Abstract
During cell division, a bipolar array of microtubules forms the spindle through which the forces required for chromosome segregation are transmitted. Interestingly, the spindle as a whole is stable enough to support these forces even though it is composed of dynamic microtubules, which are constantly undergoing periods of growth and shrinkage. Indeed, the regulation of microtubule dynamics is essential to the integrity and function of the spindle. We show here that a member of an important class of microtubule-depolymerizing kinesins, KLP10A, is required for the proper organization of the acentrosomal meiotic spindle in Drosophila melanogaster oocytes. In the absence of KLP10A, microtubule length is not controlled, resulting in extraordinarily long and disorganized spindles. In addition, the interactions between chromosomes and spindle microtubules are disturbed and can result in the loss of contact. These results indicate that the regulation of microtubule dynamics through KLP10A plays a critical role in restricting the length and maintaining bipolarity of the acentrosomal meiotic spindle and in promoting the contacts that the chromosomes make with microtubules required for meiosis I segregation.
Keywords: meiosis, Drosophila, microtubule dynamics, kinesin-like protein, chromosome orientation
ACCURATE chromosome segregation during cell division is achieved through the interaction of chromosomes with a bipolar array of microtubules that constitutes the spindle. The spindle is a stable structure that regulates and directs chromosome movements, yet is composed of microtubules that are constantly going through phases of tubulin addition and removal from their ends, a behavior referred to as dynamic instability (Mitchison and Kirschner 1984). The regulation of microtubule dynamics, therefore, is crucial to the formation and function of the spindle. Changes in the dynamic behavior of spindle microtubules can result in chromosome instability (Bakhoum and Compton 2011).
It might be expected that the dynamic behavior of microtubules would have to be modified depending on the structure of the spindle. While most studies of spindle dynamics are carried out in mitotic cells, which contain microtubule-organizing centers known as centrosomes, during oogenesis in many species such as humans and Drosophila, the meiotic spindle assembles in the absence of centrosomes (Szollosi et al. 1972; Theurkauf and Hawley 1992; Albertson and Thomson 1993). During acentrosomal meiosis, microtubules accumulate around the chromosomes and extend outward to form two spindle poles, in contrast to the centrosomal spindle assembly typical of mitotic cell divisions in which microtubules emanating from centrosomes grow inward to make contact with the chromosomes. This suggests that there is an inherent difference in the organization and regulation of microtubules between acentrosomal and centrosomal spindles.
In addition, the meiotic divisions during oogenesis are an extreme case of asymmetric cell division, resulting in the formation of a large oocyte and much smaller polar bodies. To achieve this, the spindle is positioned asymmetrically within the oocyte, and the length of the meiotic spindle is constrained to a fraction of the size of the entire cell. Increase in spindle size results in the formation of a larger-than-normal first polar body during mouse oogenesis (Dumont et al. 2007), suggesting that control of spindle length is crucial to the integrity of this asymmetric cell division. In mitotic cells, spindle length typically correlates with cell size and is dependent on several mechanisms, including the regulation of microtubule dynamics and the centrosomes (reviewed in Goshima and Scholey 2010). Whether regulating microtubule dynamics contributes to spindle length in oocytes is not known, but one might predict it would have increased importance due to the absence of centrosomes.
An important class of proteins involved in the regulation of microtubule dynamics is the kinesin-13 family of microtubule-depolymerizing enzymes. Members of the kinesin-13 family play many roles during mitotic cell division, impacting spindle bipolarity and length, the correction of chromosome–spindle attachment errors, and chromosome movement during congression and segregation (reviewed in Moores and Milligan 2006). The Drosophila melanogaster genome encodes three kinesin-13 homologs: KLP10A, KLP59C, and KLP59D. All three Drosophila kinesin-13’s promote microtubule dynamics during mitosis, albeit in different capacities (Rogers et al. 2004; Rath et al. 2009), but only loss of KLP10A results in lengthening of the mitotic spindle (Rogers et al. 2004; Goshima et al. 2007). Mammalian genomes also encode three kinesin-13 family members that function in distinct ways; however, the three Drosophila kinesin-13’s are more closely related to each other than to the mammalian kinesin-13’s (Manning et al. 2007). This suggests that, while the function of kinesin-13’s may be conserved (Bakhoum and Compton 2011), the assignment of a function to a specific kinesin-13 cannot be based on sequence comparison alone, but rather requires a functional analysis of individual kinesin-13 family members within a species.
Kinesin-13’s have been studied extensively in mitosis and in vitro, but much less is known about their function during acentrosomal meiotic cell division. MCAK, a vertebrate kinesin-13 homolog, has been shown to promote chromosome alignment and silencing of the spindle assembly checkpoint during mouse oogenesis, but no effect on spindle organization was observed (Illingworth et al. 2010; Vogt et al. 2010). Expression of an N-terminal fragment of KLP10A during Drosophila oogenesis, on the other hand, results in the shortening of meiotic spindles (Zou et al. 2008). Although interpretation of this result is complicated because the nature of the defect caused by the fragment on native KLP10A function is not clear, it does implicate KLP10A in the control of meiotic spindle length.
To investigate the regulation of microtubule dynamics and spindle length during acentrosomal meiosis, we generated a deletion allele of Klp10A. In oocytes lacking KLP10A, we find that microtubules are dramatically longer, suggesting that KLP10A functions to depolymerize or destabilize microtubules during acentrosomal meiotic cell division and to regulate the length of the meiotic spindle. In addition, we find that loss of KLP10A has a profound impact on acentrosomal meiotic spindle organization, including a loss of contact between the chromosomes and microtubules. Consistent with this, we find that homologous chromosomes do not properly orient for segregation on the spindle in the absence of KLP10A. These results show that KLP10A, and microtubule depolymerization by inference, is crucial to the organization and function of the acentrosomal meiotic spindle.
Materials and Methods
Klp10A transgene construction
Full-length coding sequence of Klp10A was amplified by PCR from the LD29208 cDNA obtained from the Drosophila Genomics Resource Center (DGRC). The amplified sequence was subcloned into pENTR4 (Gateway System, Invitrogen) via restriction sites added to the 5′ ends of the PCR primers. An expression vector encoding full-length KLP10A fused to an N-terminal 3× HA tag under control of the UASp promoter was created by a Clonase LR reaction with the pPHW vector (DGRC). Transgene lines were established through germline transformation performed by Model Systems Genomics (Duke University, Durham, NC).
