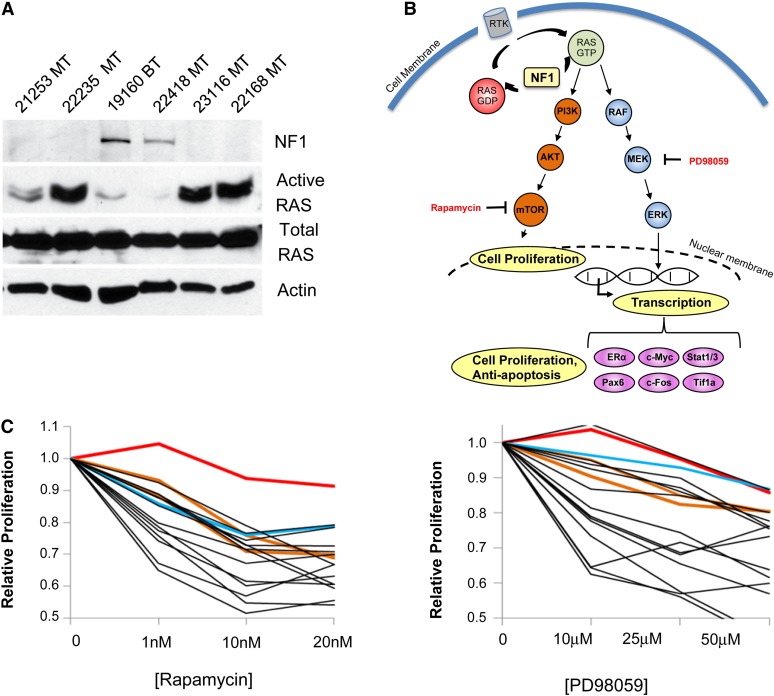
Figure 4 .
Nf1 deletion in Chaos3 mammary tumors leads to increased activated RAS and sensitivity to PI3K and MAPK inhibitors. (A) Western blot analysis of Chaos3 tumors for NF1 and active RAS levels. The mammary tumors without detectable NF1 have homozygous deletions of Nf1 (Table S4), whereas the bone tumor and mammary tumor 22418 contain no Nf1 deletions. The presence of NF1 protein is inversely correlated with the level of activated (GTP-bound RAS). (B) NF1 and the Ras pathway. NF1 loss leads to increased cell proliferation and transcription of antiapoptosis genes because of failure to negatively regulate Ras. Inhibitors used in this study to slow proliferation of NF1-deficient tumor cells are shown in red type. Not all downstream targets are shown. RTK, receptor tyrosine kinase. (C) Cell proliferation assays showing sensitivity of Chaos3 tumors to rapamycin and Mek1 inhibitor PD98059. Line colors: red, HeLa; brown, MCF-7 and MDA-MB231; blue, PyVT; and black, Chaos3. BT, bone tumor; MT, mammary tumor; MTCL, mammary tumor cell line.
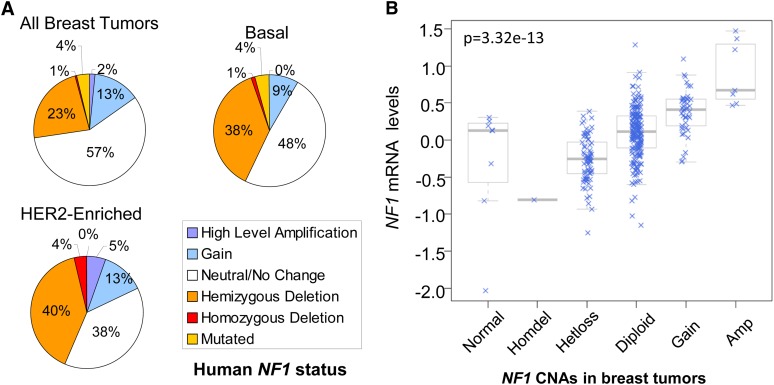
Best known for causing neurofibromas in the autosomal dominant genetic disorder neurofibromatosis type 1, women with inherited NF1 deficiency also have an increased risk of, or association with, breast cancer (Sharif et al. 2007; Salemis et al. 2010). Though there are few reports implicating spontaneous Nf1 loss in breast tumorigenesis (Guran and Safali 2005; Lee et al. 2010), upon screening TCGA breast cancer data sets we found that 27.7% of human breast tumors have NF1 deletions or mutations, most being heterozygous (Figure 5A and File S4). Furthermore, >40% of basal and HER2-enriched tumor subtypes have NF1 loss or mutations (Figure 5A). Genomic NF1 deficiency in human breast tumors significantly correlated with decreased expression levels (P = 3.32 × 10−13) (Figure 5B), both supporting the deletion calls and that the deletions impact NF1 levels.
