Abstract
Purpose: The prevalence of low birth weight (LBW) is increased in subjects born after assisted reproduction technology (ART), and defective imprinting has frequently been identified in patients with Beckwith-Wiedermann and Angelman syndromes conceived by ART. Thus, we examined methylation pattern in a girl born after ART who had Silver-Russell syndrome (SRS) which can be caused by maternal uniparental disomy for chromosome 7 and by hypomethylation of the differentially methylated region (DMR) of H19.
Methods: We examined methylation status of 31 cytosines at the CpG dinucleotides in the DMR of PEG1/MEST on 7q32.2 and 23 cytosines at the CpG dinucleotides in the DMR of H19 on 11p15, using leukocyte genomic DNA.
Results: Eight of the 31 cytosines in the patient and four of the 31 cytosines in the father were hypermethylated in the PEG1/MEST-DMR. In the H19-DMR, no abnormal methylation pattern was identified in the patient.
Conclusion: The results suggest that hypermethylation of paternally expressed genes including PEG1/MEST, which usually have growth-promoting effects, may be relevant to LBW in subjects conceived by ART.
Keywords: Hypermethylation, Imprinting, In vitro fertilization, PEG1/MEST, Silver-Russell syndrome
Introduction
Silver-Russell syndrome (SRS) is a congenital developmental disorder characterized by pre- and postnatal growth failure, body asymmetry, relative macrocephaly, triangular face, and fifth-finger clinodactyly [1]. Precocious puberty is also occasionally observed in this condition. Since maternal uniparental disomy for chromosome 7 (mUPD7) has been identified in 7–10% of patients with SRS, this implies the involvement of a single or plural imprinted genes on chromosome 7 in the development of SRS [1]. The possibility of unmasking of a recessive allele(s) by isodisomy is unlikely, because both isodisomy and heterodisomy have been found in SRS with no common isodisomic region [2]. Furthermore, molecular studies in several key patients have suggested two separate candidate regions for SRS, 7p11.2–p13 [3] and 7q31–qter [4], and several candidate genes such as GRB10 (growth factor receptor-bound protein 10) on 7p12 and PEG1/MEST (paternally expressed gene 1/mesoderm-specific transcript) on 7q32.2 have been identified [1], although the gene(s) responsible for SRS has not been identified to date. In addition, recent studies have shown hypomethylation of the differentially methylated region (DMR) located upstream of H19 (H19-DMR) in a considerable fraction of SRS patients, providing further support for the relevance of imprinting failure in SRS [5].
In vitro fertilization (IVF) with or without intracytoplasmic sperm injection (ICSI) has become a widely utilized method for infertility. However, such assisted reproduction technology (ART) has been regarded as a risk factor for low birth weight (LBW) and major birth defects [6, 7]. Furthermore, recent studies in two human imprinting disorders, Beckwith-Wiedermann syndrome and Angelman syndrome, have indicated aberrant methylation pattern in most of the affected children born after ART, suggesting that ART may perturb the epigenetic imprinting process [8–10]. In addition, the prevalence of Beckwith-Wiedermann syndrome has been reported to be significantly higher in children conceived by ART than in those conceived naturally [11].
To our knowledge, however, abnormal methylation pattern in SRS patients born after ART has been reported in only a single patient who was conceived by ICSI and found to have hypomethylation of the H19-DMR [12]. Here, we report on methylation analysis in a girl with SRS born after IVF.
Materials and methods
Case report
This Japanese girl and her twin sister were conceived with use of IVF, and delivered by a cesarean section at 37 weeks of gestation. The parents were non-consanguineous and clinically normal. The paternal height was 174.0 cm (+0.6 SD), and the maternal height 150.0 cm (−1.6 SD). They had a male infant after natural conception in the second year of their conjugal life, who had severe hydrocephalus and deceased at six months of age. Thereafter, they failed to conceive for 12 years, and received IVF. Maternal oocytes taken after gonadotropin stimulation were mixed with paternal sperms collected using a condom. Six fertilized ova were obtained, and two of them were transferred to the maternal uterus, resulting in a production of the twin sisters.
At birth, her length was 36.5 cm (−6.0 SD), weight 1.25 kg (−4.6 SD), and head circumference (HC) 30.0 cm (−2.0 SD). At 3 6/12 years of age, she was seen because of short stature. Her height was 82.1 cm (−3.6 SD), weight 7.7 kg (−3.9 SD), and HC 46.2 cm (−1.5 SD). The growth chart and photograph are shown in Fig. 1. Physical examination showed SRS-compatible features such as triangle face with micrognathia, relative macrocephaly, high arched palate, right hemihypotrophy, and bilateral clinodactyly of the 5th fingers. There were no discernible major anomalies. Psychomotor development was age-appropriate. Endocrine studies for short stature were normal, as were radiological studies. Her karyotype was 46,XX in all the 20 lymphocytes examined. She received growth hormone therapy from 4 11/12 to 8 7/12 years of age, and showed upward growth shift. She exhibited breast development from 7 1/12 years of age and pubic hair development from 8 2/12 years of age, and had menarche at 8 8/12 years of age (menarchial age of normal Japanese girls: 12.25 ± 1.25 years). She was diagnosed as having central precocious puberty by a gonadotropin releasing hormone (GnRH) test, and was placed on GnRH analog therapy from 8 9/12 years of age. On the last examination at 9 2/12 years of age, her height was 125.5 cm (−1.0 SD), weight 22.3 kg (−1.2 SD), and HC 50.3 cm (−1.5 SD).