Cytology, immunofluorescence, and microscopy
For Klp10A germline mutant analysis, late-stage oocytes were prepared using formaldehyde/cacodylate fixation (McKim et al. 2009). Briefly, 100–300 mated females were fattened on yeast for 3–5 days then pulsed in a blender to disrupt abdomens. Late-stage oocytes were separated from bulk fly tissues and then fixed in an 8% formaldehyde/100 mM cacodylate solution. Chorion and vitelline membranes were removed by rolling between the frosted part of a glass slide and a coverslip. For standard immunofluorescence, rolled oocytes were extracted in PBS/1% Triton X-100 for 1.5–2 hr and blocked in PBS/0.1% Tween 20/0.5% BSA for 1 hr, and then antibodies were added. For FISH, rolled oocytes were stepped into 20, 40, and 50% formamide solutions, followed by 1–5 hr in 50% formamide at 37°. FISH probes were added and then oocytes were incubated at 91° for 3 min, followed by overnight at 37°. Oocytes were stepped out of formamide solution, blocked for 4 hr in 10% normal goat serum, and then antibodies were added.
With our standard fixation technique described above, we saw only weak localization to the poles of the meiotic spindle with a KLP10A transgene (Supporting Information, Figure S1), while we did not observe any localization with the anti-KLP10A antibody (data not shown). Therefore, an alternative fixation technique was used, which better preserved the KLP10A signal. Late-stage oocytes were prepared using a technique similar to our standard protocol described above, but with the substitution of formaldehyde/heptane fixation (Zou et al. 2008). KLP10A localization was also observed using a methanol-based fixation, although preservation of the spindle was poor (Figure S1).
Syncytial division stage embryos were prepared by dechorionating 1.5- to 2-hr-old embryos with 50% bleach for 90 sec followed by vitelline membrane removal and fixation by agitation in 50% heptane:50% methanol. Embryos were rehydrated into PBS followed by immunostaining using the same procedure as described above for “rolled” oocytes.
Primary antibodies used for immunofluorescence were mouse anti–α-tubulin conjugated to FITC (1:50 dilution, clone DM1A, Sigma), rat anti-HA High Affinity (1:25, clone 3F10, Roche), and rabbit anti-KLP10A (1:10,000, 656) (Rogers et al. 2004). Secondary antibodies used were goat antirat (1:100) and goat antirabbit (1:250) conjugated to Cy3 (Jackson Immunoresearch) and goat antirabbit conjugated to Alexa 488 (1:200, Molecular Probes). DNA was labeled with Hoechst 33342 (1:1000, Invitrogen) or TO-PRO-3 (1:1000, Invitrogen). FISH probes used were to the AACAC satellite (2nd chromosome) and dodeca satellite (3rd chromosome). Oligonucleotides were synthesized with either Cy3 (2nd) or Cy5 (3rd) conjugated to the 5′ end (Integrated DNA Technologies) and used at 100 ng per hybridization. Images were collected on a Leica TCS SP2 or SP5 confocal microscope with a ×63, N.A. 1.3 or 1.4 lens, respectively. Images are shown as maximum projections of complete image stacks with the exception of Figure 1A. The oocyte cortex displays a strong signal with the KLP10A antibody (data not shown); therefore, the sections closest to the cortex were not included in the maximum projection to allow visualization of KLP10A localization to the meiotic spindle.
Figure 1 .
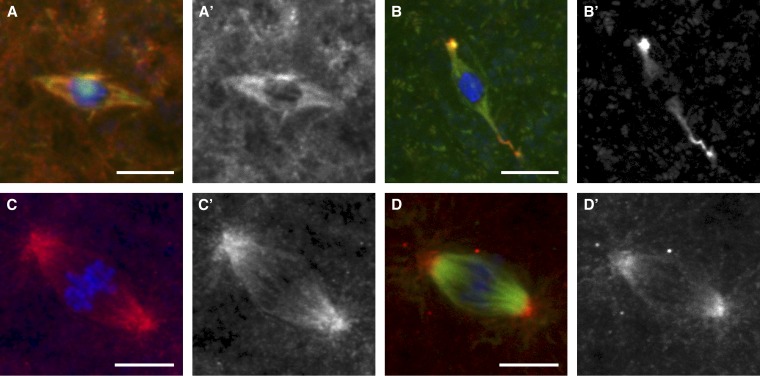
KLP10A localization in the female meiotic and embryonic mitotic spindles. Spindles from late-stage oocytes fixed with formaldehyde/heptane (A and B) and syncytial-stage embryos fixed in methanol (C and D) were examined for the localization of endogenous KLP10A (A and C) and HA-tagged KLP10A (B and D). The HA-tagged KLP10A (B and D) was expressed in a wild-type background. (A) In oocytes, endogenous KLP10A localized throughout the meiotic spindle. (B) HA-tagged KLP10A localized throughout the meiotic spindle, but was heavily concentrated at spindle poles. In addition, the “curly pole” phenotype caused by expression of the transgene is observable (see Figure S1). (C and D) In embryos, KLP10A primarily concentrates toward the spindle poles. Microtubules were not imaged in C. In all images, DNA is shown in blue and microtubules are shown in green. KLP10A is in red in merged images (A–D) and in white in single channel images (A′–D′). Bars, 5 μm.
Spindle lengths were measured by loading the Leica image stacks into Volocity image analysis software (Perkin Elmer, Waltham, MA). Microtubule endpoints were identified in three dimensions and the distance between them was determined.