Figure 5 .
Frequent NF1 deletion in human breast cancer. (A) Percentage of NF1 CNA and mutation in 511 human breast tumors, including 57 Her2-Enriched and 93 Basal breast tumors (publically available TCGA data). Note that 27.7% of human breast tumors have NF1 deletion or mutation, and HER2-enriched and basal breast tumor subtypes have ≥40% NF1 deletion or mutation. (B) Boxplot of NF1 mRNA expression vs. copy number in human breast cancer. Data are from TCGA Research Network. P-value is for ANOVA between het-loss and diploid groups, indicating expression levels significantly correlate with genomic deletion status.
Chaos3 mammary tumors have hyperactivated RAS
NF1 is a negative regulator of the RAS signaling pathway that stimulates the GTPase activity of RAS, pushing it to the inactive state. NF1 is important for negatively regulating the progrowth factor mTOR, which is stimulated by RAS (Figure 4B). Tumor cells of patients with acute myelogenous leukemia (AML) having NF1 deficiency demonstrate an elevated level of activated RAS and sensitivity to the mTOR inhibitor rapamycin (Parkin et al. 2010). To assess the functional impact of Nf1 deletion, we examined the level of activated RAS and found it to be dramatically higher in Chaos3 mammary tumor cells deleted for Nf1 (Figure 4A). We hypothesized that if the elevation of RAS signaling in Nf1-deleted mammary tumor cells is important for their maintenance, then inhibition of downstream pathways would compromise the growth of these cells. Chaos3 mammary tumor cell lines were markedly sensitive to MAPK/MEK1 and/or mTOR inhibitors, PD98059 and rapamycin, respectively (Figure 4C).
Discussion
Cancer genome resequencing studies are finding evidence that NF1 is mutated (either by deletion or intragenic mutation) at significant rates in different cancers. The two major studies published by TCGA Research Network on glioblastoma multiforme and ovarian carcinoma both found that NF1 and the PI3K/RAS pathway were mutated at significantly elevated rates (The Cancer Genome Atlas Research Network 2008, 2011). Though the TCGA Research Network has yet to publish its breast cancer genome data, the correlation between lower NF1 expression and copy number (as called by their implementation of the GISTIC algorithm) supports their calls of NF1 heterozygosity in the tumors they examined. Canonically, tumor suppressors are thought to require loss of both copies to have functional impact. However, there is accumulating evidence that haploinsufficiency or reduced expression of tumor suppressor genes can influence carcinogenesis (Berger et al. 2011). Indeed heterozygosity for Nf1 causes cancer-like cellular phenotypes in astrocytes (Gutmann et al. 1999; Bajenaru et al. 2001; Gutmann et al. 2001). We note that a recent cancer genome study did not report a high frequency of NF1 CNAs (Curtis et al. 2012). It is likely that the differences are related to different analytical methods of CNA calling and they highlight the need for follow-up analyses such as the qPCR we performed on the mouse tumors.
