Abstract
Capsella rubella is an inbreeding annual forb closely related to Arabidopsis thaliana, a model species widely used for studying natural variation in adaptive traits such as flowering time. Although mutations in dozens of genes can affect flowering of A. thaliana in the laboratory, only a handful of such genes vary in natural populations. Chief among these are FRIGIDA (FRI) and FLOWERING LOCUS C (FLC). Common and rare FRI mutations along with rare FLC mutations explain a large fraction of flowering-time variation in A. thaliana. Here we document flowering time under different conditions in 20 C. rubella accessions from across the species’ range. Similar to A. thaliana, vernalization, long photoperiods and elevated ambient temperature generally promote flowering. In this collection of C. rubella accessions, we did not find any obvious loss-of-function FRI alleles. Using mapping-by-sequencing with two strains that have contrasting flowering behaviors, we identified a splice-site mutation in FLC as the likely cause of early flowering in accession 1408. However, other similarly early C. rubella accessions did not share this mutation. We conclude that the genetic basis of flowering-time variation in C. rubella is complex, despite this very young species having undergone an extreme genetic bottleneck when it split from C. grandiflora a few tens of thousands of years ago.
Keywords: flowering time, mapping-by-sequencing, Arabidopsis thaliana, Capsella rubella, FLOWERING LOCUS C (FLC)
FOR 2 decades, Arabidopsis thaliana has been the preeminent model for mechanistic studies of many aspects of plant development and physiology. In addition, it is extensively used to investigate the genetic basis of natural variation for adaptive traits such as the onset of flowering (Bergelson and Roux 2010; Koornneef and Meinke 2010; Weigel 2012). As in other species, flowering of A. thaliana is influenced by prolonged exposure to cold, which signals winter, by ambient temperature and long days, both of which indicate spring, as well as the age of the plant. An indication of the complexity of its regulation is that well over 100 genes affecting flowering time in the laboratory have been identified through mutant analysis (Srikanth and Schmid 2011).
A. thaliana is found throughout much of the Northern hemisphere, with a native range that extends from North Africa to the Arctic Circle, and from the Atlantic coast of Western Europe to Central Asia. Accordingly, A. thaliana accessions show a wide range of flowering-time behaviors when grown in climate chambers or in common gardens (Gazzani et al. 2003; Lempe et al. 2005; Werner et al. 2005; Brachi et al. 2010; Li et al. 2010; Hancock et al. 2011; Méndez-Vigo et al. 2011; Strange et al. 2011). In contrast to the multitude of laboratory-induced mutations that change flowering behavior, genetic analysis has identified only a handful of loci responsible for flowering variation among natural accessions. Chief among these are FRIGIDA (FRI) and FLOWERING LOCUS C (FLC), with FRI repressing flowering by allowing expression of FLC. Vernalization stably reduces the FLC transcription, thus reversing the action of FRI. Together, allelic differences at FRI and FLC can account for more than half of flowering-time variation when A. thaliana accessions are grown in constant, strongly flower-promoting conditions and without vernalization (Johanson et al. 2000; Le Corre et al. 2002; Gazzani et al. 2003; Michaels et al. 2003; Lempe et al. 2005; Shindo et al. 2005; Werner et al. 2005; Atwell et al. 2010; Brachi et al. 2010; Li et al. 2010; Méndez-Vigo et al. 2011; Salomé et al. 2011; Strange et al. 2011).
The ancestors of the two extant genera Arabidopsis and Capsella separated from each other about twice as long ago as A. thaliana split from other species in the genus Arabidopsis (Koch and Kiefer 2005). Reduced genetic diversity and the pattern of allele sharing suggest that Capsella rubella was founded about 30,000 to 50,000 years ago when a single C. grandiflora individual became self-compatible. The general picture is that most C. rubella alleles can be easily found in the C. grandiflora population and that loci typically have only one or two different major alleles (Foxe et al. 2009; Guo et al. 2009). Compared with C. grandiflora, C. rubella has a much wider distribution (Hurka and Neuffer 1997), which raises the question of how C. rubella could adapt so seemingly rapidly to a much wider range of environmental conditions than encountered by the genetically much more diverse C. grandiflora. In addition to new mutations, recombination could expose new epistatic interactions, allowing selection to act on standing variation (Barrett and Schluter 2008).
To address the extent and genetic basis of variation in a model adaptive trait, we have examined flowering-time variation in a set of C. rubella accessions. Using mapping-by-sequencing, we have identified a rare mutation in an FLC homolog as being responsible for rapid flowering in one particularly early accession. We demonstrate by transformation in A. thaliana that a splice site mutation greatly reduces the flowering-repressing activity of this allele.
Materials and Methods
Plant material
The C. rubella accessions studied (Table 1) have been described (Guo et al. 2009); they included the reference accessions MTE, for which a draft genome sequence has been produced by the U.S. Department of Energy Joint Genome Institute (DOE-JGI). The A. thaliana strain in the Col-0 background with the FRISf2 and flc-3 alleles has been described (Lee et al. 1993; Michaels and Amasino 1999).
Table 1 . Flowering times of C. rubella accessions.