P-element excision
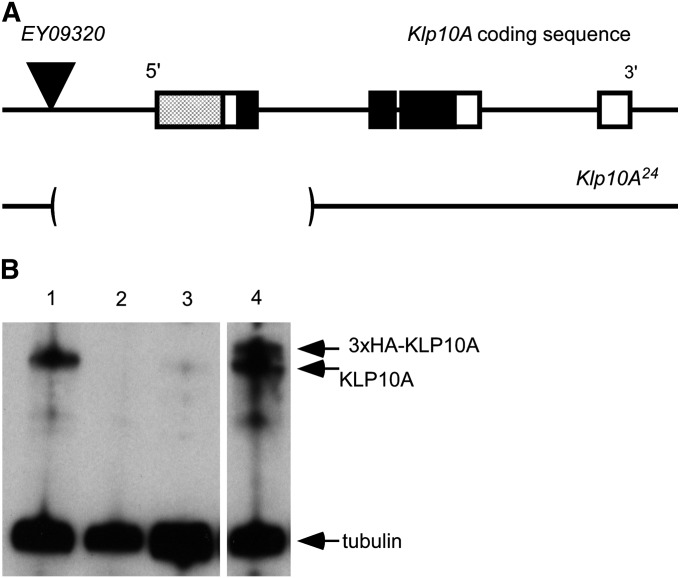
To create deletions of Klp10A coding sequence, we generated excisions of the P{EPgy2}EY09320 transposable element (Bloomington Stock Center), which is 1114 bp upstream of the start of Klp10A coding sequence. Because Klp10A is on the X chromosome, and we expected that deletions of Klp10A were likely to be homo- and hemizygous lethal, excisions were selected in heterozygous females. Excisions were screened for deletion of Klp10A coding sequence by PCR. DNA for PCR was prepared from adult flies for viable excisions as described (Gloor et al. 1993). Embryos homozygous for lethal excision chromosomes were selected over a GFP-tagged X chromosome balancer (Casso et al. 2000) and DNA for PCR was prepared by the same method as adult flies.
Western blotting
Protein samples were prepared by collecting stage 14 oocytes by the same method used for immunostaining, but instead of fixation, oocytes were weighed and SDS gel loading buffer was added to obtain a final concentration of 1 mg oocytes/8 μl total volume. The mixture was boiled for 5 min and 4 μl was loaded per lane on a SDS–PAGE gel. Primary antibodies used were rabbit anti-KLP10A (1:10,000,000) and rabbit anti–α-tubulin (1:5000, ab15246, Abcam). Secondary antibody used was goat rabbit anti-HRP (1:5000, Jackson Immunoresearch), detected using ECL Plus (Amersham).
Drosophila stocks and genetics
Flies were reared on standard media at 25°. Genetic loci not described in the text are described on FlyBase (FlyBase.org, Tweedie et al. 2009). To test whether the inviability of Klp10A24 mutants was due to the loss of KLP10A, we crossed y w Klp10A24/Bwinscy females heterozygous for a 2nd or 3rd chromosome insertion of the transgene encoding HA-tagged KLP10A to y w/y+Y; tubP-GAL4/TM3, Sb males. Progeny were scored for the presence of the Klp10A mutation (B+) and presence of the tubP-GAL4 driver (Sb+). Female B+ progeny are heterozygous for Klp10A24 and display no inviability relative to female B progeny (data not shown). Male B+ progeny are hemizygous for Klp10A24 and are inviable unless the tubP-GAL4 driver is present (data not shown). Although the Klp10A transgene carries a w+ marker, this could not be reliably scored in the presence of the w+ carried by tubP-GAL4. Because half of all male progeny receive the Klp10A transgene, percentage of viability was calculated as twice the number of B+ Sb+ males divided by B Sb+ males.
Klp10A germline mutants
The Klp10A24 allele was crossed onto a chromosome bearing an FLP recombination target (FRT) sequence inserted at 14A-B near the centromere of the X chromosome (FRT101, Bloomington Stock Center). Females with this recombinant chromosome (or a wild-type FRT chromosome for controls) were crossed in vials to males with a matching FRT chromosome carrying the dominant female sterile mutation ovoD1 and a heat-shock–inducible FLP recombinase. After 3–4 days, the parents were transferred to new vials and progeny were heat shocked in a 37° water bath for 1 hr. Females carrying both FRT chromosomes and the FLPase were selected among the progeny for examination as germline clones.
For RNAi depletion of KLP10A in the female germline, we obtained a fly stock from the Transgenic RNAi Project (TRiP, Harvard Medical School, Boston, MA) in which a Klp10A RNAi short hairpin (HMS00920) is under the control of the GAL4/UAS expression system. For RNAi depletion of Subito, we obtained short hairpin GL00583 from the TRiP project. The RNAi short hairpins were expressed in the germline using either the nanos-GAL4:VP16 or matα4-GAL-VP16 drivers (Rorth 1998; Sugimura and Lilly 2006).
Results
KLP10A localizes throughout the acentrosomal meiotic spindle at metaphase I
To gain insight into the function of Drosophila kinesin-13’s during meiosis, we analyzed the spindle localization of KLP10A. In Drosophila females, meiosis arrests at metaphase I late in oogenesis and does not resume until just prior to egg laying (King 1970). The localization of KLP10A on meiosis I spindles from late-stage Drosophila oocytes was analyzed using an anti-KLP10A antibody (Rogers et al. 2004) and by expression of HA-tagged KLP10A. To drive expression of HA-tagged KLP10A, a transgene was made under the control of the GAL4/UASP system (Brand and Perrimon 1993), and we used the nanos-GAL4:VP16 driver, which expresses throughout oogenesis (Rorth 1998).
Both endogenous KLP10A and HA-tagged KLP10A localized throughout the meiotic spindle at metaphase I (Figure 1, A and B), while HA-tagged KLP10A also concentrated toward the spindle poles, consistent with previous work in Drosophila oocytes (Zou et al. 2008). It is important to note that this localization pattern is dependent on the tissue fixation method (see Materials and Methods and Figure S1 for details). It might be expected that Klp10A transgene expression in a wild-type background would result in increased depolymerase activity and shorter spindles. This was not observed. Instead, we observed spindles with normal length but an abnormal “curly pole” phenotype (Figure 1B, Figure S1). A similar phenotype was observed by Zou et al. (2008) using a different Klp10A transgene. These authors concluded that KLP10A localizes to a discrete structure, the “spindle pole body” (Zou et al. 2008). Because we do not observe endogenous KLP10A in this localization pattern, we instead conclude that overexpression of KLP10A results in aberrant pole morphology and the accumulation of KLP10A. While we do not understand why overexpression of KLP10A causes abnormal spindle poles, these results suggest that minimum spindle length is regulated by additional factors.
