Abstract
Unaccounted population stratification can lead to spurious associations in genome-wide association studies (GWAS) and in this context several methods have been proposed to deal with this problem. An alternative line of research uses whole-genome random regression (WGRR) models that fit all markers simultaneously. Important objectives in WGRR studies are to estimate the proportion of variance accounted for by the markers, the effect of individual markers, prediction of genetic values for complex traits, and prediction of genetic risk of diseases. Proposals to account for stratification in this context are unsatisfactory. Here we address this problem and describe a reparameterization of a WGRR model, based on an eigenvalue decomposition, for simultaneous inference of parameters and unobserved population structure. This allows estimation of genomic parameters with and without inclusion of marker-derived eigenvectors that account for stratification. The method is illustrated with grain yield in wheat typed for 1279 genetic markers, and with height, HDL cholesterol and systolic blood pressure from the British 1958 cohort study typed for 1 million SNP genotypes. Both sets of data show signs of population structure but with different consequences on inferences. The method is compared to an advocated approach consisting of including eigenvectors as fixed-effect covariates in a WGRR model. We show that this approach, used in the context of WGRR models, is ill posed and illustrate the advantages of the proposed model. In summary, our method permits a unified approach to the study of population structure and inference of parameters, is computationally efficient, and is easy to implement.
Keywords: genome-wide association studies, GenPred, whole-genome regression, population stratification, shared data resources
GENOME-WIDE association studies (GWAS) have successfully identified a large number of single nucleotide polymorphisms (SNPs) related to complex disease traits (Donnelly 2008). In addition to potentially increasing the understanding of the physiology of the trait, information from multiple SNPs used together with environmental risk factors holds the promise of more accurately predicting the risk of disease.
It has long been established that a potential problem in population-based association studies is the presence of undetected substructure that can result in false-positive or negative associations and in distorted inferences in general (Lander and Schork 1994; Marchini et al. 2004). A substantial amount of literature has been devoted to methods to account for unobserved population substructure in the context of GWAS, including genomic control (Devlin and Roeder 1999), mixed models (Yu et al. 2006; Kang et al. 2008, 2010; Zhang et al. 2010), and principal components (Patterson et al. 2006; Price et al. 2006); a review can be found in Price et al. (2010).
Typically, the focus of GWAS is to detect significant SNP effects using extremely low P-values derived from single-marker regressions. Testing SNPs for association one at a time can be a sensible option when traits show simple Mendelian inheritance with one or few loci involved. However, there is increasing evidence that a number of important traits and diseases are affected by a very large number of genes (McClellan and King 2010), as well as environmental factors. In this situation, a better false-positive and false-negative performance is achieved by analyzing all SNPs jointly (Hoggart et al. 2008) using whole-genome random regression (WGRR) models, as in de los Campos et al. (2010a) and Yang et al. (2010). For a recent review of different linear models in the context of WGGR see de los Campos et al. (2012).
These methods, largely developed in the field of animal breeding (e.g., Meuwissen et al. 2001), were proposed as a way of confronting the so-called missing heritability problem and have been used for estimation of the proportion of variance accounted for by regression on common SNPs (genomic heritability), for prediction of genetic values of complex traits, and for prediction of genetic risk to diseases. The problem of stratification also emerges in WGRR; however, existing proposals to account for stratification in the context of WGRR models (Yang et al. 2010, 2011; Stahl et al. 2012) are unsatisfactory. Here we address this problem and describe a reparameterization of a Bayesian WGRR model that can fit a vast number of genetic markers jointly and that in a unified manner can estimate parameters and quantify and account for unobserved population structure. With the proposed parameterization, when individuals cluster due to population stratification, the total genomic variance can be partitioned into two independent within- and between-cluster components. Two decompositions are possible: one that depends on the distribution of the marker genotypes only and one that is trait dependent. This enables investigation into the circumstances for which existing unobserved structure can affect parameter estimates, such as genomic heritability, marker effects, and genomic values. The properties of the model are illustrated using grain yield in wheat from a population that is known to show considerable substructure, and HDL cholesterol, systolic blood pressure and height from the British 1958-cohort study, data which were reported not to show signs of structure (Wellcome Trust Case Control Consortium 2007). The latter includes registrations on approximately 3000 nominally unrelated individuals genotyped for 1 million SNPs. The traits were chosen as classic examples of continuous phenotypes.
A joint analysis involves including hundreds of thousands or millions of SNPs, typically in thousands or tens of thousands of individuals. While this task is computationally feasible, there is still a need for parameterizations and algorithms that facilitate implementation and lead to satisfactory numerical behavior. We show that the proposed reparameterized Bayesian WGRR model fulfills these needs.
This article is organized as follows. A Whole-Genome Random Regression Model defines the WGRR model in its standard parameterization and An Equivalent Probability Model describes the proposed parameterization. The topic of population structure and the subdivision of the genomic variance into components between and within populations is presented in Decomposition of the Genomic Variance. The decomposition leads to a natural definition of between and within populations estimators of genomic heritability, SNP effects and genomic values. A brief description of the data, the traits, and results are in Analysis of the Wheat and British 1958-Cohort Data and the article concludes with a discussion. Some technical details are deferred to the Appendix. These include a method to retrieve posterior means of SNP effects from posterior means of genomic values and the Markov chain Monte Carlo (McMC) algorithm.
A Whole-Genome Random Regression Model
Consider the model for the record of individual i, yi, with observed marker genotype j labeled Wij
| (1) |
where μi is the mean, bj is the effect of marker genotype j, and there are m markers. The variable μi is the intercept (a scalar) in the case of wheat or the ith element in Zs, where Z is an observed incidence matrix of ones and zeroes, and s is a column vector with effects of sex, smoking status, and social class, in the case of the human data. In matrix notation the model is written as
| (2) |
where y is a column vector of records of length n, μ is a vector of length n with elements μi,, and W is an n × m matrix with elements Wij. The m × 1 column vector of unobserved SNP effects is assumed to have the normal distribution
| (3) |
and residuals (uncorrelated with b) are assumed to have the normal distribution
| (4) |
Above, reflects prior uncertainty in the distribution of each element of b. In other words, is the a priori variance of the effect of one SNP, the same for all m SNPs. The parameter is the residual variance.