| Accession | Country of origin | Latitude (°)a | Longitute (°)a | Elevation (m) | 16LD |
16LDv |
23LD |
23SD |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Range | Mean | Median | n | Range | Mean | Median | n | Range | Mean | Median | n | Range | Mean | Median | |||||
| 690 | Esp | +36.15 | −5.58 | 50 | 6 | 65–74 | 71.8 | 74.0 | 6 | 32-35 | 34.5 | 35.0 | — | — | — | — | — | — | — | — |
| 697 | Ita | N/A | N/A | 950 | 6 | 56–56 | 56.0 | 56.0 | 6 | 35-35 | 35.0 | 35.0 | 6 | 36–40 | 38.7 | 39.0 | 6 | 63–71 | 68.7 | 69.5 |
| 698 | Ita | N/A | N/A | 1180 | 6 | 70–74 | 72.0 | 72.0 | 6 | 49-49 | 49.0 | 49.0 | — | — | — | — | — | — | — | — |
| 762 | Gre | +37.58 | +23.43 | N/A | 6 | 72–72 | 72.0 | 72.0 | 6 | 32-32 | 32.0 | 32.0 | — | — | — | — | — | — | — | — |
| 844 | Gre | N/A | N/A | 200 | 6 | 60–64 | 62.5 | 63.0 | 6 | 37-37 | 37.0 | 37.0 | — | — | — | — | — | — | — | — |
| 879 | Gre | +35.29 | +24.42 | 1200 | 6 | 51–51 | 51.0 | 51.0 | 6 | 32-32 | 32.0 | 32.0 | 6 | 28–29 | 28.3 | 28.0 | 6 | 63–77 | 69.8 | 69.5 |
| 907 | Gre | +39.40 | +19.48 | 40 | 6 | 61–61 | 61.0 | 61.0 | 5 | 51-51 | 51.0 | 51.0 | — | — | — | — | — | — | — | — |
| 925 | Gre | +39.40 | +20.51 | 450 | 6 | 98–100 | 98.3 | 98.0 | 6 | 51-51 | 51.0 | 51.0 | — | — | — | — | — | — | — | — |
| 1204 | Esp | +28.19 | −16.34 | 1000 | 6 | 72–74 | 72.3 | 72.0 | 6 | 37-37 | 37.0 | 37.0 | — | — | — | — | — | — | — | — |
| 1207 | Esp | +28.19 | −16.34 | 1100 | 6 | 56–58 | 57.7 | 58.0 | 6 | 29-29 | 29.0 | 29.0 | 6 | 70–85 | 77.3 | 78.0 | 6 | 58–103 | 75.2 | 70.5 |
| 1208 | Esp | +28.19 | −16.34 | 100 | 6 | 51–56 | 53.5 | 53.5 | 6 | 29-29 | 29.0 | 29.0 | 4 | 70–95 | 78.5 | 74.5 | 6 | 68–106 | 88.7 | 87.5 |
| 1209 | Esp | +28.19 | −16.34 | 1200 | 6 | 70–72 | 70.3 | 70.0 | 6 | 32-32 | 32.0 | 32.0 | — | — | — | — | — | — | — | — |
| 1215 | Esp | +28.19 | −16.19 | 0 | 6 | 60–65 | 62.3 | 63.0 | 6 | 27-29 | 28.7 | 29.0 | 6 | 44–67 | 56.2 | 59.5 | 1 | 103 | 103 | 103 |
| 1311 | Fra | +42.53 | −0.06 | N/A | 6 | 70–81 | 76.5 | 77.0 | 6 | 27-29 | 28.0 | 28.0 | — | — | — | — | — | — | — | |
| 1377 | Arg | −34.40 | −58.30 | 10 | 6 | 65–70 | 66.7 | 65.0 | 6 | 32-32 | 32.0 | 32.0 | 5 | 36–72 | 48.6 | 36.0 | 5 | 52–91 | 70.0 | 68.0 |
| 1408 | Gre | +35.29 | +24.42 | 170 | 6 | 51–52 | 51.3 | 51.0 | 6 | 29-29 | 29.0 | 29.0 | 4 | 65–72 | 68.0 | 67.5 | 4 | 56–56 | 56.0 | 56.0 |
| 1453 | Ita | +43.28 | +11.02 | N/A | 6 | 74–77 | 75.5 | 75.5 | 6 | 42-44 | 43.0 | 43.0 | 6 | 60–79 | 71.5 | 79.0 | 1 | 77 | 77.0 | 77.0 |
| 1482 | Aus | −31.56 | +115.50 | N/A | 6 | 77–79 | 77.7 | 77.0 | 6 | 32-35 | 33.0 | 32.0 | — | — | — | — | — | — | — | — |
| MTE | Ita | N/A | N/A | N/A | 6 | 100–106 | 102.5 | 103.0 | 6 | 42-44 | 43.3 | 44.0 | — | — | — | — | — | — | — | — |
| RIAH | Ita | N/A | N/A | N/A | 6 | 70–74 | 71.3 | 71.0 | 6 | 32-32 | 32.0 | 32.0 | — | — | — | — | — | — | — | — |
N/A, information not available.
+ or − indicates Northern and Southern latitudes, and Eastern and Western longitudes.
For flowering-time measurements, siblings from the same maternal family were used. Seeds were stratified for 7 days at 4° in 0.1% agar. For each treatment, we sowed six plants, most of which survived. Trays were moved to random positions in the growth rooms every 2 days to reduce positional effects. Flowering time was measured as total leaf number (TLN) and days to flowering (DTF).
Mapping-by-sequencing
Early flowering F2 plants from a cross of accession 1408 and the reference accession MTE were selected, and pooled genomic DNA was sequenced to 30-fold coverage on the Illumina GenomeAnalyzer IIx platform with 101-bp paired-end reads (59 million alignable reads, 11 Gb of sequence). SHORE (Ossowski et al. 2008) was used to discover single-nucleotide polymorphisms (SNPs) in the pool by alignment to a preliminary assembly of the MTE genome (CR_stitch_Feb15th.fasta) kindly provided by Jeremy Schmutz (DOE-JGI and HudsonAlpha), Stephen Wright and Khaled Hazzouri (University of Toronto), and Adrian Platts (McGill University). For annotation of variants, we used the Capsella v. 1.0 gene set released with Phytozome v8.0 (http://www.phytozome.net/capsella/). Using SHORE’s scoring matrix approach optimized for heterozygous SNP detection and stringent filtering (requiring uniqueness of reads, minimum 10× coverage, minimum 20% allele frequency, SHORE SNP quality score >25), 354,080 SNPs could be identified (Supporting Information, Table S2). These SNPs were used as markers in SHOREmap (Schneeberger et al. 2009) to identify genomic regions with an excess of homozygous 1408 alleles. The final mapping interval was defined as having an allele frequency of at least 90% on both sides for 40 contiguous SNPs and fewer than 10 contiguous SNPs with allele frequency below 50% within this region.
Targeted DNA sequence analysis
Individual young leaves were collected for DNA extraction using the cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle 1987), followed by polymerase chain reaction (PCR) amplification with Pfu polymerase (Fermentas, St. Leon-Rot, Germany). Products from two independent reactions were mixed and directly Sanger sequenced on an ABI3730XL (Applied Biosystems, Foster City, CA). Primers for FRI were designed based on sequence of a C. rubella BAC (GenBank accession no. JX003248), and a 2.3-kb fragment covering the entire transcription unit was sequenced. Primers for FLC were designed based on A. thaliana genome sequence, and a 5.8-kb fragment encompassing most of the first exon through the 3′ UTR was sequenced. Sequences were assembled and inspected with Lasergene Seqman (DNASTAR, Madison, WI). See Table S1 for oligonucleotide primers.
Sequences were aligned using ClustalX v. 1.81 (Thompson et al. 1997), and alignments were refined manually. All polymorphic sites were individually confirmed again based on the original traces. DnaSP v. 4.10.9 (Rozas et al. 2003) was used to perform population analyses: levels of nucleotide diversity per site (π) (Nei 1987).