In mitotic metaphase of the syncytial divisions of Drosophila embryogenesis, both endogenous KLP10A and HA-tagged KLP10A were more concentrated toward the spindle poles than in the oocytes (Figure 1, C and D), although we did not observe aberrant pole morphology. Because the localization of KLP10A differs between meiosis and mitosis, this suggests that the function of KLP10A during meiotic and mitotic cell division may not be identical. Alternatively, the different localization patterns may reflect the difference in spindle organization between acentrosomal meiotic and centrosomal mitotic spindles. In both the oocytes and embryos, however, we found no evidence for centromere localization of KLP10A, such as enrichment in foci on the chromosomes as observed in Drosophila S2 cells (Rogers et al. 2004).
Klp10A is an essential gene
There have been no previous studies of the Drosophila kinesin-13’s using loss-of-function mutations. We excised a P transposable element (EY09320) that is inserted 1114 bp upstream of the start of Klp10A coding sequence and screened by PCR for flanking deletions (Figure 2A). Several deletions were obtained, including one that we designated Klp10A24 in which 2742 bp of genomic sequence are deleted. By Western blot, we did not observe any full-length Klp10A protein expression in ovaries from the Klp10A24 germline mutants described below (Figure 2B). Because the KLP10A antibody was generated against the N terminus of the protein (Rogers et al. 2004), the coding sequence of which is deleted in the Klp10A24 mutant, we cannot be certain that a shortened form of KLP10A is not expressed in this mutant. This allele removes part of the kinesin motor domain coding sequence, however, making it likely that any shortened KLP10A that might be expressed would be nonfunctional.
Figure 2 .
Generation and characterization of Klp10A germline mutants. (A) Klp10A coding sequence is shown with boxes representing exons. The UTRs are not shown. The hatched box indicates the region encoding the portion of KLP10A used to raise the anti-KLP10A antibody (Rogers et al. 2004). The black box indicates the region encoding the motor domain of KLP10A. The P-element (EY09320) used to generate deletions of Klp10A coding sequence is depicted by a black triangle. The sequence deleted by the Klp10A24 allele is shown below with brackets surrounding the deleted region. (B) Western blot showing KLP10A expression in late-stage oocytes. Endogenous expression of full-length KLP10A is eliminated in Klp10A24 germline clones (lane 2) and severely knocked down in Klp10A RNAi (lane 3) compared to wild type (lane 1). HA-tagged KLP10A is expressed at levels comparable to endogenous KLP10A (lane 4). Tubulin serves as a loading control in all lanes.
Klp10A24 hemizygous mutants are inviable (Table 1), arresting development prior to the third larval instar stage (data not shown). This phenotype is consistent with the proposed function of KLP10A in mitotic cell division (Rogers et al. 2004). To test whether the lethal phenotype of the Klp10A24 mutation is due to loss of KLP10A, we expressed HA-tagged KLP10A using the ubiquitously expressed tubP-GAL4 driver (Lee and Luo 1999) in males hemizygous for Klp10A24 (Table 1). In the absence of transgene expression, either due to no transgene (Table 1) or no driver (data not shown), Klp10A24 males were inviable. In the presence of transgene expression, the inviability was rescued (Table 1). We tested several different lines in which the transgene is inserted at different locations in the genome and saw a high level of rescue in all lines (from 68 to 112%). This indicates that the inviability of Klp10A24 mutants is caused by the deletion of the Klp10A gene.
Table 1. Rescue of Klp10A24 inviability by Klp10A transgene expression with tubP-GAL4 driver.
| Klp10A transgene | % viability (n) |
|---|---|
| None | 0 (1088) |
| A | 100 (1192) |
| B | 92 (499) |
| C | 88 (960) |
| D | 68 (423) |
| E | 112 (1168) |
| F | 111 (676) |
| H | 92 (1050) |
y w Klp10A24/Bwinscy females heterozygous for a transgene encoding HA-tagged KLP10A were crossed to y w/y+Y; tubP-GAL4/TM3, Sb males. Percentage of viability was calculated as 2(B+ Sb+ males)/ B Sb+ males.
Early embryogenesis is disrupted in embryos from Klp10A germline mutants
Because Klp10A24 mutants are inviable, to study the role of KLP10A in meiosis and early embryogenesis, we employed two methods: (1) generating homozygous mutant germline cells through induced mitotic recombination in a heterozygous animal (germline clones) (Chou and Perrimon 1992) and (2) RNAi-mediated depletion in the germline (see Materials and Methods for details, Ni et al. 2011). We observed identical phenotypes using either method, consistent with both Klp10A24 and RNAi drastically reducing KLP10A protein levels in oocytes (Figure 2B). Therefore, we have combined the results and will refer to these experiments collectively as Klp10A germline mutants. In Klp10A germline mutants, oogenesis is completed and eggs are successfully laid; however, the embryos fail to hatch into larvae. To determine when the block in Klp10A germline mutant development occurs, we examined embryos cytologically.
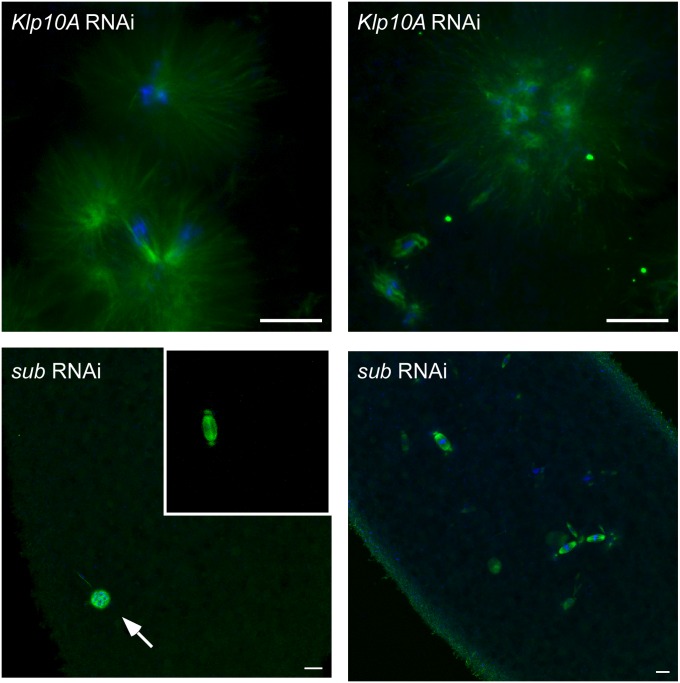
In wild-type embryos, the male and female pronuclei fuse and immediately begin synchronized syncytial nuclear divisions (see Figure 1, C and D, for example). In contrast, no structures identifiable as pronuclear fusion or the syncytial divisions were observed in embryos from Klp10A germline mutant females (Figure 3). These mutant embryos exhibited severe disorganization of both the DNA and microtubules, suggesting that the maternal contribution of KLP10A is essential for the earliest steps in embryogenesis. Mutants in two other Drosophila kinesin genes, subito and Klp3A, produce embryos that fail to develop because of pronuclear fusion failure (Williams et al. 1997; Giunta et al. 2002). Failure of pronuclear fusion typically results in a block to initiating the syncytial divisions of the early embryo (Figure 3). Klp10A germline mutant embryos have additional problems. The mutant embryos exhibit dispersed chromosomes and large microtubule arrays, suggesting that loss of Klp10A affects more than just pronuclear fusion. This is, however, reminiscent of the microtubule arrays observed after MCAK knockdown in Xenopus egg extracts (Walczak et al. 1996), suggesting that the microtubule-depolymerizing function of these two kinesin-13 homologs is similarly required during mitotic cell division.