The mechanistic basis behind frequent deletion of NF1/Nf1 as opposed to intragenic mutation is an intriguing question that could yield insight into etiology of cancers with NF1 deletion. A combination of factors may contribute to NF1 CNAs, including fragile sites in the vicinity (Figure S2), a complex chromatin structure, and/or the gene’s large size. Replication fork stalling near Nf1 has been noted at a 5-kb isochore transition zone conserved between human and mouse, separating early and late replicating chromatin (Schmegner et al. 2005). Furthermore, collisions between replication and transcription complexes cause instability at fragile sites in the longest human genes (Helmrich et al. 2011). In the case of Chaos3 cancers, there may be a predisposition for replication fork stalling at particular genomic regions that are problematic for the destabilized Chaos3 helicase (Chuang et al. 2012), coupled with selective growth advantage conferred to cells by such mutations. Another curious issue has to do with strain specificity of Chaos3 mammary tumors, which occur on the C3H but not C57BL/6J background. One possible explanation may be related to differential DNA replication mechanics per se. Chaos3 cells have reduced Mcm2-7 mRNA and protein levels that lead to a decreased number of backup (“dormant”) replication origins (Kawabata et al. 2011a; Chuang et al. 2012), and these two strains differ in the density of licensed origins (Kawabata et al. 2011b). Backup origins are important for rescue of stalled or collapsed DNA replication forks that may otherwise lead to chromosomal aberrations (Blow et al. 2011). The Nf1 region may be particularly susceptible to stalled replication forks, and thus strain differences in backup origin density could have an impact. Alternatively, the two strains may have differential chromatin structure near Nf1 and/or other key tumor drivers in mammary epithelial cells, leading to an increased likelihood of CNAs in those regions. Finally, there may be genetic differences between the strains that confer biological resistance or susceptibility of an unknown nature. These differences would be mappable by standard genetic crosses, lending insight into heritable factors in breast cancer.
The C3H-Chaos3 mouse has important features that make it a highly relevant breast cancer model. First, because it is not genetically engineered to lack a tumor suppressor or express an oncogene, the cancers are driven by spontaneous events. Presumably, these events are mutations arising as a secondary consequence of the Mcm4Chaos3 genomic instability allele. Second, tumor differentiation score analyses of mRNA levels indicated that Chaos3 mammary tumors more closely resemble human luminal precursors than all other characterized mouse models. Finally, the secondary mutations arising in this model are primarily interstitial CNAs that are limited in number, relatively small in size, and often have nested breakpoints that refine the “critical regions” containing potential cancer driver genes. Together, the mouse and TCGA human data indicate that NF1 loss in conjunction with other CNAs is important for initiation and maintenance of mammary tumorigenesis in Chaos3 mice and a substantial subset of human patients. Among the other CNAs, Ube4b and Kif1b were frequently deleted in Chaos3 mammary tumors specifically, and human breast tumors also show frequent deletion (26%; Table 3, Table S2, and Figure S3). Genes in these and other recurrently altered regions are candidates for future studies of potential susceptibility genes underlying spontaneous or heritable forms of breast cancer.
Identification of NF1 as a potential tumor driver in a subset of breast cancers can provide guidance for patient treatment. First, suppression of the RAS pathway would be an appropriate target. However, loss or decrease of NF1 may trigger more than RAS pathway activation, as NF1 has been shown to bind to focal adhesion kinase (FAK) and has multiple isoforms of unknown functions (Kweh et al. 2009). Second, there is reason to believe that tamoxifen, the estrogen receptor (ER) inhibitor that is standard treatment for ER+ breast cancers, may not be appropriate for women whose cancers involve NF1 mutations. NF1 depletion was reported to confer resistance of human breast cancer (MCF7) cells to tamoxifen, and tamoxifen-treated patients whose tumors had lower NF1 expression levels had poorer clinical outcomes (Mendes-Pereira et al. 2012).
Based on cancer incidence estimates (Jemal et al. 2011; Siegel et al. 2012) and the frequency of NF1 deletions (Figure 5A), ∼63,450 patients in the United States and 383,230 worldwide will develop breast cancer with an NF1 deficiency annually. Our results demonstrate a consistent pattern of spontaneous CNAs associated with mouse and human mammary tumor carcinogenesis, particularly the importance of Nf1 deletion and provide a model for validation of these genes and drugs that target them.
Supplementary Material
Acknowledgments
We thank R. Weiss for providing the MMTV-Neu tumor samples and the MDA-MB231 cell line, R. Davisson for providing the MCF-7 and HeLa cell lines, A. Nikitin for continuous advice, the Cornell Genomics Core Facility for processing our genomic samples, and the Cornell Computational Biology Service Unit for consultation on data analysis. This study was supported by National Institutes of Health training grants IT32HDO57854 and 5T32GM007617 that supported M.D.W., Empire State Stem Cell Fund contract nos. C026442 and C024174 to J.C.S., and C.M.P. and A.D.P. were supported by National Cancer Institute Breast SPORE program (P50-CA58223-09A1), U24-CA143848, and by the Breast Cancer Research Foundation.