Marker labels are centered and scaled random variables defined as
| (5) |
where the random variable Xij can take values 0, 1, or 2 according to the number of the arbitrarily chosen allele of SNP j in individual i. Therefore E(Wij) = 0 and Var(Wij) = 1. Let g denote the n × 1 vector of genomic values, defined as
| (6) |
The genomic values are proxies for the true (unobserved) genetic values of the causal genotypes. The conditional variance of g given W is
The term is the average (over SNPs) realized additive genetic relationship among the n individuals and is the unconditional (with respect to W) variance of an element in g (Hayes et al. 2009). This is evident from the fact that
because E(gi|Wi) = 0. Labeling as the ith row of matrix W, the ith diagonal term of WW′ is . Then
| (7) |
because . A genomic heritability or proportion of variance accounted for by the SNPs can be defined as
| (8) |
The reparameterized WGRR model is based on assigning improper uniform prior distributions to the elements of μ and scaled inverse chi-squared distributions to and to .
From standard normal theory, given and , the posterior distribution of g and μ is normal, with mean equal to the best linear unbiased predictor (BLUP) of g and best linear unbiased estimator (BLUE) of μ (Lindley and Smith 1972; Henderson 1984).
An Equivalent Probability Model
Consider the factorization (eigenvalue or spectral decomposition) of the symmetric, nonnegative definite matrix WW′ of order (n × n), n being the number of genotyped individuals,
| (9) |
where U = [U1, U2, …, Un], of order n × n is the matrix of eigenvectors of WW′, Uj is the jth column (dimension n × 1), and D is a diagonal matrix with elements equal to the eigenvalues λ1, λ2, …, λn associated with the n eigenvectors. Properties of the eigenvalues are λi ≥ 0, i = 1, 2, …, n (because WW′ is nonnegative definite; due to the centering, rank is equal to n − 1, and one of the eigenvalues is equal to zero). The eigenvectors satisfy U′U = UU′ = I.
Model (2) can be written as
| (10) |
where is an n × 1 column vector with scalar elements αi. Then,
Since Uα and Wb are both Gaussian, with the same mean and variance, (2) and (10) represent two parameterizations of the same probability model, with g = Wb = Uα. Premultiplying by U′,
| (11) |
The transformation U′g is known as the principal component transformation in the literature (Mardia et al. 1979; Anderson 1984; Jolliffe 2002) and the ith principal component of g is the ith element of the vector α, namely
where Ui is the ith column of U whose elements are the principal component loadings.
The conditional expectation of a datum is
where Uij is the element in the ith row and jth column of matrix U. Note that the vector of SNP effects, b, is of order m × 1, whereas α is of order n × 1. The order of Wb and of Uα is n × 1. For the ith individual (i = 1, 2 …, n),
| (12) |
This equivalent form of the WGRR model has two attractive properties; one is computational and the other is conceptual. Computationally, as pointed out by de los Campos et al. (2010b), due to the orthogonality of the eigenvectors the fully conditional posterior distribution of vector α is multivariate normal with diagonal covariance matrix. In an McMC environment this means that the elements of vector α can be updated jointly. This improves mixing behavior and convergence of the chain, relative to the standard single-site updating Gibbs sampler. Details are shown in the Appendix. Conceptually, the alternative parameterization leads to a natural decomposition of the genomic variance into orthogonal components. This property can be used to investigate the existence of unobserved substructure in the data and to study how it affects inferences.
Decomposition of the Genomic Variance
The orthogonal decomposition of the genomic variance that is possible using parameterization (10) can be used to investigate the existence of unobserved substructure in the data from two sources: one that is only a function of the marker genotypes and the other that is trait dependent.
Trait-independent decomposition
Consider the conditional variance of the ith element of the vector g given in (12). The average genomic variance over the n individuals in the sample is
| (13) |
because . (Strictly, averaging over eigenvectors involves division by (n − 1), because one λ is equal to zero, and its corresponding α is zero a posteriori, with probability 1. This is ignored here and in the rest of the article.) This average variance admits the following partition. First write (10) as
| (14) |
With this formulation, given U and using , (13) can be decomposed as
| (15) |
where the first term in the right-hand side represents the genomic variance explained by the first d eigenvectors (i.e., those associated to the first d largest eigenvalues) and the second the part explained by the remaining n − d eigenvectors. The proportion of the genomic variance explained by the first d eigenvectors (d = 1, 2, …, n) is
| (16) |
which is a function of the structure of the marker genotypes only.
Trait-dependent decomposition
The regression of genomic values on eigenvectors is given by the α’s. The variance decomposition (15) involves an integration over the distribution of the α’s and is therefore a function of markers only. However, an eigenvector may explain a large proportion of the genomic variance (16) but may not covariate with the genomic values because its α is close to zero. To obtain further insight into the relative contribution of each of the eigenvectors to interindividual differences in realized genomic values for a particular trait, we propose the following trait-specific variance decomposition. Along the same lines as in Sorensen et al. (2001), consider first the parameter defined as the variance of a genomic value randomly sampled from the population of n genomic values that constitute vector g. This random variable gi can take n possible values Wb = Uα = {gi}, each with probability 1/n. By definition the variance of gi (ith element of the n × 1 vector g) is
| (17) |
where is the expected value of gi. Although both (17) and (7) express variability of genomic effects, there is an important conceptual difference between the two quantities. The variance (7) is a parameter of the distribution of g and represents variation in conceptual replications of a particular element of vector g (given W and ). In other words, the index i is fixed. On the other hand, the stochastic element associated with (17) is the index i, and the inference is conditional on the particular realization of the n elements of g. We use the symbol to distinguish (17) from the parameter of the distribution of g, in (7).