Figure 3 .
Microtubule and DNA disorganization in Klp10A germline mutant embryos. Embryos produced by Klp10A germline mutants show severely disorganized DNA and microtubule structures. Chromosomes are dispersed throughout the cytoplasm, and microtubules form large asters surrounding the dispersed chromosomes. See Figure 1 for wild-type embryo spindles. Also shown are two examples of embryos lacking Subito (by RNAi, see Materials and Methods). About half of the embryos show only the female polar body (arrow) and the male pronucleus (inset). Drosophila female meiosis does not segregate chromosomes into a separate polar body. In the other half of the embryos, there are nuclei attempting to divide, which may have originated from the haploid male genome. DNA is in blue and microtubules are in green. Bars, 10 μm.
Klp10A germline mutants have disorganized meiotic metaphase I spindles
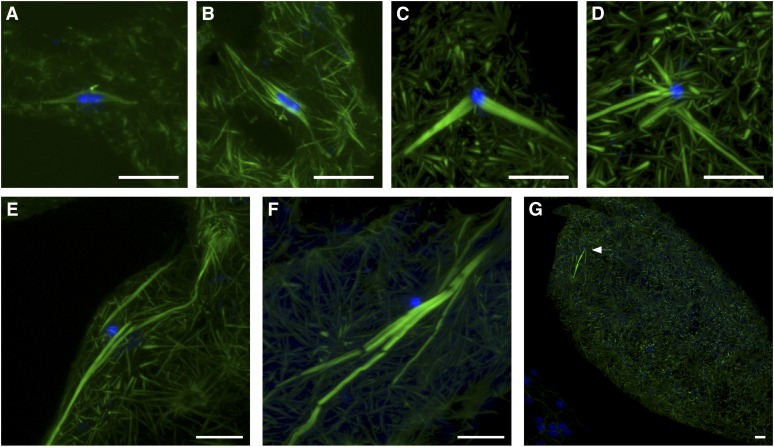
The failure to begin embryonic divisions in Klp10A germline mutants is consistent with a requirement for KLP10A during mitotic cell division, but does not address whether there is a defect in meiosis. To investigate this, we examined the cytology of late-stage oocytes from Klp10A germline mutants. In wild-type, late-stage oocytes are characterized by a bipolar spindle around a condensed mass of chromosomes, termed a karyosome (Figure 4A). In Klp10A germline mutants, we observed two dramatic phenotypes. First, spindle microtubule bundles were much longer than in wild type (Figure 4, B–F). We could not determine an average spindle length in Klp10A germline mutants because the microtubule bundles around the karyosome were extremely disorganized, making it impossible to choose which microtubule ends to measure from for an accurate spindle length. Instead, we measured the length of microtubule bundles, typically from the karyosome to the end of the bundle, in several representative Klp10A germline mutants, and these bundles ranged in length from 12.0 to 60.0 μm (Figure S2). In comparison, the average wild-type half-spindle was 6.7 μm (SD ± 4.0 μm; n = 45, longest 16.6 µm). Second, while wild-type ooplasm contains short microtubule fibers surrounding the meiotic spindle (Figure 4A), in Klp10A germline mutants, these ooplasmic microtubules were more numerous and longer than in wild type, and arranged in a “starburst” pattern (Figure 4G). The spindle microtubules appear to interact with or originate from these starburst structures (Figure 4E). These results show that KLP10A is required to maintain length control over both spindle and ooplasmic microtubules in oocytes, which is consistent with its proposed function as a microtubule-depolymerizing kinesin.
Figure 4 .
Spindle disorganization in late-stage oocytes from Klp10A germline mutants. (A) In wild type, a bipolar spindle surrounds the karyosome. Short microtubule fragments are present throughout the ooplasm. (B–G) In oocytes from Klp10A germline mutants, spindles are disorganized. Microtubule fragments in the ooplasm are much longer than wild type and are often arranged in a starburst pattern. (B) Bipolar spindle that is long and frayed. (C) Bipolar spindle in which the two half spindles are not connected by a central spindle. (D) Multipolar spindle. (E and F) Extremely long, disorganized spindles in which the contact between the karyosome and microtubules appears to be lacking. In addition, the spindle in E appears to connect to one of the starburst structures in the ooplasm. (G) Long microtubule fragments in starburst patterns are present throughout the ooplasm of the entire oocyte, not just near the karyosome (arrow). DNA is in blue and microtubules are in green. Bars, 10 μm.
The organization of the spindle is also dramatically affected in Klp10A germline mutants. Spindle disorganization ranged from slight (bipolar with some fraying or bending, Figure 4B) to extreme (long microtubule bundles with no apparent poles, Figure 4, E and F). In addition, wild-type oocytes normally accumulate microtubules between the two half spindles in the region surrounding the DNA, a region termed the central spindle. This region is often lacking in Klp10A germline mutants even in a bipolar spindle, resulting in two disconnected half spindles (Figure 4C) and multipolar spindles (Figure 4D). In wild type, the central spindle accumulates several proteins, including Incenp (Jang et al. 2005). In Klp10A germline mutants, Incenp was not detected on some spindles, correlating with the presence and absence of the central spindle (data not shown). Furthermore, frequently the karyosome and spindle were not in proper contact in Klp10A germline mutants (Figure 4, E and F). These results show that KLP10A is essential for the integrity of the meiotic metaphase I spindle, affecting microtubule length and organization, and the interaction of the spindle with the chromosomes.