Expression analysis
RNA was extracted from 2-week-old plants with the Plant RNeasy kit (Qiagen, Hilden, Germany) or with the TRIzol reagent (Invitrogen, Carlsbad, CA) and treated with RNase-free DNase I (Fermentas). Two to four micrograms of RNA was reverse transcribed using RevertAid first-strand cDNA Synthesis kit (Fermentas). PCRs were carried on in the presence of SYBR Green (Invitrogen, Carlsbad, CA) in 20-µl reactions. Amplification was monitored in real time with the Opticon continuous fluorescence detection system (MJ Research, Reno, NV). Threshold cycles (cT) were based on a reaction reaching a specific fluorescence intensity in the log-linear phase of the amplification curve.
For the analysis of splice forms, a fragment of about 200 bp was amplified by PCR from pooled complementary DNA (cDNA) obtained from four primary transformants for each transgenic line and digested for 2 hr with 1 μl of Fast Digest BstXI (Fermentas). See Table S1 for oligonucleotide primers.
Transgenic lines
FLC genomic fragments covering the entire open reading frame and introns were amplified using Phusion polymerase (Thermo Fisher Scientific, Waltham, MA). These fragments were placed under the control of the constitutive CaMV 35S promoter in the pGREEN-derived (Hellens et al. 2000) vector pFK210 and introduced into different A. thaliana genotypes by Agrobacterium tumefaciens-mediated transformation (Weigel and Glazebrook 2002).
Statistical analyses
Mean, median, and standard deviation were calculated using Microsoft Excel (Microsoft, Redmond, WA). Differences between groups in Figure 7B were tested by ANOVA using Tukey’s test, as implemented in Excel. Spearman’s rank-order correlation coefficient rs was determined in R (http://www.r-project.org/).
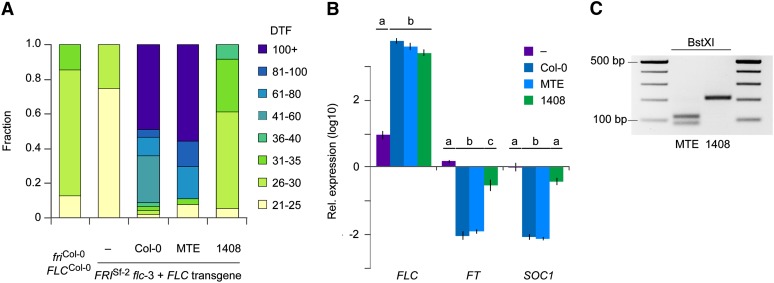
Figure 7 .
Functional analysis of C. rubella FLC alleles in A. thaliana. (A) Flowering time of plants grown in 23LD. The three left strains are controls. friCol-0 FLCCol-0 is the Col-0 reference strain, which carries a functional FLC allele, but a naturally inactive FRI allele (Johanson et al. 2000). FRISf-2 flc-3 carries an introgression of the functional FRI allele from the Sf-2 accession and has also an induced mutation at the FLC locus (Michaels and Amasino 1999). The three right strains are transgenic lines, expressing the indicated FLC allele from the constitutive CaMV 35S promoter. The effect of the C. rubella MTE allele is similar to that of the fully functional A. thaliana Col-0 allele. n ≥ 30. (B) Expression of FLC and of two downstream flowering regulators in transgenic and non-transgenic FRISf-2 flc-3 plants, determined by qRT–PCR using the 2−ΔΔcT method. Expression levels were normalized to those of wild-type Col-0 plants. Averages for four individuals for controls and eight for transgenic lines are shown. Error bars indicate standard errors of the mean. Differences between groups are significant at P < 0.05. (C) cDNA analysis of MTE and 1408 alleles expressed in transgenic A. thaliana plants. The splicing variant generated by the SNP in 1408 abolishes a BstXI restriction site in the amplified cDNA fragment.
Results
Flowering-time variation in C. rubella
We analyzed the response of C. rubella accessions to long-day (16 hr light) and short-day (8 hr light) photoperiods and to vernalization for 6 weeks at 4° in short-day photoperiods, after seeds had been germinated on soil in 16°. Twenty accessions were grown in 16° long days without vernalization (16LD) and in 16° long days with vernalization (16LDv) (Table 1). A subset of eight accessions was grown in two 23° environments with contrasting photoperiods, long days (23LD), and short days (23SD) (Table 1). As in A. thaliana (Koornneef et al. 1991; Gazzani et al. 2003; Lempe et al. 2005), DTF and TLN were correlated (16LD, rs = 0.62, P = 0. 003; 16LDv, rs = 0.80, P << 0.001). Below, we report only DTF data.
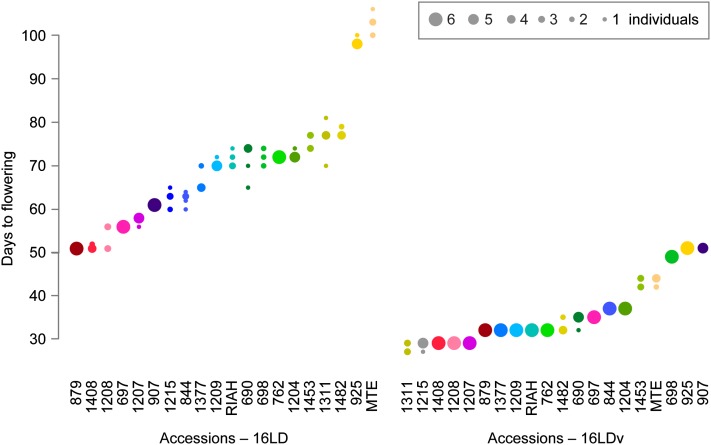
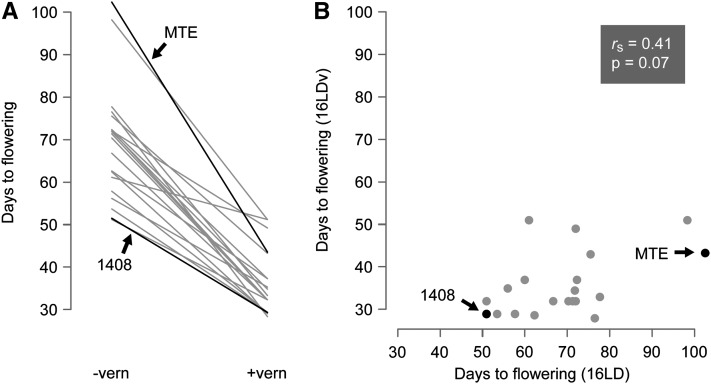
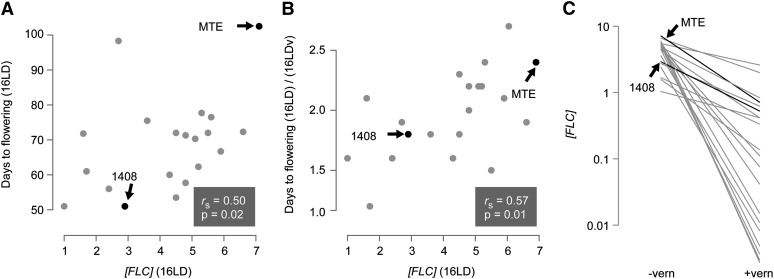
Accessions flowered on average much earlier after vernalization (Figure 1). In 16LD, onset of flowering ranged from 51 to 103 days (mean, 69; median, 71). In 16 LDv, it ranged from 28 to 51 days (mean, 36; median, 33). While all accessions responded to vernalization, there was considerable variation in the extent of the response, as seen most clearly in a reaction norm representation (Figure 2A). Late-flowering accessions tended to respond more strongly to vernalization than early flowering ones, but DTF variation in 16LD explained about 17% of DTF variation in 16LDv (Figure 2B). In addition to vernalization, both longer photoperiods and increased ambient temperature tended to accelerate flowering (Table 1).