Replacing g = Uα in expression (17),
| (18) |
because when matrix W is centered, the second term in the right hand side of (18) vanishes. The α’s are unobserved and inferred from their posterior distribution. This leads to a trait-dependent partition of the realized genomic variance along the same lines as in (16). The proportion explained by the first d eigenvectors is
| (19) |
Inferences accounting for population structure
When individuals cluster due to population substructure it may be of interest to define a within-component genomic heritability
| (20) |
The parameter can be interpreted as the proportion of genomic variance in the sample of n individuals, after accounting for variation explained by the largest d eigenvectors. This is relevant when the latter represents artifact variation. A classical case is the substructure that arises as a consequence of population admixture.
In a similar way, inferences of SNP effects accounting for variation explained by the largest d eigenvectors can be obtained using (29) and (30) in the Appendix. This results in a Monte Carlo estimate of the posterior means given by
| (21) |
The posterior means of genomic values accounting for variation explained by the largest d eigenvectors are directly retrieved from (14). The Monte Carlo estimate is
| (22) |
The question that remains is the choice of the number of eigenvectors d whose variation one wishes to account for.
Measuring the importance of an eigenvector
The standard literature on principal components analysis suggests various ways to select the number of eigenvectors, but all are to some extent arbitrary (Mardia et al. 1979; Jolliffe 2002). The most common are: (i) plot λj vs. j to see where “large” eigenvalues cease and “small” start; (ii) include enough components to explain a given percentage of the total variance. Again, exact choice of “given percentage” must be decided by the investigator; (iii) exclude those principal components whose eigenvalues are less than the average (less than 1 when W has been centered and scaled). The typical objective in the standard literature is to seek parsimony and this is achieved by removing the eigenvectors associated with the smallest eigenvalues. In contrast, geneticists often wish to keep these and remove the eigenvectors that may describe stratification. These are typically those with the largest eigenvalues. A formal approach is presented in Patterson et al. (2006) and is based on the sampling distribution of the largest eigenvalue from which a P-value can be computed.
On the other hand, in the context of inference of genomic parameters, the directly relevant parameters are not the eigenvalues but the regression coefficients α. Therefore a rationale for choosing an eigenvector j could be based on the posterior probability that its contribution to the genomic variance is larger than a threshold, chosen by the user. The choice of this threshold is a matter of judgement, context specific, driven by knowledge of what is causing population substructure and the relevance of correcting for it.
As an example, suppose one wishes to base the assessment on the posterior probability that the jth eigenvector has a contribution to genomic variance greater than the average eigenvector. Formally,
| (23) |
In an McMC environment, the posterior probabilities of these hypotheses are estimated as follows. For the kth draw, noting that the denominator (n) in each of the hypothesis cancels out, set δjk = 1 if and δjk = 0 otherwise. Here, αj(k) and are the kth draws of the jth regression coefficient and of the trait-dependent genomic variance, respectively, from their marginal posterior distributions. Averaging the over the McMC samples leads to Monte Carlo estimates of Pr(Hj1|y) and Pr(Hj2|y) = 1 − Pr(Hj1|y). When the output from implementing (10) is stored, only one McMC run is needed to perform these computations.
Analysis of the Wheat and British 1958-Cohort Data
The decomposition of the genomic variance with the proposed parameterization of the WGRR model is illustrated using data from two contrasting populations. The wheat population, consisting of 599 highly inbred lines, is characterized by a strong degree of relationship among individuals and marked population substructure. The human population includes nominally unrelated individuals of homogeneous background (supporting information for both data sets is in Acknowledgments and in File S1 and File S2).
The wheat data comprise grain yield from 599 pure lines typed for 1279 genetic markers, from Centro Internacional de Mejoramiento de Maiz y Trigo’s (CIMMYT) Global Wheat Breeding program. The data set is publicly available within the BLR package of R (de los Campos and Perez 2010). Further details are given in Crossa et al. (2010). A display of the wheat data (standardized to null mean and unit variance) did not reveal signs of asymmetry (not shown).
The British 1958-cohort data consist of longitudinal records from individuals born during a single week in 1958 in England, Scotland, and Wales. A detailed description and sources of access to the data can be found in Power and Elliott (2006). The present study uses a subset of the original data consisting of records from approximately 3000 individuals that have been genotyped for 1 million SNPs using the 1M Affymetrix chip. After standard editing, the final number of markers amounted to 696,823. From the 3000 individuals, records on height, systolic blood pressure, and HDL cholesterol were also extracted, together with a number of environmental covariates. The latter were chosen on the basis of their effect on the dependent variables determined from preliminary analyses.
The raw means and standard deviations (in brackets) for height, systolic blood pressure, and HDL cholesterol in males are 176.2 (6.7), 134.7 (16.5), 1.43 (0.32) and in females 162.5 (6.2), 121.1 (17.1), 1.69 (0.41). A graphical display of residuals for the three human traits from a standard least-squares analysis of a linear model that includes the effects of sex, social status, and a covariate for smoking status did not show signs of asymmetry or important departures from normality (not shown).