Klp10A germline mutants misorient homologous chromosomes during meiosis
In Klp10A germline mutants there is an apparent lack of proper contact between the chromosomes and microtubules (Figure 4, E and F), although the integrity of the karyosome is maintained (Figure 4, B–F). Because accurate homolog segregation requires the interaction of chromosomes with spindle microtubules, we investigated the orientation of homologous chromosomes using fluorescent in situ hybridization (FISH) of probes to centromeric heterochromatin. Prior to nuclear envelope breakdown and meiotic spindle assembly in wild-type Drosophila females, centromeres are homologously paired (Dernburg et al. 1996), which we also observe in Klp10A germline mutants (Figure 5B). Concomitant with assembly of a bipolar spindle in wild type, the homologous centromeres separate toward opposite spindle poles (Figure 5A). This is referred to as biorientation and indicates the homologous pairs of chromosomes are poised to segregate correctly at anaphase I.
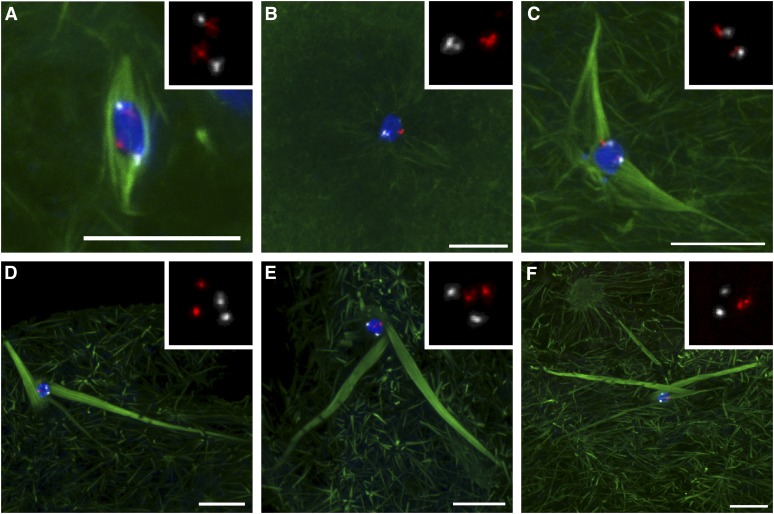
Figure 5 .
Biorientation of homologous chromosomes is defective in Klp10A germline mutants. (A) In wild type, both the 2nd (red) and 3rd (white) chromosome centromeric FISH probes show biorientation toward opposite spindle poles. All 17 centromere pairs scored were separated (two FISH signals) and oriented correctly. (B) Klp10A germline mutant oocyte prior to or during nuclear envelope breakdown. Homologous centromeres are paired. (C) Klp10A germline mutant oocyte with a long and disorganized spindle, which is nonetheless bipolar and has properly oriented centromeres. (D) Klp10A germline mutant oocyte with a disorganized spindle, loosely “bioriented” chromosomes, but “mono-oriented” 3rd chromosomes. (E and F) Klp10A germline mutant oocytes with disorganized spindles and randomly oriented centromeres. Centromeres in E are not associated with microtubule bundles. DNA is in blue and tubulin is in green. Insets show only the FISH signals. Bars, 10 μm.
In most Klp10A germline mutants, it is difficult to apply conventional definitions of homolog biorientation because the spindles are severely disorganized (Figure 5, D–F). Nonetheless, two observations suggest that Klp10A mutant oocytes have defects in the connections between the chromosomes and the microtubules. First, the position of homologous centromeres relative to each other is disturbed. In wild type, centromeres point in two directions, one member of each pair pointing in each direction (Figure 5A). In Klp10A germline mutants, however, centromeres do not always point in two directions, but rather appear to be positioned randomly within the karyosome (Figure 5, E and F). Second, whether centromeres appeared “bioriented” or not, the relationship to microtubules is disturbed. In wild type, centromeres are positioned at the edge of the karyosome, close to the microtubule bundles that make up a spindle pole. In Klp10A germline mutants, centromeres are not always positioned in close proximity to the end of microtubule bundles (Figure 5, D and E). In addition, both centromeres of a homologous pair can be positioned close to the same microtubule bundle, appearing mono-oriented (Figure 5, D and F).
This discordance between centromere orientation and spindle organization may reflect the unstable nature of the meiotic spindle in Klp10A germline mutants. Whereas wild-type spindles maintain their length and bipolarity, the mutant spindle length and organization may be unstable (see Discussion). The microtubule–chromosome connections may be ephemeral in the absence of KLP10A. The presence of well-separated centromeres or pairs of centromeres at the edge of the karyosome suggests that the microtubule–chromosome connections are made and the centromeres move. In fact, in the rare bipolar spindles in Klp10A germline mutants, biorientation of homologous centromeres appears normal (Figure 5C). The observation of centromeres that are not associated with microtubules in Klp10A germline mutants suggests these connections are easily broken. These results demonstrate that functional connections between centromeres and spindle poles are lacking in the absence of KLP10A.
Discussion
During cell division, a stable bipolar spindle is crucial for the accurate distribution of genetic material to daughter cells. How the stability of this structure is achieved when the spindle is composed of dynamically unstable microtubules is an important question. We have shown here that KLP10A, a member of the kinesin-13 family of microtubule-depolymerizing proteins, is essential to the organization of the acentrosomal meiotic spindle in Drosophila oocytes.
An interesting feature of acentrosomal meiosis is that microtubule ends appear to be distributed throughout the spindle (Burbank et al. 2006; Liang et al. 2009). This has implications for the regulation of microtubule dynamics by kinesin-13 family members, which have been shown in vitro to act at the ends of microtubules to induce depolymerization (Desai et al. 1999). We observed that KLP10A localizes throughout the meiotic spindle at metaphase I. This distribution may reflect the binding of KLP10A along the entire length of microtubules. Kinesin-13’s are known to bind along the length of microtubules in vitro, diffusing to the ends before becoming active (Helenius et al. 2006). This seems unlikely, however, given the propensity for kinesin-13’s in general, and KLP10A specifically, to localize to the regions of the mitotic spindle with the highest concentrations of microtubule ends—spindle poles and kinetochores (Rogers et al. 2004).