Figure 1 .
Distribution of flowering time among 20 C. rubella accessions in 16LD and16LDv. Note the differences in scale. Accessions, which are ordered by mean flowering time in each condition, are color coded for comparison. For each accession, the number of individuals that flowered on a given day is indicated.
Figure 2 .
Relationship of flowering-time variation and vernalization response. (A) Reaction norms of flowering time in 16LD without and with vernalization (−vern, +vern). (B) Correlation of mean flowering times with and without vernalization; same data as in Figure 2A.
FLC underlies a major effect locus for flowering time
The reference accession MTE, from Italy, was the last accession to flower in 16LD, after 103 days on average, which was twice as late as accession 1408, from Greece (Table 1). Following vernalization in 16LDv, MTE was still late, flowering after 43 days, but it was no longer the last accession to flower. A robust vernalization response was also seen in accession 1408, which flowered in 16LDv after 29 days, as did several other accessions.
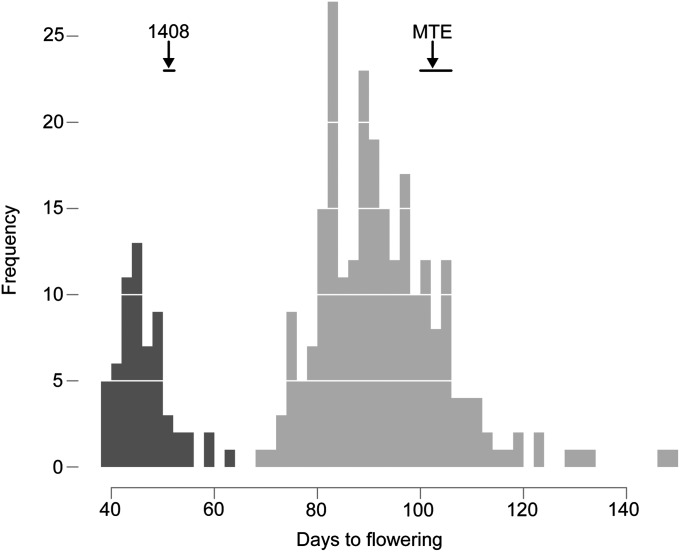
To begin to investigate the genetic basis of flowering-time variation in C. rubella, we crossed the late-flowering accession MTE with the early flowering accession 1408. In an F2 population, about one-quarter of plants flowered early, similar to 1408, and three- quarters flowered late, similar to MTE (Figure 3), suggesting that a major, recessively acting gene controlled early flowering of accession 1408.
Figure 3 .
Flowering time in 16LD among 305 F2 individuals from the MTE × 1408 cross. Average and range of flowering times of grandparents are indicated. Darker shading indicates the 61 individuals used for bulked segregant mapping.
MTE is the reference accession for the ongoing C. rubella genome sequencing effort (collaboration with Jeremy Schmutz from the Joint Genome Institute of the U.S. Department of Energy and the HudsonAlpha Institute for Biotechnology, Stephen Wright and Khaled Hazzouri of the University of Toronto, and Adrian Platts of McGill University). The MTE draft genome sequence in turn enables the discovery of polymorphisms between MTE and 1408, including polymorphisms that are potentially causal for the flowering-time differences between the two accessions. With this rationale in mind, we selected from 305 F2 plants of the MTE × 1408 cross 61 individuals that had flowered after 65 days in 16 LD (Figure 3 and Table S3). We extracted a single pool of genomic DNA from these plants and sequenced the pooled DNA on the Illumina GAIIx platform.
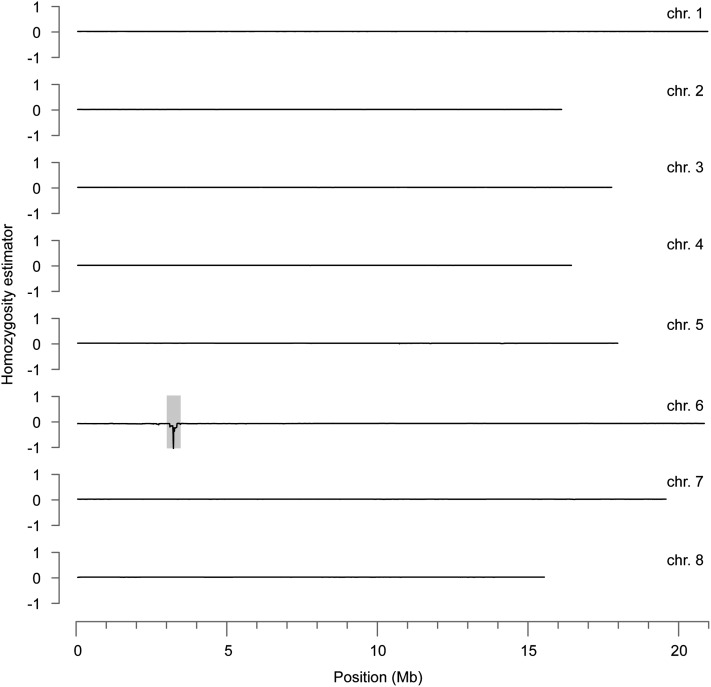
Sequencing reads were aligned to a preliminary genome assembly of C. rubella MTE. The proportion of sequence reads that could not be mapped on the MTE genome was 7.3%, which is similar to what we have found with A. thaliana accessions (Cao et al. 2011). As described (Schneeberger et al. 2009), polymorphisms that can be used for genetic mapping, including SNPs, can be identified directly from F2 data, if one allows for both homo- and heterozygous SNPs. Accordingly, SNPs characteristic for 1408 were discovered from the bulked segregant data. These SNPs were then used to identify regions enriched for either 1408 or MTE SNPs (Schneeberger et al. 2009). Rearrangements and indels were not considered in this step. With this strategy, we identified on chromosome 6 of the genome assembly a region of 333 kb that contained over 600 positions where over 90% of reads supported a nonreference polymorphism (Figure 4 and Table S4). The overall density of 2.0 SNPs/kb was close to the genome-wide average of 2.3 SNPs/kb.