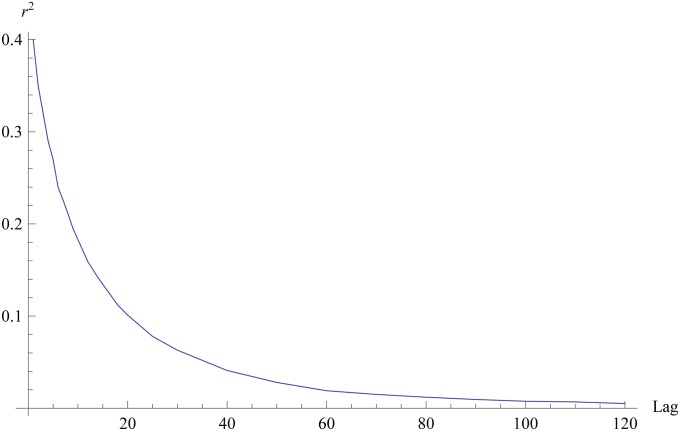
Figure 1 shows the lag−x, x = 1, 2, …120, average squared correlation between SNP genotypes. When the lag is equal to 1 (adjacent loci), r2 = 0.4, it falls to 0.012 at lag−80, to 0.0076 at lag−100, and to ∼0.0051 at lag−120. The average distance between genotypes 80, 100, and 120 SNPs apart is 336, 420, and 504 kb. At these distances, the value of r2 in humans tends to be very close to zero (Hartl and Clark 2007). Unfortunately for the wheat data set a similar figure cannot be generated because the markers are not mapped.
Figure 1 .
Human data. Lag-x, x = 1, 2, … 120, linkage disequilibrium (average squared correlation between SNP genotypes). For adjacent loci, x = 1, and x = 120 indicates that loci are 120 genotypes apart.
Results
Inferences about variance parameters and SNP effects
The posterior mean of the genomic heritability (8) for grain yield in wheat (posterior standard deviation in brackets) is 0.49 (0.05). In the human data the figures for height, systolic blood pressure, and HDL cholesterol are 0.40 (0.11), 0.15 (0.09), and 0.21 (0.10), respectively. For height, our estimate is of the same order of magnitude as that reported by Yang et al. (2010), who also used nominally unrelated individuals. These estimates differ from the estimate of 0.83 reported by Makowski et al. (2011) obtained from a sample of related individuals.
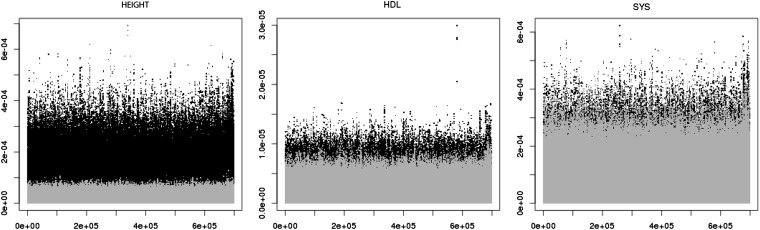
Posterior means of SNP effects, computed using Equation 30 in the Appendix, are shown in Figure 2 for the three human traits. The figure also shows the SNP effects obtained by fitting the model to the data in which phenotypes, together with the parameters representing effects of sex, smoking status, and social class, were randomly assigned to genotypes. The row vectors with the marker genotypes for each record were reshuffled, whereas phenotypes and the associated effects of sex, smoking status, and social class were kept together. In this way, the model still accounts for the effects of sex, smoking status, and social class. This reshuffling leads to a null distribution of SNP effects. Despite the fact that the present model does not allow for differential shrinkage of SNP effects, the values of posterior means are clearly larger for height in the original (not reshuffled) data, signaling clearly an association between markers and phenotypes. For HDL cholesterol and systolic blood pressure, the signals from the marker effects are weaker but clearly discernible. The figures suggest that different genetic architectures may be responsible for the three traits. Height shows many small marker effects scattered across the whole genome, in agreement with results from Yang et al. (2010). On the other hand, systolic blood pressure and particularly LDL cholesterol show marker effects of very different magnitude in particular regions of the genome.
Figure 2 .
Posterior means of marker effects (y-axis) obtained using expression (30) vs. marker loci, labeled from 1 to total number (x-axis) for the three traits. Black regions correspond to effects estimated with the original data, and shaded regions correspond to effects estimated from data in which the rows of matrix W were reshuffled and therefore randomized with respect to the phenotypes and their conditional means μi.
Population substructure and decomposition of the genomic variance
Marker-dependent decomposition:
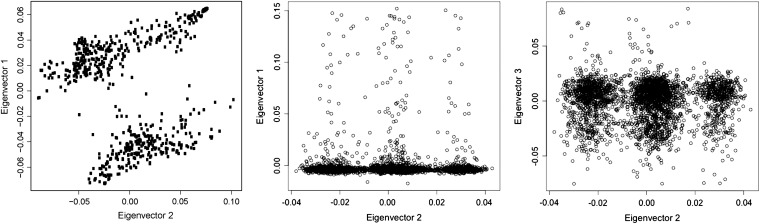
The first leftmost plot in Figure 3 displays loadings of the first two eigenvectors for the wheat data. The two rightmost plots display loadings of the first two eigenvectors and those corresponding to the third vs. the second for the human data. The leftmost figure gives a clear indication of the presence of substructure in the wheat data. For the human data, the first of the two plots shows that the first eigenvector represents a feature common to the majority of the individuals, whereas the third and particularly the second, cluster individuals in three groups. From the fifth or sixth eigenvector onward, similar plots do not reveal any form of structure (not shown). The analysis indicates the existence of substructure in both sets of data. The results for the human data are in contrast with a previous analysis that reported absence of detectable substructure using a nonparametric approach (Wellcome Trust Case Control Consortium 2007).
Figure 3 .
Left: The first vs. the second largest axes of variation in wheat. Middle: The first vs. the second largest axes of variation for the human marker data. Right: The third vs. the second largest axes of variation for the human marker data.