Instead, we suggest that KLP10A localizes to microtubule ends that are present throughout the spindle, which implies that microtubule depolymerization occurring throughout the meiotic spindle may be a normal part of spindle assembly and stability. Indeed, Domnitz et al. (2012) have shown that MCAK activity at the tips of nonkinetochore microtubules regulates mitotic spindle length. Alternatively, KLP10A may be present at ends throughout the spindle, but maintained in an inactive state in most locations. If the microtubule end to which KLP10A is bound is near a spindle location where depolymerization is needed, such as near the chromosomes or poles, then KLP10A may become active. There is a large body of evidence that the activity and localization of kinesin-13’s are regulated during mitotic cell division by phosphorylation and protein interactions (reviewed in Ems-McClung and Walczak 2010), but whether these mechanisms are active during meiotic cell division remains to be examined.
Surprisingly, we found no evidence for enrichment of KLP10A at centromeres as observed in S2 cells (Rogers et al. 2004). This is in contrast to the conclusions by Zou et al. (2008), although it should be noted that the interpretation of centromere localization in this previous study was not confirmed with a centromere marker. Indeed, the overall pattern of KLP10A localization in the two studies is similar. Our finding that its localization in embryos is enriched toward the poles is consistent with the conclusion that KLP10A is required for poleward flux, which depends on microtubule depolymerization at the poles (Rogers et al. 2004). Thus, while we cannot rule out that KLP10A localizes to the centromeres during female meiosis, there is no conclusive evidence for it.
Our results show that loss of KLP10A dramatically impacts spindle organization. The spindles assembled in the absence of KLP10A present widely varying organizational arrangements, which may result from an imbalance in the dynamic nature of microtubules during spindle assembly and maintenance. In wild-type Drosophila oocytes, a bipolar spindle assembles, and its organization and length is stably maintained in metaphase I for extended periods of time (Gilliland et al. 2007; Colombie et al. 2008). In contrast, in mutants that affect spindle organization, the spindle can dramatically change shape over the course of live imaging (Matthies et al. 1996; Colombie et al. 2008). We propose that the deregulation of microtubule dynamics in Klp10A germline mutants results in the formation of unstable meiotic spindles because of the loss of the ability to shorten microtubules. This implies that the regulation of microtubule dynamics by KLP10A is required to maintain a stable bipolar spindle in Drosophila oocytes.
The central spindle comprises a band of antiparallel microtubules that extends across the chromosomes to connect the two half spindles. While many spindles from Klp10A germline mutants are severely disorganized, even in some cases of only mild spindle disorganization, the central spindle is missing. This suggests that the integrity of the central spindle depends on the regulation of microtubule dynamics. Several proteins including the chromosomal passenger complex (CPC) and Subito localize to the central spindle and are required for meiotic spindle assembly (Radford et al. 2012) and bipolarity (Jang et al. 2005), respectively. Thus, the central spindle is important for organizing the meiotic spindle, and the instability of this structure in the absence of KLP10A may contribute to the spindle organization defects. Loss of the central spindle cannot explain all of the spindle defects, however, because the central spindle is absent in subito mutants, but this results primarily in monopolar and tripolar spindles with no effect on spindle length (Jang et al. 2005).
Interestingly, the spindle defects we observed in Klp10A germline mutants differ from previous kinesin-13 loss-of-function studies. Knockdown of kinesin-13 homologs in human cells, Xenopus laevis egg extracts, and Drosophila S2 cells primarily results in monopolar spindles, chromosome congression and segregation defects, and long astral microtubules (Walczak et al. 1996; Ganem and Compton 2004; Rogers et al. 2004; Ganem et al. 2005; Manning et al. 2007; Ohi et al. 2007; Rath et al. 2009). RNAi knockdown of KLP10A in Drosophila S2 cells does result in an increase in spindle length (Goshima et al. 2007); however, the magnitude of the effect is modest in comparison to the effect on microtubule length that we observed in Klp10A germline mutants. One obvious explanation for the different effects is the different organization of the centrosomal mitotic and acentrosomal meiotic spindles. The need for KLP10A to maintain spindle length in mitotic spindles may be tempered by the presence of centrosomes and astral microtubules, whereas in acentrosomal spindles, the determination of spindle length is dominated by a balance between microtubule depolymerization by KLP10A and spindle elongation by a mechanism that is not yet known. At this point, however, it is possible that there are other differences between the mitotic and oocyte spindles that make acentrosomal spindle length hypersensitive to loss of KLP10A. Whether KLP10A plays an additional role in spindle organization or whether the spindle disorganization in Klp10A germline mutants results from overgrowth of microtubules also remains to be determined.
The loss of KLP10A also impacts the interaction of the spindle with the chromosomes. KLP10A could be required to regulate interactions between microtubules and chromosomes because kinesin-13’s have been shown to play an important role in correcting improper kinetochore–microtubule attachments in mitosis (Kline-Smith et al. 2004). In Drosophila female meiosis, however, KLP10A appears to have the opposite effect, promoting or maintaining contact between chromosomes and spindle poles, suggesting that the mechanism by which KLP10A regulates chromosome–microtubule attachments differs in mitotic vs. acentrosomal meiotic spindles. Interestingly, Domnitz et al. (2012) argue that MCAK depolymerizing activity at the tips of microtubules actually promotes robust kinetochore attachments. It is also possible, however, that this function could be indirect, through the maintenance of microtubule length and spindle organization. Importantly, however, these results demonstrate that the microtubule-depolymerizing kinesin KLP10A is essential for the establishment of an acentrosomal meiotic spindle with the capacity to properly segregate homologous chromosomes.
Supplementary Material
Acknowledgments
We are grateful to Li Nguyen for technical assistance, David Sharp for the anti-KLP10A antibody, and Hiro Ohkura for advice on the methanol fixation of oocytes. We also thank Arunika Das and Kathryn Landy for comments on the manuscript. We thank the TRiP at Harvard Medical School (National Institutes of Health (NIH)/National Institute of General Medical Sciences R01-GM084947) for providing transgenic RNAi fly stocks used in this study. Some stocks used in this study were obtained from the Bloomington Stock Center. S.J.R. was supported by a Helen Hay Whitney Foundation postdoctoral fellowship. This work was supported by a grant from the NIH (GM 067142) to K.S.M.
Note added in proof: See S. J. Radford et al. (pp. 417–429) in this issue, for a related work.