Fig. 1.
Growth charts depicted on the standard growth curves for the Japanese girls and photographs of the twin sisters at 4 11/12 of age. GH: growth hormone; and GnRH: gonadotropin releasing hormone
The twin sister was clinically normal (Fig. 1). Her birth length was 43.0 cm (−3.1 SD), weight 2.3 kg (−2.0 SD), and HC 31.3 cm (−1.0 SD). She showed catch-up growth during infancy, and her height remained around −0.8 SD in childhood. At 9 2/12 years of age, he exhibited breast budding, and her height was 128.7 cm (−0.3 SD), weight 24.7 kg (−0.8 SD), and HC 51.9 cm (−0.5 SD).
One and half years after the birth of the twins, a younger son with normal phenotype was born after natural conception.
Methylation analysis
This study has been approved by the Institutional Review Board Committee at National Center for Child Health and Development. Methylation pattern was first analyzed for the DMR of PEG1/MEST, using leukocyte genomic DNA of the twins and the parents (Fig. 2A). In brief, after bisulphite treatment that converts all the cytosines except for methylated cytosines at the CpG islands into uracils and subsequently thymines [13], a genomic sequence containing 31 CpG dinucleotides was amplified by polymerase chain reaction (PCR) with a methylated allele specific primer pair (MET) hybridizing to a region containing unconverted methylated cytosines and an unmethylated allele specific primer pair (UNMET) hybridizing to a region containing thymines converted from unmethylated cytosines [14]. Subsequently, the PCR products were subjected to direct sequencing on a CEQ 8000 autosequencer (Beckman Coulter, Fullerton, CA). Since maternally derived cytosines at the CpG dinucleotides are normally methylated, they should appear as cytosines in the PCR products obtained with MET; conversely, since paternally derived cytosines at the CpG dinucleotides are normally unmethylated, they should be detected as thymines in the PCR products obtained with UNMET. Furthermore, sequencing analysis was also performed with another primer pair (COMMON) that hybridizes to a region lacking CpG dinucleotides [15]. Since PCR with COMMON amplifies both methylated and unmethylated alleles, cytosines at the CpG dinucleotides are normally delineated as heterozygous peaks for cytosines and thymines after bisulphite treatment. For a control, the DNA samples of a previously described SRS patient with mUPD7 [14] and 50 normal subjects were similarly examined with permission.
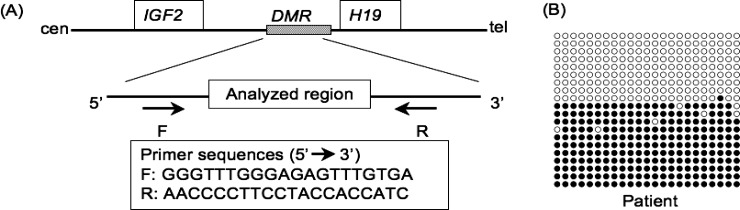
Fig. 2.
Methylation analysis of the PEG1/MEST-DMR (differentially methylated region). (A) Methylation pattern analysis employed in the present study. The upper part: a schematic representation indicating the analyzed region within the DMR encompassing E1 (exon 1) of PEG1/MEST. The positions of the primers utilized for the polymerase chain reaction (PCR) are also shown. The lower left part: genomic sequence of the analyzed region containing 31 CpG dinucleotides. The cytosine residues surrounded by squares are methylated on the maternally derived alleles and unmethylated on the paternally derived alleles. The shaded nucleotide sequence is shown in the electrochromatograms. The lower right part: primer sequences utilized in this study. MET-F and MET-R are designed to hybridize the sequence harboring cytosines, so that they specifically amplify the maternally derived alleles with methylated cytosines at the CpG dinucleotides after bisulphite treatment. UNMET-F and UNMET-R are designed to hybridize the sequence harboring thymines, so that they specifically amplify the paternally derived alleles with thymines converted from unmethylated cytosines via uracils after bisulphite treatment. The nucleotides specific to the methylated and unmethylated alleles are surrounded by squares. COMMON-F and COMMON-R are designed to hybridize the sequence lacking CpG dinucleotides, so that they amplify both paternally and maternally derived alleles after bisulphite treatment. (B) Representative electrochromatograms of a region containing two CpG dinucleotide sequences obtained after bisulphite treatment. The cytosines at the CpG nucleotides are delineated as cytosines (arrows) in the PCR products obtained with MET-F and MET-R (Methylated) in the mother and the patient. In the PCR products obtained with UNMET-F and UNMET-R (Unmethylated), the cytosines at the CpG nucleotides are detected as thymines in the mother and as cytosines in the patient (arrows). (C) Summary of the methylation status of the cytosine residues at the 31 CpG dinucleotides examined after bisulphite treatment. The black circles represent cytosine residues, and the open circles denote thymine residues. The sequence results obtained with MET-F and MET-R are shown on the upper array of circles, and those obtained with UNMET-F and UNMET-R are indicated on the lower array of circles. Hypermethylation, as indicated by the presence of cytosine residues in the PCR products obtained with UNMET-F and UNMET-R, has been identified in 4 of the 31 cytosines in the father and in 8 of the 31 cytosines in the patient. In a subject with maternal disomy for chromosome 7, the PCR products have been obtained only with MET-F and MET-R, and cytosines only have been delineated. The methylation pattern is normal in the mother and the sister, as well as in control subjects
Next, methylation analysis was performed for the H19-DMR which is methylated after paternal transmission and unmethylated after maternal transmission (Fig. 3A). In brief, after the bisulphite treatment, a 319 bp sequence containing 23 CpG dinucleotides was amplified with primers that hybridize to a region lacking CpG dinucleotides. Subsequently, the PCR products were subcloned with TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA), and 20 clones were subjected to direct sequencing.