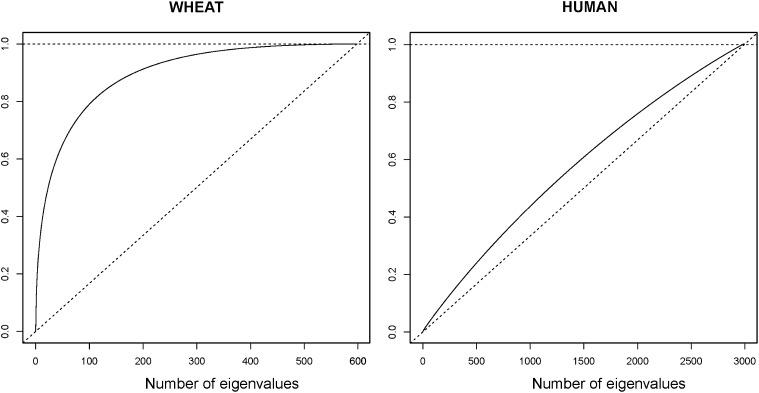
The effect of population clustering on inferences about genomic variance can be studied using expressions such as (16) and (19). The trait-independent decomposition based on (16) is displayed in Figure 4, for d = 1, 2, …, n for both data sets. The left plot is based on the wheat data set and shows that the first 100 eigenvalues explain ∼80% of the genomic variance. For the human data, the relationship between the proportion of variance explained with increasing number of eigenvectors is linear (close to the 45° line) and reveals that the proportion of the variance explained is similar for all the n eigenvectors. This indicates that in contrast with the wheat data the eigenvalues are all small and of similar magnitude. Figures 3 and 4 represent features that are functions of only the marker information and not of the traits.
Figure 4 .
Proportion of variance explained by the eigenvectors given in (16) in the y-axis for increasing number of eigenvectors d. Left: Wheat data. Right: Human data.
Trait-dependent decomposition:
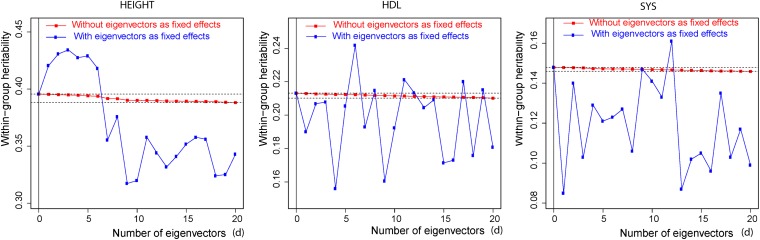
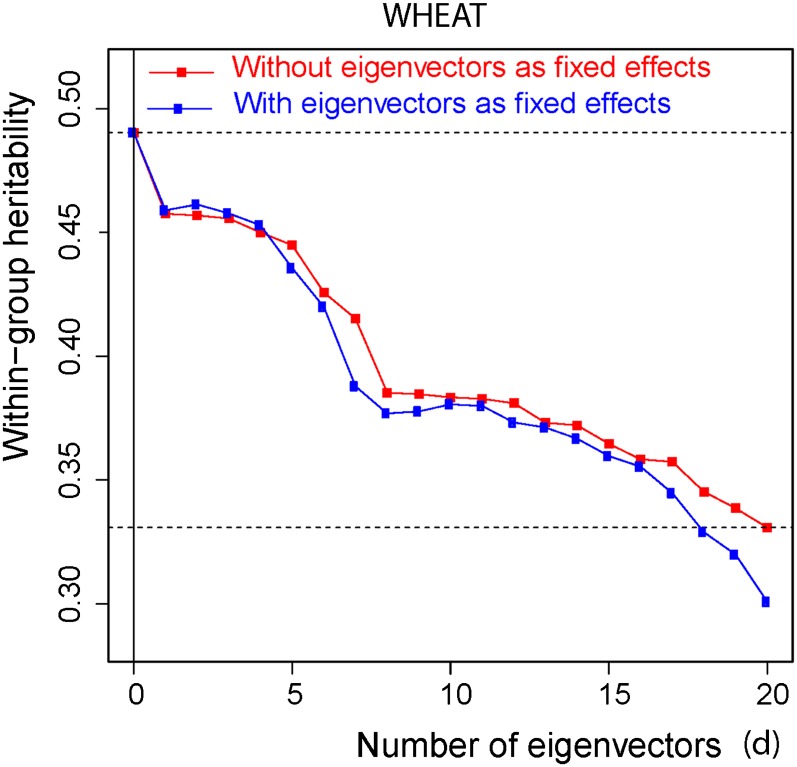
The trait-dependent decomposition of genomic variance is based on expression (19) and (20). Posterior means of (20) plotted against increasing number of eigenvectors, d = 0, 1, …, 20, for the three human traits are shown in Figure 5 (in red). The figure illustrates that the 20 eigenvectors with the largest eigenvalues account for <2% of the genomic heritability for height [that is, (0.396 − 0.388)/0.396] and ∼1.4% for the other two traits. On the other hand, Figure 6 shows a different pattern in the case of wheat. The within-group genomic heritability (20) falls sharply from 0.49 when d = 0 to a little over 0.25 when variation due to the 20 eigenvectors with the largest eigenvalues is accounted for. In this case the first 20 eigenvectors account for ∼49% of the genomic heritability. The different results in wheat and in humans are due to the different sizes of the regression coefficients α of genomic values on eigenvectors. This affects the variance partition given in (19), since, as shown in (18), the contribution to the genomic variance from eigenvector j is . In the case of wheat the posterior means of the α’s associated with the largest eigenvalues are markedly larger than in the three traits in the human data sets.
Figure 5 .
Human data. Red: Posterior means of within population genomic heritability in the y-axis [expression 20)] computed using the WGRR model (14) after accounting for the proportion of variance due to the number of eigenvectors (d) with the largest eigenvalues, in the x-axis. Blue: Genomic heritability (8) computed using model (2) with the addition of the d eigenvectors with the largest eigenvalues treated as fixed effects. The horizontal dotted lines emphasize the range of values of the posterior means of between d = 0 and d = 20, obtained with the WGRR model.
Figure 6 .