Footnotes
Communicating editor: S. E. Bickel
Literature Cited
- Albertson D. G., Thomson J. N., 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1: 15–26 [DOI] [PubMed] [Google Scholar]
- Bakhoum S. F., Compton D. A., 2011. Kinetochores and disease: keeping microtubule dynamics in check! Curr. Opin. Cell Biol. 24: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Burbank K. S., Groen A. C., Perlman Z. E., Fisher D. S., Mitchison T. J., 2006. A new method reveals microtubule minus ends throughout the meiotic spindle. J. Cell Biol. 175: 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso D., Ramirez-Weber F., Kornberg T. B., 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 91: 451–454 [DOI] [PubMed] [Google Scholar]
- Chou T. B., Perrimon N., 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombie N., Cullen C. F., Brittle A. L., Jang J. K., Earnshaw W. C., et al. , 2008. Dual roles of Incenp crucial to the assembly of the acentrosomal metaphase spindle in female meiosis. Development 135: 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., Sedat J. W., Hawley R. S., 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 85: 135–146 [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T. J., Walczak C. E., 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell 96: 69–78 [DOI] [PubMed] [Google Scholar]
- Domnitz S. B., Wagenbach M., Decarreau J., Wordeman L., 2012. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J. Cell Biol. 197: 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Petri S., Pellegrin F., Terret M. E., Bohnsack M. T., et al. , 2007. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J. Cell Biol. 176: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung S. C., Walczak C. E., 2010. Kinesin-13s in mitosis: key players in the spatial and temporal organization of spindle microtubules. Semin. Cell Dev. Biol. 21: 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Compton D. A., 2004. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 166: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Upton K., Compton D. A., 2005. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 15: 1827–1832 [DOI] [PubMed] [Google Scholar]
- Gilliland W. D., Hughes S. E., Cotitta J. L., Takeo S., Xiang Y., et al. , 2007. The multiple roles of mps1 in Drosophila female meiosis. PLoS Genet. 3: e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta K. L., Jang J. K., Manheim E. M., Subramanian G., McKim K. S., 2002. subito encodes a kinesin-like protein required for meiotic spindle pole formation in Drosophila melanogaster. Genetics 160: 1489–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Preston C. R., Johnson-Schlitz D. M., Nassif N. A., Phillis R. W., et al. , 1993. Type I repressors of P element mobility. Genetics 135: 81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Scholey J. M., 2010. Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 26: 21–57 [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., et al. , 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J., Brouhard G., Kalaidzidis Y., Diez S., Howard J., 2006. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature 441: 115–119 [DOI] [PubMed] [Google Scholar]
- Illingworth C., Pirmadjid N., Serhal P., Howe K., Fitzharris G., 2010. MCAK regulates chromosome alignment but is not necessary for preventing aneuploidy in mouse oocyte meiosis I. Development 137: 2133–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. K., Rahman T., McKim K. S., 2005. The kinesin-like protein Subito contributes to central spindle assembly and organization of the meiotic spindle in Drosophila oocytes. Mol. Biol. Cell 16: 4684–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. C., 1970. Ovarian Development in Drosophila melanogaster, Academic Press, New York [Google Scholar]
- Kline-Smith S. L., Khodjakov A., Hergert P., Walczak C. E., 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell 15: 1146–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L., 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461 [DOI] [PubMed] [Google Scholar]
- Liang Z. Y., Hallen M. A., Endow S. A., 2009. Mature Drosophila meiosis I spindles comprise microtubules of mixed polarity. Curr. Biol. 19: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A. L., Ganem N. J., Bakhoum S. F., Wagenbach M., Wordeman L., et al. , 2007. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell 18: 2970–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H. J., McDonald H. B., Goldstein L. S., Theurkauf W. E., 1996. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Joyce E. F., Jang J. K., 2009. Cytological analysis of meiosis in fixed Drosophila ovaries. Methods Mol. Biol. 558: 197–216 [DOI] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M., 1984. Dynamic instability of microtubule growth. Nature 312: 237–242 [DOI] [PubMed] [Google Scholar]
- Moores C. A., Milligan R. A., 2006. Lucky 13-microtubule depolymerisation by kinesin-13 motors. J. Cell Sci. 119: 3905–3913 [DOI] [PubMed] [Google Scholar]
- Ni J. Q., Zhou R., Czech B., Liu L. P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Burbank K., Liu Q., Mitchison T. J., 2007. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr. Biol. 17: 953–959 [DOI] [PubMed] [Google Scholar]
- Radford S. J., Jang J. K., McKim K. S., 2012. The chromosomal passenger complex is required for meiotic acentrosomal spindle assembly and chromosome biorientation. Genetics 192: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath U., Rogers G. C., Tan D., Gomez-Ferreria M. A., Buster D. W., et al. , 2009. The Drosophila kinesin-13, KLP59D, impacts Pacman- and Flux-based chromosome movement. Mol. Biol. Cell 20: 4696–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., et al. , 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427: 364–370 [DOI] [PubMed] [Google Scholar]
- Rorth P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78: 113–118 [DOI] [PubMed] [Google Scholar]
- Sugimura I., Lilly M. A., 2006. Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev. Cell 10: 127–135 [DOI] [PubMed] [Google Scholar]
- Szollosi D., Calarco P., Donahue R. P., 1972. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11: 521–541 [DOI] [PubMed] [Google Scholar]
- Theurkauf W. E., Hawley R. S., 1992. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 116: 1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., et al. , 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt E., Sanhaji M., Klein W., Seidel T., Wordeman L., et al. , 2010. MCAK is present at centromeres, midspindle and chiasmata and involved in silencing of the spindle assembly checkpoint in mammalian oocytes. Mol. Hum. Reprod. 16: 665–684 [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Mitchison T., Desai A., 1996. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 84: 37–47 [DOI] [PubMed] [Google Scholar]
- Williams B. C., Dernburg A. F., Puro J., Nokkala S., Goldberg M. L., 1997. The Drosophila kinesin-like protein KLP3A is required for proper behavior of male and female pronucleii at fertilzation. Development 124: 2365–2376 [DOI] [PubMed] [Google Scholar]
- Zou J., Hallen M. A., Yankel C. D., Endow S. A., 2008. A microtubule-destabilizing kinesin motor regulates spindle length and anchoring in oocytes. J. Cell Biol. 180: 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.