Footnotes
Communicating editor: D. W. Threadgill
Literature Cited
- Arima Y., Hayashi H., Kamata K., Goto T. M., Sasaki M., et al. , 2009. Decreased expression of neurofibromin contributes to epithelial-mesenchymal transition in neurofibromatosis type 1. Exp. Dermatol. 19: e136–e141 [DOI] [PubMed] [Google Scholar]
- Bajenaru M. L., Donahoe J., Corral T., Reilly K. M., Brophy S., et al. , 2001. Neurofibromatosis 1 (NF1) heterozygosity results in a cell-autonomous growth advantage for astrocytes. Glia 33: 314–323 [DOI] [PubMed] [Google Scholar]
- Berger A. H., Knudson A. G., Pandolfi P. P., 2011. A continuum model for tumour suppression. Nature 476: 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J. J., Ge X. Q., Jackson D. A., 2011. How dormant origins promote complete genome replication. Trends Biochem. Sci. 36: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L. A., Perou C. M., Livasy C. A., Dressler L. G., Cowan D., et al. , 2006. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295: 2492–2502 [DOI] [PubMed] [Google Scholar]
- Chang S. B., Miron P., Miron A., Iglehart J. D., 2007. Rapamycin inhibits proliferation of estrogen-receptor-positive breast cancer cells. J. Surg. Res. 138: 37–44 [DOI] [PubMed] [Google Scholar]
- Chuang C. H., Wallace M. D., Abratte C., Southard T., Schimenti J. C., 2010. Incremental genetic perturbations to MCM2–7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stress. PLoS Genet. 6: e1001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C. H., Yang D., Bai G., Freeland A., Pruitt S. C., et al. , 2012. Post-transcriptional homeostasis and regulation of MCM2–7 in mammalian cells. Nucleic Acids Res. 40: 4914–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C., Shah S. P., Chin S. F., Turashvili G., Rueda O. M., et al. , 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Getz G., Wheeler D. A., Mardis E. R., McLellan M. D., et al. , 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goettel J. A., Liang D., Hilliard V. C., Edelblum K. L., Broadus M. R., et al. , 2011. KSR1 is a functional protein kinase capable of serine autophosphorylation and direct phosphorylation of MEK1. Exp. Cell Res. 317: 452–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., et al. , 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guran S., Safali M., 2005. A case of neurofibromatosis and breast cancer: loss of heterozygosity of NF1 in breast cancer. Cancer Genet. Cytogenet. 156: 86–88 [DOI] [PubMed] [Google Scholar]
- Gutmann D. H., Loehr A., Zhang Y., Kim J., Henkemeyer M., et al. , 1999. Haploinsufficiency for the neurofibromatosis 1 (NF1) tumor suppressor results in increased astrocyte proliferation. Oncogene 18: 4450–4459 [DOI] [PubMed] [Google Scholar]
- Gutmann D. H., Wu Y. L., Hedrick N. M., Zhu Y., Guha A., et al. , 2001. Heterozygosity for the neurofibromatosis 1 (NF1) tumor suppressor results in abnormalities in cell attachment, spreading and motility in astrocytes. Hum. Mol. Genet. 10: 3009–3016 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A., 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Helmrich A., Ballarino M., Tora L., 2011. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44: 966–977 [DOI] [PubMed] [Google Scholar]
- Herschkowitz J. I., Simin K., Weigman V. J., Mikaelian I., Usary J., et al. , 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz J. I., Zhao W., Zhang M., Usary J., Murrow G., et al. , 2012. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc. Natl. Acad. Sci. USA 109: 2778–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube F., Mutawe M., Leygue E., Myal Y., 2004. Human small breast epithelial mucin: the promise of a new breast tumor biomarker. DNA Cell Biol. 23: 842–849 [DOI] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M. M., Ferlay J., Ward E., et al. , 2011. Global cancer statistics. CA Cancer J. Clin. 61: 69–90 [DOI] [PubMed] [Google Scholar]
- Kan Z., Jaiswal B. S., Stinson J., Janakiraman V., Bhatt D., et al. , 2010. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466: 869–873 [DOI] [PubMed] [Google Scholar]