Wheat data. Red: Posterior mean of within population genomic heritability in the y-axis [expression 20)] computed using the WGRR model (14) after accounting for the proportion of variance due to the d eigenvectors with the largest eigenvalues, vs. d, in the x-axis. Blue: Genomic heritability (8) computed using model (2) with the addition of the d eigenvectors with the largest eigenvalues treated as fixed effects. The horizontal dotted lines emphasize the range of values of the posterior means of between d = 0 and d = 20, obtained with the WGRR model.
Insight into the magnitude of the α’s can be revealed by inspecting their conditional posterior distribution given in (31) in the appendix. For the ith regression coefficient,
| (24) |
where is the 1 × n row vector whose elements are the loadings of the ith eigenvector Ui. This shows that the magnitude of the α’s is determined by two factors,
the (unpenalized) regression of phenotype on the ith eigenvector and the extent of shrinkage, which is controlled by the size of . When is large shrinkage is weak and approaches , the ordinary least-squares regression of phenotype on the ith eigenvector. On the other hand, approaches zero for small values of . The variance of the conditional posterior distribution (31) is
| (25) |
The variance is governed by . It becomes negligible when tends to a very small quantity and approaches when is large. It is precisely this term that differs between the wheat and the human data. For example, in the case of SYS and for the largest λ, the regression term in (24) is 0.54 and the square root of (25) is 10.5. For the wheat data set and for the largest λ these values are 0.015 and 0.73, respectively. The largest values of λi/m for the human and wheat data are 4.8 and 67.8, respectively. These expressions indicate that if an eigenvector is associated with an eigenvalue close to zero, its regression coefficient α approaches zero with probability one, regardless of the amount of data. In this case there is no Bayesian learning.
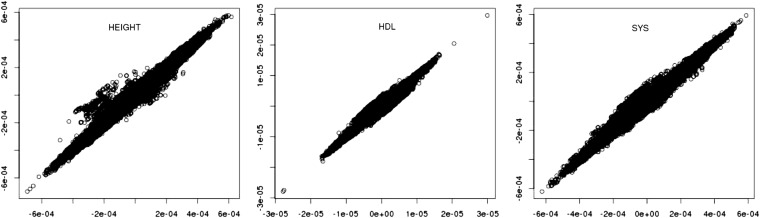
The effect of population substructure on inferences about SNP effects is illustrated in Figure 7 for the three human traits. Posterior means of SNP effects corrected for population substructure [y-axis, given by (21) with d = 20] are plotted against those uncorrected for population substructure [x-axis, given by (30)]. There is an overall strong association, but for height and to a lesser extent for systolic blood pressure, intermediate SNP effects are relatively more affected by the effect of population substructure than extreme ones.
Figure 7 .
Human data. Posterior means of SNP effects corrected for population substructure (y-axis, given by 21, with d = 20), vs. posterior means of SNP effects uncorrected for population substructure (x-axis, given by 30), for the three traits.
Including the dominating eigenvectors as fixed effects in the WGRR model to account for substructure:
As an illustration, Figure 5 also shows the results obtained by fitting a model similar to (2), with the addition of d = 0, 1, 2, …, 20, dominating eigenvectors with the α’s treated as fixed effects (blue). This model, recently reported in the literature (Yang et al. 2010, 2011) is not well posed, because the same eigenvectors whose coefficients are treated as fixed enter implicitly in the random part of the model. When the degree of shrinkage in (24) is large, if the variance component is known the addition of a “fixed” α has the effect of reducing the error sum of squares [relative to the value obtained with model (10)] and as a result, the estimate of genomic heritability is inflated. When the variance components are unknown the consequences are more difficult to predict. Figure 5 displays the erratic behavior of inferences based on this model for the human data. For the wheat data (Figure 6, blue), the consequences of adding the superfluous “fixed” α’s on the genomic heritability are very different. Due to the very small size of the shrinkage parameter in (24), estimates of the α’s are very similar, but not identical, to least-squares estimates. Adding the α’s associated with the largest eigenvalues and treating them as fixed effects causes a very small change in the error sum of squares and a small proportion of the genomic variation is removed. As a result, inferences of genomic heritability (8) with the ill-posed model are similar to those from model (10).
Measuring the contribution of an eigenvector to genomic variance:
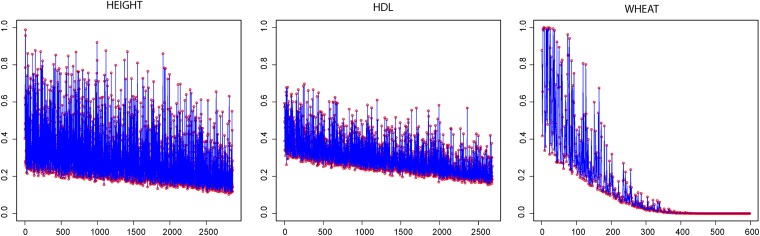
Figure 8 displays the posterior probabilities Pr(Hj1|y), j = 1, 2, …, n, defined in (23), for height and HDL cholesterol in humans, and for yield in wheat. The wheat data set is characterized by large differences in the sizes of the eigenvalues associated with the eigenvectors (see Figure 4, left). The largest lead to less informative prior distributions of the α’s; this allows for Bayesian learning and results in fluctuating contributions to genomic variability and in extreme posterior probabilities (23). As the eigenvalues tend to zero, the prior distribution of α becomes more informative, limiting the possibilities for Bayesian learning; the contribution to genomic variability is reduced and the posterior probabilities become small. In humans, the range of values of the eigenvalues is markedly narrower than in wheat (Figure 4, right). Therefore the variance of the posterior distribution of the α’s is smaller and declines at a small rate. This induces a more uniform and narrower fluctuation of the posterior probabilities Pr(Hj1|y) and a milder rate of their overall decay. The degree of covariation between the eigenvectors and phenotype is larger in the case of height, than in HDL (which shows a similar pattern as systolic blood pressure, not shown).
Figure 8 .