Figure 4 .
SHOREmap analysis of early flowering QTL. The homozygosity estimator is 0 at even allele frequencies of both parents, 1 when homozygous for late-flowering accession MTE, and −1 when homozygous for early flowering accession 1408 (Schneeberger et al. 2009). Sliding windows of 100 kb with step size 10 kb were used. The region of chromosome 6 enriched at over 600 markers indicative of 1408 is indicated in gray. The eight largest scaffolds of a preliminary C. rubella genome assembly are shown, corresponding to the majority of the eight chromosomes.
Using all reads from the bulked segregant pools that mapped to this region, we annotated 1408 variants relative to the Capsella v. 1.0 gene set (http://www.phytozome.net/capsella/), which contained 90 protein-coding genes in the final mapping interval. The algorithms implemented in SHORE v. 0.8 (Ossowski et al. 2008) and pindel v. 0.2.4s (Ye et al. 2009) detected 652 SNPs, 184 small indels of length 1 to 3 bp, 25 longer structural variants (SVs) of length 7 to 679 bp, and 109 highly variable regions, recognizable as simultaneous insertions and deletions (Table S4). Copy number variants (CNVs) and inversions were not detected. Fifty-one SNPs were predicted to cause nonsynonymous amino acid substitutions in 35 genes, with one SNP altering a stop codon. Five SVs affected the coding regions of four genes, and 14 highly variable regions affected the coding regions of 11 genes. Close to the middle of the final mapping interval from position 2.968 to 3.301 Mb, at position 3.112 to 3.120 Mb, was the only C. rubella homolog of A. thaliana FLC, a well-known flowering repressor (Sheldon et al. 1999; Michaels et al. 2003). None of the other genes in this interval were closely related to homologs of known flowering-time genes.
In A. thaliana, there is large variation in FLC levels, and accessions with weak FLC alleles often have very little FLC expression (Lempe et al. 2005; Shindo et al. 2005). Variation in FLC expression was more moderate in C. rubella accessions. MTE had the highest FLC levels, consistent with it being the latest accession, but accession 1408 did not have the least FLC expression (Figure 5A). Across all accessions, there was no clear correlation of FLC expression and flowering in unvernalized plants (Figure 5A), but a modest correlation of FLC and vernalization response (Figure 5B). As expected, expression of FLC was reduced after vernalization, although to quite a different extent in different accessions, which is clearly seen in a reaction norm representation (Figure 5C).
Figure 5 .
Relationship between FLC expression measured by qRT–PCR and flowering time of accessions. (A) Correlation between FLC expression and flowering without vernalization. The lowest value was arbitrarily set to 1. (B) Correlation between FLC expression and vernalization response. (C) Effect of vernalization (vern) on FLC expression. The difference between FLC and BETA-TUBULIN, ΔcT, was used to calculate FLC expression, assuming PCR efficiency was 100% for both genes. Two biological replicates, each with two technical replicates, were analyzed.
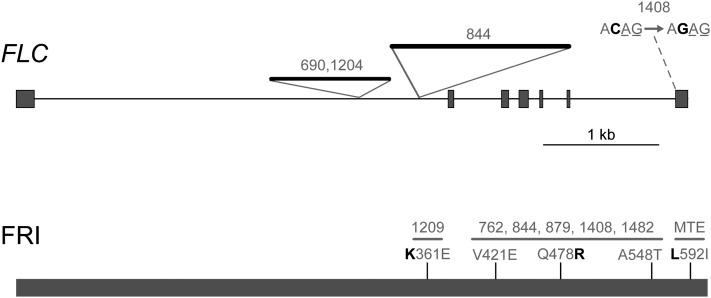
Because it was unclear whether differences of FLC expression were likely to be the cause for earlier flowering of accession 1408 compared to MTE, we compared the FLC DNA sequence of the two accessions. There is an ACAG-to-AGAG substitution at the very end of intron 6 that shifts the splice acceptor site so that two bases are inserted into the FLC mRNA of accession 1408, as revealed by sequencing of cDNA. This leads to a frame shift and truncation of the open reading frame, removing the last 35 of 198 amino acids (Figure 6).
Figure 6 .
Sequence variation of FLC and FRI in C. rubella. In addition to the FLC point mutation at the end of the last intron in accession 1408, two insertions that are segregating in the population, are shown. Thin lines indicate introns; thick lines protein-coding sequences. The C → G substitution creates a new splice acceptor site. PCR amplification and sequencing of cDNA confirmed that 2 bases are inserted into the FLC mRNA in accession 1408. There was no apparent heterogeneity in the sequence, suggesting that the canonical splice form, if it exists at all, is rare. For FRI, amino acid residues that appear to be ancestral based in several Brassicaceae (Irwin et al. 2012) are shown in boldface type; numbers indicate position in the peptide sequence. Shaded numbers indicate affected accessions.
To test whether this polymorphism could indeed be responsible for the flowering-time difference between MTE and 1408, we introduced both FLC alleles into A. thaliana FRISf-2 flc-3 plants. Because we did not know the extent of regulatory sequences in C. rubella, we expressed the FLC alleles under the control of the constitutive cauliflower mosaic virus 35S promoter. The MTE allele strongly delayed flowering, as did overexpression of a wild-type FLC copy from A. thaliana, and both were similarly effective in repressing the flowering activators FT and SOC1 (Figure 7, A and B). In contrast, the 1408 allele delayed flowering by only a few days, from 25.2 to 29.6 days, and it had a much smaller effect on FT and SOC1 expression (Figure 7, A and B). FLC transcript levels were similar in all three transgenic lines (Figure 7B). An analysis of cDNA obtained from plants expressing either the MTE or 1408 allele showed that the 1408 allele produced only the misspliced version of the FLC transcript (Figure 7C).
FLC and variation in flowering-time and vernalization response
Several other accessions flowered as early as 1408 both with and without vernalization (Table 1 and Figure 2). A survey of FLC sequences, however, did not reveal any additional accessions with the 1408 mutation. Compared to C. grandiflora, there were very few FLC polymorphisms in C. rubella. All FLC sequences from C. rubella clustered together and formed a group that was distinct from C. grandiflora FLC alleles (Guo et al. 2009). This strongly suggested that the 1408 mutation arose very recently, after the split of C. rubella and C. grandiflora.