- Kawabata T., Luebben S. W., Yamaguchi S., Ilves I., Matise I., et al. , 2011a Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell 41: 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T., Yamaguchi S., Buske T., Luebben S. W., Wallace M., et al. , 2011b A reduction of licensed origins reveals strain-specific replication dynamics in mice. Mamm. Genome 22: 506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klijn C., Holstege H., de Ridder J., Liu X., Reinders M., et al. , 2008. Identification of cancer genes using a statistical framework for multiexperiment analysis of nondiscretized array CGH data. Nucleic Acids Res. 36: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweh F., Zheng M., Kurenova E., Wallace M., Golubovskaya V., et al. , 2009. Neurofibromin physically interacts with the N-terminal domain of focal adhesion kinase. Mol. Carcinog. 48: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Wang J., Torbenson M., Lu Y., Liu Q. Z., et al. , 2010. Loss of SDHB and NF1 genes in a malignant phyllodes tumor of the breast as detected by oligo-array comparative genomic hybridization. Cancer Genet. Cytogenet. 196: 179–183 [DOI] [PubMed] [Google Scholar]
- Leong D. T., Lim J., Goh X., Pratap J., Pereira B. P., et al. , 2010. Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res. 12: R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. C., Schimenti J. C., Tye B. K., 2009. Aneuploidy and improved growth are coincident but not causal in a yeast cancer model. PLoS Biol. 7: e1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P., Holm N. V., Verkasalo P. K., Iliadou A., Kaprio J., et al. , 2000. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343: 78–85 [DOI] [PubMed] [Google Scholar]
- Lieberman H. B., Bernstock J. D., Broustas C. G., Hopkins K. M., Leloup C., et al. , 2011. The role of RAD9 in tumorigenesis. Mol. cell Biol. 3: 39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano J., Xing R., Cai Z., Jensen H. L., Trempus C., et al. , 2003. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Res. 63: 4232–4238 [PubMed] [Google Scholar]
- Mendes-Pereira A. M., Sims D., Dexter T., Fenwick K., Assiotis I., et al. , 2012. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc. Natl. Acad. Sci. USA 109: 2730–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munirajan A. K., Ando K., Mukai A., Takahashi M., Suenaga Y., et al. , 2008. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J. Biol. Chem. 283: 24426–24434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin B., Ouillette P., Wang Y., Liu Y., Wright W., et al. , 2010. NF1 inactivation in adult acute myelogenous leukemia. Clin. Cancer Res. 16: 4135–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C. M., Sorlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., et al. , 2000. Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Prat A., Parker J. S., Karginova O., Fan C., Livasy C., et al. , 2010. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 12: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D., 2011. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer 11: 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemis N. S., Nakos G., Sambaziotis D., Gourgiotis S., 2010. Breast cancer associated with type 1 neurofibromatosis. Breast Cancer 17: 306–309 [DOI] [PubMed] [Google Scholar]
- Schmegner C., Berger A., Vogel W., Hameister H., Assum G., 2005. An isochore transition zone in the NF1 gene region is a conserved landmark of chromosome structure and function. Genomics 86: 439–445 [DOI] [PubMed] [Google Scholar]
- Sharif S., Moran A., Huson S. M., Iddenden R., Shenton A., et al. , 2007. Women with neurofibromatosis 1 are at a moderately increased risk of developing breast cancer and should be considered for early screening. J. Med. Genet. 44: 481–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., et al. , 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N., Alcaraz A., Liachko I., Buske T. R., Andrews C. A., et al. , 2007. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 39: 93–98 [DOI] [PubMed] [Google Scholar]
- Siegel R., Naishadham D., Jemal A., 2012. Cancer statistics, 2012. CA Cancer J. Clin. 62: 10–29 [DOI] [PubMed] [Google Scholar]
- Stephens P. J., Tarpey P. S., Davies H., Van Loo P., Greenman C., et al. , 2012. The landscape of cancer genes and mutational processes in breast cancer. Nature 486: 400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network, 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network, 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Pomeroy S. L., Ferreira M., Teider N., Mariani J., et al. , 2011. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat. Med. 17: 347–355 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.