Monte Carlo estimates of posterior probabilities Pr(Hj1|y) defined in (23), in y-axis, against eigenvectors j, j = 1, 2, …, n, labeled in decreasing order according to the size of their eigenvalues, x-axis. Left and middle: Human data. Right: Wheat data.
Discussion
In structured populations genomic variability can be partitioned into components within, and between clusters. The focus of inference is typically the within-cluster component, which is interpreted as the genomic variability available after accounting for population stratification, often considered as contributing artifact variation. For example, one of the methods used in GWAS to correct for differences between groups (EIGENSTRAT, Price et al. 2006) consists of expanding the regression model that defines the relationship between markers and phenotypes with the addition of marker-derived eigenvectors whose coefficients are treated as fixed effects and describe differences between groups.
The need to account for population structure also emerges in the context of estimation of genomic heritability and prediction problems in WGRR, where phenotypes are regressed simultaneously on hundreds of thousands of genetic markers. Drawing on ideas largely developed in the field of single-marker regressions, Yang et al. (2010) propose to account for population structure by adding to the WGRR model the dominating eigenvectors with coefficients treated as fixed effects. The model could be of the form
| (26) |
where d is the number of the eigenvectors with the largest eigenvalues, often 10 or 20, and the α’s in are estimated by least squares (i.e., estimated without shrinkage). However, this approach, recently applied in the literature (Stahl et al. 2012), is not advisable, because model (26) suffers from “double counting” since the same eigenvectors whose coefficients are included as fixed effects enter, implicitly, as random effects in the random part of the model. The consequences on inferences of fitting this model are highly dependent on the distribution of marker genotypes in the data, as was illustrated in Figures 5 and 6. This ill-posed model can be avoided because as was shown in A Whole-Genome Random Regression Model and An Equivalent Probability Model, model (2) can be expressed as model (10). In this way the marker-derived eigenvectors enter naturally in a WGRR model that can be used to infer variance components and genomic heritability, estimation of marker effects, and prediction of genetic risk accounting jointly for population structure in a single analysis. The model and the eigenvalue parameterization can be easily extended for analyzing binary outcomes, such as disease status (healthy, diseased), using for example, threshold models. Bayesian McMC implementations of threshold models have been described in Albert and Chib (1993) and in Sorensen et al. (1995).
The duality between the WGRR model and the principal components regressions can also be exploited to develop efficient algorithms and the method proposed here is also computationally attractive. The Gibbs sampling algorithm described in the Appendix showed excellent mixing behavior and for the human data set it took 30 min to generate 60,000 draws from the posterior distribution in an Intel Xeon E5450 3.0 GHz Linux cluster, including the calculation of posterior means of the 696,823 SNP effects. The cost of the eigenvalue decomposition of WW′, equivalent to the calculation of its inverse, amounted to <1 min of CPU, and the computation of WW′ took a little under 5 hr. This part of the computation can be more demanding as the number of genotyped individuals and covariates becomes larger. An attractive feature of the algorithm is that with little computing effort, one can retrieve posterior means of marker effects from posterior means of genomic values (see Appendix).
The model we have proposed is a simple alternative to carrying out preliminary investigations into the genetics of complex traits. However, the Gaussian assumptions adopted induce a homogeneous degree of shrinkage across all markers. This may not be appropriate for the analysis of traits affected by genes with sizable effects, traits affected by rare variants (Mathieson and McVean 2012), and data from populations with short span linkage disequilibrium. In such cases, models using priors that induce marker-specific shrinkage such as the Bayesian Lasso (e.g., Park and Casella 2008; de los Campos et al. 2009) or various forms of finite mixture models (e.g., George and Mcculloch 1993; Meuwissen et al. 2001; Habier et al. 2011) may be more appropriate. However, in these models, the use of orthogonal representations such as those based on the eigenvalue decomposition presented here cannot be easily implemented because the implied covariance structure of genomic values, which in the model presented here is G = WW′, depends on model unknowns that are updated at every iteration of the sampler. Therefore, an important and challenging task is to develop statistical procedures that can combine both features in a unified manner.
Supplementary Material
Acknowledgments
This work made use of data and samples generated by the 1958 Birth Cohort (National Child Development Study). Access to these resources was enabled via the 58READIE Project funded by the Wellcome Trust Medical Research Council (grant nos. WT095219MA and G1001799). We also acknowledge financial support from the Leverhulme Trust (Research fellowship RF/9/RFG/2009/0062). The wheat data set is publicly available with the BLR R-package (de los Campos and Perez 2010). Description of the genotypes, phenotypes, and pedigree information can be found in Crossa et al. (2010). Phenotype and genotype data from the British 1958 Birth Cohort are freely available to research scientists worldwide on application to the Access Committee for CLS cohorts. Information on the application procedure can be found on the website: http://www2.le.ac.uk/projects/birthcohort. The freely available software PLINK (Purcell et al. 2007) was used to edit the human genotype data and to compute linkage disequilibrium statistics. The freely available software GCTA (genome-wide complex trait analysis) (Yang et al. 2011) was used to compute the genomic relationship matrix (GRM). The R function ’eigen’ (R Development Core Team, 2012) was used to compute the eigenvalue decomposition of the GRM. The WGRR (whole-genome random regression) model was run in the software bayz (http://www.bayz.biz) where this model is available with option “-m wgrr.”
Appendix
Retrieving SNP effects from genomic values:
Parameterization (10) yields inferences at the level of genomic values g = Uα and one may wish to draw inferences at the level of SNP effects b. This can be readily obtained from the McMC output as follows.