In A. thaliana, large insertions into the first intron are a common cause for reduced FLC activity (Gazzani et al. 2003; Michaels et al. 2003; Liu et al. 2004; Lempe et al. 2005; Shindo et al. 2005; Strange et al. 2011; Sánchez-Bermejo et al. 2012). Three C. rubella accessions had over 1-kb-long insertions into the first intron, but none of them stood out as being particularly early flowering or having a particularly weak vernalization response (Table 1 and Figure 6). We also surveyed the accessions for changes in the open reading frame of FRI, which is located in a region syntenic to chromosome 5 of A. thaliana (Figure S1) (Irwin et al. 2012). The open reading frames were intact in all 20 accessions. However, there were several nonsynonymous substitutions (Figure 6), and π was much higher for FRI than for FLC (0.004 compared with 0.0004).
Discussion
Flowering-time variation in the Brassicaceae
Apart from disease resistance, flowering time is perhaps the most comprehensively studied adaptive trait in A. thaliana. In both laboratory and field conditions, there is a wide range in the onset of flowering. Much of this variation can be explained by allelic variation at FRI along with FLC and its homologs. Consistent with the role of FRI and FLC in repressing flowering in unvernalized plants, much of the flowering variation disappears after vernalization (recently reviewed by Weigel 2012).
Because allelic variation at FRI and FLC is so common in A. thaliana, their homologs have been prime candidates for loci affecting flowering time in other Brassicaceae (Axelsson et al. 2001; Tadege et al. 2001; Lou et al. 2007; Zhao et al. 2010a; Uptmoor et al. 2012). There is evidence for FRI or FLC homologs being responsible for quantitative trait loci (QTL) affecting flowering in A. lyrata, Brassica napus, and B. rapa, with the situation being less clear for B. oleracea (Schranz et al. 2002; Pires et al. 2004; Long et al. 2007; Okazaki et al. 2007; Razi et al. 2007; Kuittinen et al. 2008; Zhao et al. 2010b; Wang et al. 2011). While the potential molecular causes for a change in FLC function in these cases remain uncertain, splice-site mutations in FLC homologs have been linked to earlier flowering in B. rapa and Capsella bursa-pastoris (Slotte et al. 2009; Yuan et al. 2009; Hu et al. 2011). However, in all these cases there has been little direct proof for a causal role of FLC or FRI alleles in flowering-time variation. An induced mutation in the FLC homolog PERPETUAL FLOWERING 1 (PEP1) changes the perennial flowering habit of Arabis alpina. Its response to vernalization is different from the one of A. thaliana FLC, pointing to diversification in the function of FLC homologs in the Brassicaceae (Wang et al. 2009).
Flowering-time variation in C. rubella
Capsella is more distantly related to A. thaliana than the other Arabidopsis species, but more closely than Brassica and Arabis. Capsella is a small genus with only five species, including one tetraploid species, C. bursa-pastoris, one of the most frequent and widespread cosmopolitan weeds (Hurka and Neuffer 1997; Ceplitis et al. 2005; Hurka et al. 2012). The four diploid species include the self-compatible C. rubella, which originated only a few tens of thousands of years ago from the self-incompatible C. grandiflora (Foxe et al. 2009; Guo et al. 2009). C. grandiflora has a restricted distribution, in Western Greece and Albania and, rarely, Northern Italy, while C. rubella has spread across the Mediterranean, and followed European settlers into the New World and Australia (Hurka and Neuffer 1997). In this work, we have documented flowering-time variation in C. rubella under laboratory conditions. Similar to A. thaliana, flowering of many C. rubella accessions is accelerated by vernalization, longer photoperiods, and increased ambient temperature. As in A. thaliana, later-flowering C. rubella accessions tend to respond more strongly to vernalization than early flowering ones (Lempe et al. 2005; Shindo et al. 2005). However, the correlation between flowering-time and vernalization response, or between flowering-time and FLC expression, is less pronounced in C. rubella. This could be due to biased sampling, or it might reflect functional differences between C. rubella and A. thaliana. Given the different ranges of the two species, the second hypothesis is well worth further testing.
FRI alleles with premature stop codons explain a large fraction of flowering-time variation in A. thaliana (Le Corre et al. 2002; Lempe et al. 2005; Shindo et al. 2005; Werner et al. 2005; Atwell et al. 2010) In C. rubella, we did not find any potential loss-of-function allele at FRI, even though the C. rubella accessions surveyed flower faster than many A. thaliana accessions. An indication for the functionality of FRI is that all C. rubella accessions analyzed responded to vernalization. In A. thaliana, structural variation at FLC itself appears to be often responsible for differences in the magnitude of the vernalization response (Michaels et al. 2003; Lempe et al. 2005; Shindo et al. 2006; Sánchez-Bermejo et al. 2012). In C. rubella, we did not observe an obvious correlation between structural variation at FLC and vernalization response.
Flowering-time variation through independent FLC mutations
The reference accession MTE turned out to be one of the latest-flowering C. rubella accessions we surveyed. The segregation pattern in the F2 of a cross to one of the earliest flowering accession in our collection, 1408, suggested the presence of a single major effect QTL responsible for much of the flowering-time difference between the two accessions. To rapidly fine map this QTL, we exploited recent advances in mapping-by-sequencing methods that do not require separate sequence analysis of the parental genomes (Schneeberger et al. 2009). We found that the gene most likely to be responsible for the QTL is the C. rubella homolog of FLC. While we cannot exclude that other genes in the interval contribute to this QTL, none of the 89 other genes in the mapping interval is closely related to a known flowering-time regulator. Moreover, we note that it is difficult to draw firm conclusions about the quantitative differences in the activity of the MTE and 1408 alleles from the A. thaliana experiments, since there might be additional trans- and cis-factors that modify FLC activity between the two species.
The likely causal polymorphism in FLC affects splicing, which in turn removes the C-terminal 35 amino acids. That most of the open reading frame remains intact may explain why accession 1408 retains a robust vernalization response. In A. thaliana, reduction- or loss-of-function alleles of FLC either have insertions in the first, large intron, or they carry premature stop codons that can lead to alternative splicing (Michaels et al. 2003; Liu et al. 2004; Lempe et al. 2005; Shindo et al. 2005; Werner et al. 2005; Méndez-Vigo et al. 2011; Sánchez-Bermejo et al. 2012). However, alleles with splice-site mutations have, to our knowledge, not been described in A. thaliana. It is striking that FLC alleles with splice-site mutations have now been found in three other species, in B. rapa, C. bursa-pastoris, and C. rubella (this work; Slotte et al. 2009; Yuan et al. 2009; Hu et al. 2011).
Although several other accessions show a similar flowering behavior as in 1408, the 1408 FLC allele is not shared by any other accession. Moreover, the pattern of polymorphisms in FLC alleles suggests that the 1408 mutation arose only after the split from C. grandiflora (Figure S2). Because of the strong recent bottleneck experienced by C. rubella (Foxe et al. 2009; Guo et al. 2009), there is only limited standing variation in this species, and new mutations likely play an important role in the adaptation to different environments. There is precedence for FLC, and to a lesser extent, FRI, loss-of-function alleles being rare. Although few individual loss-of-function alleles segregate at appreciable frequency in A. thaliana flowering, collectively, such alleles are quite frequent. There is good evidence that the more common FRI loss-of-function alleles have increased in frequency due to selection for early flowering (Toomajian et al. 2006), a conclusion that is supported by experimental selection experiments (Scarcelli and Kover 2009). In animals, most cases of parallel evolution are from closely related species, but there are also examples of the same gene being responsible for genetic variation in more distantly related taxa, the most notorious one being the melanocortin-1 receptor gene (Mc1r), which underlies changes in pigmentation in reptiles, birds, and mammals (Wood et al. 2005; Hoekstra 2006; Arendt and Reznick 2008; Protas and Patel 2008; Gompel and Prud’homme 2009; Elmer and Meyer 2011). Similarly, there are several cases of parallel evolution in flower pigmentation (Quattrocchio et al. 1999; Schwinn et al. 2006; Whittall et al. 2006; Hoballah et al. 2007; Streisfeld and Rausher 2009; Des Marais and Rausher 2010; Smith and Rausher 2011). How broadly FLC contributes to flowering-time variation in C. rubella is, however, not known yet. Thus, further studies are needed to determine whether it is appropriate to speak of parallel evolution when comparing the basis of flowering-time variation in A. thaliana and C. rubella.
In summary, we have found substantial flowering-time variation in C. rubella, although its extent is smaller than in A. thaliana, perhaps reflecting the more restricted geographic range of C. rubella (Hurka and Neuffer 1997; Hoffmann 2002). In one accession, a major-effect QTL affecting flowering maps FLC. In experimental crosses of A. thaliana, a small number of major effect QTL, including FRI, FLC, the FLC homolog FLOWERING LOCUS M (FLM)/MADS AFFECTING FLOWERING 1 (MAF1), the MAF2-5 cluster, and FLOWERING LOCUS T (FT), explain much of the variation in flowering time (Brachi et al. 2010; Salomé et al. 2011; Strange et al. 2011). Given the reduced genetic diversity in C. rubella (Foxe et al. 2009; Guo et al. 2009), it will be interesting to determine how many additional loci are responsible for flowering-time variation.
Supplementary Material
Acknowledgments
We thank Jeremy Schmutz (DOE-JGI and HudsonAlpha), Stephen Wright and Khaled Hazzouri (University of Toronto), and Adrian Platts (McGill University) for access to a preliminary version of the C. rubella genome assembly. We thank Sureshkumar Balasubramanian, Patrice Salomé, Jia-Wei Wang, Xuan Zhao, and Zhong Zhao for advice, Dan Koenig and Patrice Salomé for comments on the manuscript, Korbinian Schneeberger for help with SHOREmap analyses, and Christa Lanz for sequencing support. This work was supported by Bundesministerium für Bildung und Forschung (BMBF) grant Next Generation Forward Genetics for Crop Plants (NUGGET), a Gottfried Wilhelm Leibniz Award to the Deutsche Forschungsgemeinschaft (DFG), and the Max Planck Society.
Footnotes
Communicating editor: J. Borevitz
Literature Cited
- Arendt J., Reznick D., 2008. Convergence and parallelism reconsidered: What have we learned about the genetics of adaptation? Trends Ecol. Evol. 23: 26–32 [DOI] [PubMed] [Google Scholar]
- Atwell S., Huang Y. S., Vilhjalmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson T., Shavorskaya O., Lagercrantz U., 2001. Multiple flowering time QTLs within several Brassica species could be the result of duplicated copies of one ancestral gene. Genome 44: 856–864 [PubMed] [Google Scholar]
- Barrett R. D., Schluter D., 2008. Adaptation from standing genetic variation. Trends Ecol. Evol. 23: 38–44 [DOI] [PubMed] [Google Scholar]
- Bergelson J., Roux F., 2010. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat. Rev. Genet. 11: 867–879 [DOI] [PubMed] [Google Scholar]
- Brachi B., Faure N., Horton M., Flahauw E., Vazquez A., et al. , 2010. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6: e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Schneeberger K., Ossowski S., Gunther T., Bender S., et al. , 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Ceplitis A., Su Y., Lascoux M., 2005. Bayesian inference of evolutionary history from chloroplast microsatellites in the cosmopolitan weed Capsella bursa-pastoris (Brassicaceae). Mol. Ecol. 14: 4221–4233 [DOI] [PubMed] [Google Scholar]
- Des Marais D. L., Rausher M. D., 2010. Parallel evolution at multiple levels in the origin of hummingbird pollinated flowers in Ipomoea. Evolution 64: 2044–2054 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 19: 11–15 [Google Scholar]
- Elmer K. R., Meyer A., 2011. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol. Evol. 26: 298–306 [DOI] [PubMed] [Google Scholar]
- Foxe J. P., Slotte T., Stahl E. A., Neuffer B., Hurka H., et al. , 2009. Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl. Acad. Sci. USA 106: 5241–5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S., Gendall A. R., Lister C., Dean C., 2003. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., 2009. The causes of repeated genetic evolution. Dev. Biol. 332: 36–47 [DOI] [PubMed] [Google Scholar]
- Guo Y. L., Bechsgaard J. S., Slotte T., Neuffer B., Lascoux M., et al. , 2009. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock A. M., Brachi B., Faure N., Horton M. W., Jarymowycz L. B., et al. , 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86 [DOI] [PubMed] [Google Scholar]
- Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M., 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hoballah M. E., Gubitz T., Stuurman J., Broger L., Barone M., et al. , 2007. Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra H. E., 2006. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97: 222–234 [DOI] [PubMed] [Google Scholar]
- Hoffmann M. H., 2002. Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J. Biogeogr. 29: 125–134 [Google Scholar]
- Hu G. L., Hu Z. L., Li Y., Gu F., Zhao Z. P., et al. , 2011. A splicing site mutation in BrpFLC1 and repressed expression of BrpFLC genes are associated with the early flowering of purple flowering stalk. Russ. J. Plant Physiol. 58: 431–438 [Google Scholar]
- Hurka H., Neuffer B., 1997. Evolutionary processes in the genus Capsella (Brassicaceae). Plant Syst. Evol. 206: 295–316 [Google Scholar]
- Hurka H., Friesen N., German D. A., Franzke A., Neuffer B., 2012. ‘Missing link’ species Capsella orientalis and Capsella thracica elucidate evolution of model plant genus Capsella (Brassicaceae). Mol. Ecol. 21: 1223–1238 [DOI] [PubMed] [Google Scholar]