Write the degenerate distribution
| (27) |
where . The conditional distribution [b|g] is normal with conditional mean E(b|g) = W′ (WW′)−1g and conditional variance . Factorizing WW′ = UDU′ and using
then
| (28) |
and the vector E(b|g) takes the form
| (29) |
where Ui is the ith column of matrix U and the scalar αi is the ith element of α. In general
But from the variance structure of (27) it can be shown that E(b|α, y) = E(b|α). Therefore
where E(b|α) is given in (29). In an McMC environment, g or α are draws from [g|y] or from [α|y], respectively. A Monte Carlo estimate of E(b|y) can be obtained by first averaging vector α over realizations of [α|y]. That is,
where α[i] is the ith draw of vector α from [α|y], i = 1, 2, …, K. Then
| (30) |
Computational properties of the transformed model:
Models (2) and (10) are completely standard and can be implemented using a Gibbs sampler (e.g., Sorensen and Gianola 2002). Computationally, (10) is simpler to work with and as shown here, vector α can be updated jointly, resulting in better mixing and convergence behavior. If , and , then
| (31) |
where C = U′U + D−1k, , and . Since U′U = I, C is a diagonal matrix with the ith element
where λi is the ith eigenvalue. Expression (31) is the fully conditional posterior distribution of α. Each variance component is updated from a scaled inverse chi-square distribution.
Footnotes
Communicating editor: I. Hoeschele
Literature Cited
- Albert J. H., Chib S., 1993. Bayesian analysis of binary and polychotomous response data. J. Am. Stat. Assoc. 88: 669–679 [Google Scholar]
- Anderson T. W., 1984. An Introduction to Multivariate Statistical Analysis. Wiley, New York [Google Scholar]
- Crossa J., de los Campos G., Perez P., Gianola D., Burgueño J., et al. 2010. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186: 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Perez P., 2010. BLR: Bayesian linear regression. R package v. 1.2 (http://cran.r-project.org/web/packages/BLR/index.html) [DOI] [PMC free article] [PubMed]
- de los Campos G., Naya H., Gianola D., Crossa J., Legarra A., et al. 2009. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Allison D. B., 2010a Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 12: 880–886 [DOI] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Rosa G. J. M., Weigel K. A., Crossa J., 2010b Semi-parametric genomic-enabled prediction of genetic values using reproducing kernel Hilbert spaces methods. Genet. Res. 92: 295–308 [DOI] [PubMed] [Google Scholar]
- de los Campos G., Hickey J., Pong-Wong R., Daetwyler H. D., Calus M. P. L., 2012. Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Roeder K., 1999. Genomic control for association studies. Biometrics 55: 997–1004 [DOI] [PubMed] [Google Scholar]
- Donnelly P., 2008. Progress and challenges in genome-wide assocation studies in humans. Nature 456: 728–731 [DOI] [PubMed] [Google Scholar]
- George E. I., McCulloch R. E., 1993. Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 8: 881–889 [Google Scholar]
- Habier D., Fernando R., Kizilkaya K., Garrik D. J., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Clark A. G., 2007. Principles of Population Genetics. Sinauer, Sunderland, MA [Google Scholar]
- Hayes B., Visscher P. M., Goddard M. E., 2009. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60 [DOI] [PubMed] [Google Scholar]
- Henderson C. R., 1984. Applications of Linear Models in Animal Breeding. University of Guelph, Guelph, Ontario, Canada [Google Scholar]
- Hoggart C. J., Whittaker J. C., De Lorio M., Balding D. J., 2008. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genet. 4: e1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe I. T., 2002. Principal Component Analysis. Springer, Berlin [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Servide S. K., Zaitlen N. A., Kong S., et al. 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Schork N. J., 1994. Genetic dissection of complex traits. Science 265: 2037–2048 [DOI] [PubMed] [Google Scholar]
- Lindley D. V., Smith A. F. M., 1972. Bayesian estimates for the linear model. J. R. Stat. Soc. B 34: 1–41 [Google Scholar]
- Makowski R., Pajewski N. M., Klimentidis Y. C., Vazquez A. I., Duarte C. W., et al. 2011. Beyond missing heritability: prediction of complex traits. PLoS Genet. 7: e1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Cardon L. R., Phillips M. S., Donnelly P., 2004. The effects of human population structure on large genetic association studies. Nat. Genet. 36: 512–517 [DOI] [PubMed] [Google Scholar]
- Mardia K. V., Kent J. T., Bibby J. M., 1979. Multivariate Analysis. Academic Press, San Diego [Google Scholar]
- Mathieson I., McVean G., 2012. Differential confounding of rare variants in spatially structured populations. Nat. Genet. 44: 243–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J., King M. C., 2010. Genetic heterogeneity in human disease. Cell 16: 210–217 [DOI] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T., Casella G., 2008. The Bayesian LASSO. J. Am. Stat. Assoc. 103: 681–686 [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: 2074–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C., Elliott J., 2006. Cohort profile: 1958 British birth cohort (National Child Development Study). Int. J. Epidemiol. 35: 34–41 [DOI] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., et al. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909 [DOI] [PubMed] [Google Scholar]
- Price A. L., Zaitlen N. A., Reich D., Patterson N., 2010. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. 2007. PLINK: a tool set for whole-genome association and population-based analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2012. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Sorensen D., Gianola D., 2002. Likelihood, Bayesian, and MCMC Methods in Quantitative Genetics Springer-Verlag, Berlin (reprinted with corrections, 2006).
- Sorensen D., Andersen S., Gianola D., Korsgaard I. R., 1995. Bayesian inference in threshold models using Gibbs sampling. Genet. Sel. Evol. 27: 229–249 [Google Scholar]
- Sorensen D., Fernando R. L., Gianola D., 2001. Inferring the trajectory of genetic variance in the course of artificial selection. Genet. Res. 77: 83–94 [DOI] [PubMed] [Google Scholar]
- Stahl E. A., Wegmann D., Trynka G., Gutierrez-Achury J., Do R., et al. 2012. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 44: 483–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium, 2007. Genome-wide association study of 14, 000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. 2010. Common SNP’s explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee H., Goddard M. E., Visscher P. M., 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Bi I. V., Yamasaki M., et al. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C. Q., Todhunter R. J., Tiwari H. K., et al. 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.