- Irwin J. A., Lister C., Soumpourou E., Zhang Y., Howell E. C., et al. , 2012. Functional alleles of the flowering time regulator FRIGIDA in the Brassica oleracea genome. BMC Plant Biol. 12: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., et al. , 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Koch M. A., Kiefer M., 2005. Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species—Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana. Am. J. Bot. 92: 761–767 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Meinke D., 2010. The development of Arabidopsis as a model plant. Plant J. 61: 909–921 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C. J., van der Veen J. H., 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kuittinen H., Niittyvuopio A., Rinne P., Savolainen O., 2008. Natural variation in Arabidopsis lyrata vernalization requirement conferred by a FRIGIDA indel polymorphism. Mol. Biol. Evol. 25: 319–329 [DOI] [PubMed] [Google Scholar]
- Le Corre V., Roux F., Reboud X., 2002. DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol. Biol. Evol. 19: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Lee I., Bleecker A., Amasino R., 1993. Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237: 171–176 [DOI] [PubMed] [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., et al. , 2005. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang Y., Bergelson J., Nordborg M., Borevitz J. O., 2010. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., He Y., Amasino R., Chen X., 2004. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 18: 2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y., Shi J., Qiu D., Li R., Zhang C., et al. , 2007. Flowering time quantitative trait loci analysis of oilseed brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177: 2433–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou P., Zhao J., Kim J. S., Shen S., Del Carpio D. P., et al. , 2007. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 58: 4005–4016 [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B., Picó X. F., Ramiro M., Martinez-Zapater J. M., Alonso-Blanco C., 2011. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 157: 1942–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M., 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., He Y., Scortecci K. C., Amasino R. M., 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular Evolutionary Genetic. Columbia University Press, New York [Google Scholar]
- Okazaki K., Sakamoto K., Kikuchi R., Saito A., Togashi E., et al. , 2007. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 114: 595–608 [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R. M., Lanz C., Warthmann N., et al. , 2008. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires J. C., Zhao J., Schranz M. E., Leon E. J., Quijada P. A., et al. , 2004. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. 82: 675–688 [Google Scholar]
- Protas M. E., Patel N. H., 2008. Evolution of coloration patterns. Annu. Rev. Cell Dev. Biol. 24: 425–446 [DOI] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J., van der Woude K., Souer E., de Vetten N., et al. , 1999. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi H., Howell E. C., Newbury H. J., Kearsey M. J., 2007. Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor. Appl. Genet. 116: 179–192 [DOI] [PubMed] [Google Scholar]
- Rozas J., Sánchez-DelBarrio J. C., Messeguer X., Rozas R., 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497 [DOI] [PubMed] [Google Scholar]
- Salomé P. A., Bomblies K., Laitinen R. A., Yant L., Mott R., et al. , 2011. Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Bermejo E., Méndez-Vigo B., Picó F. X., Martinez-Zapater J. M., Alonso-Blanco C., 2012. Novel natural alleles at FLC and LVR loci account for enhanced vernalization responses in Arabidopsis thaliana. Plant Cell Environ 35: 1672–1684 [DOI] [PubMed] [Google Scholar]
- Scarcelli N., Kover P. X., 2009. Standing genetic variation in FRIGIDA mediates experimental evolution of flowering time in Arabidopsis. Mol. Ecol. 18: 2039–2049 [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A. H., et al. , 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Schranz M. E., Quijada P., Sung S. B., Lukens L., Amasino R., et al. , 2002. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K., Venail J., Shang Y., Mackay S., Alm V., et al. , 2006. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Burn J. E., Perez P. P., Metzger J., Edwards J. A., et al. , 1999. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Aranzana M. J., Lister C., Baxter C., Nicholls C., et al. , 2005. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Lister C., Crevillen P., Nordborg M., Dean C., 2006. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Huang H. R., Holm K., Ceplitis A., Onge K. S., et al. , 2009. Splicing variation at a FLOWERING LOCUS C homeolog is associated with flowering time variation in the tetraploid Capsella bursa-pastoris. Genetics 183: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. D., Rausher M. D., 2011. Gene loss and parallel evolution contribute to species difference in flower color. Mol. Evol. Biol. 28: 2799–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., Schmid M., 2011. Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange A., Li P., Lister C., Anderson J., Warthmann N., et al. , 2011. Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS ONE 6: e19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisfeld M. A., Rausher M. D., 2009. Genetic changes contributing to the parallel evolution of red floral pigmentation among Ipomoea species. New Phytol. 183: 751–763 [DOI] [PubMed] [Google Scholar]
- Tadege M., Sheldon C. C., Helliwell C. A., Stoutjesdijk P., Dennis E. S., et al. , 2001. Control of flowering time by FLC orthologues in Brassica napus. Plant J. 28: 545–553 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G., 1997. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian C., Hu T. T., Aranzana M. J., Lister C., Tang C., et al. , 2006. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptmoor R., Li J., Schrag T., Stutzel H., 2012. Prediction of flowering time in Brassica oleracea using a quantitative trait loci-based phenology model. Plant Biol. 14: 179–189 [DOI] [PubMed] [Google Scholar]
- Wang N., Qian W., Suppanz I., Wei L., Mao B., et al. , 2011. Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA.FRI.a. J. Exp. Bot. 62: 5641–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Farrona S., Vincent C., Joecker A., Schoof H., et al. , 2009. PEP1 regulates perennial flowering in Arabis alpina. Nature 459: 423–427 [DOI] [PubMed] [Google Scholar]
- Weigel D., 2012. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J., 2002. Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Werner J. D., Borevitz J. O., Uhlenhaut N. H., Ecker J. R., Chory J., et al. , 2005. FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittall J. B., Voelckel C., Kliebenstein D. J., Hodges S. A., 2006. Convergence, constraint and the role of gene expression during adaptive radiation: floral anthocyanins in Aquilegia. Mol. Ecol. 15: 4645–4657 [DOI] [PubMed] [Google Scholar]
- Wood T. E., Burke J. M., Rieseberg L. H., 2005. Parallel genotypic adaptation: when evolution repeats itself. Genetica 123: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. X., Wu J., Sun R. F., Zhang X. W., Xu D. H., et al. , 2009. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 60: 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Artemyeva A., Del Carpio D. P., Basnet R. K., Zhang N., et al. , 2010a Design of a Brassica rapa core collection for association mapping studies. Genome 53: 884–898 [DOI] [PubMed] [Google Scholar]
- Zhao J., Kulkarni V., Liu N., Del Carpio D. P., Bucher J., et al. , 2010b BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 61: 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.