Fig. 3.
Methylation analysis of the H19-DMR (differentially methylated region). (A) A schematic representation indicating the analyzed region. PCR was performed after bisulphite treatment with the primers that amplify both the methylated and unmethylated alleles. (B) The results of methylation analysis. Each lane indicates a single cloned allele, and each circle denotes a CpG island; filled and open circles represent methylated and unmethylated cytosines, respectively. Both of the methylated and unmethylated alleles are identified with a similar frequency in the patient
Furthermore, methylation status was similarly analyzed for a genomic sequence containing 10 CpG dinucleotides within the DMR of SNRPN (small nuclear ribonucleoprotein polypeptide N) on 15q11.2, where cytosines at the CpG dinucleotides are unmethylated on the paternally derived allele and methylated on the maternally derived allele [13, 16].
Microsatellite analysis
Microsatellite analysis was performed for 14 loci widely dispersed on chromosome 7. In brief, leukocyte genomic DNA of the twins and the parents was amplified by PCR with fluorescently labeled forward primers and unlabeled reverse primers, and the PCR products were determined for the fragment size on an ABI PRISM 310 autosequencer using GeneScan software (Applied Biosystems, Perkin Elmer, Foster City, CA). The primer sequences were as described in Genome Database (http://www.gdb.org/).
Results
Methylation analysis
The results of the PEG1/MEST-DMR are shown in Fig. 2B and C. In the patient and the father, while all the 31 cytosines at the CpG dinucleotides were detected as cytosines in the PCR products amplified with MET, eight of the 31 cytosines in the patient and four of the 31 cytosines in the father were found to be cytosines, not thymines, in the PCR products amplified with UNMET. In the mother and the sister as well as in the 50 control subjects, all the 31 cytosines at the CpG dinucleotides were delineated as cytosines and thymines in the PCR products amplified with MET and UNMET, respectively. In the SRS patient with mUPD7, PCR products were obtained only with MET, and all the 31 cytosines at the CpG dinucleotides were detected as cytosines. The sequences of the PCR products with COMMON confirmed the results, and showed the absence of abnormal methylation in the MET and UNMET hybridizing region.
The results of the H19-DMR are indicated in Fig. 3B. The methylated and the unmethylated alleles were delineated with a similar frequency in the patient. The methylation pattern of the DMR at SNRPN was also normal (data not shown).
Microsatellite analysis
Genotyping results were consistent with biparental origin of the chromosome 7 in the patient as well as in the twin sister (Fig. 4). Furthermore, the genotype comparison between the twins indicated that, for six of the seven loci examined in the sister, the twins inherited different alleles from the father and the same allele from the mother, although the results of D7S634 were not informative. In particular, the paternal origin of the alleles for D7S515 and D7S684 flanking PEG1/MEST was different between the twins.
Fig. 4.
Summary of microsatellite analysis. The ideogram of chromosome 7 is shown with the cytogenetic localization of the microsatellite loci examined. The locus position is based on Ensembl Human Genome Browser (http://www.ensembl.org). N.E.: not examined
Discussion
The present study revealed a partial hypermethylation (8/31) at the examined DMR of PEG1/MEST in the patient. Furthermore, four of the eight abnormally methylated cytosines were also identified in the father, and the microsatellite analysis indicated the different paternal allelic origin of PEG1/MEST between the twins. It is inferred, therefore, that the paternal PEG1/MEST allele with mildly hypermethylated DMR was further methylated and transmitted to the patient. Although the mechanism leading to such enhanced methylation remains to be clarified, it may be due to some environmental factor(s) inherent to IVF such as in vitro culture or media used, or to some genetic factor(s) relevant to the mild hypermethylation in the father that may also be involved in the subfertility in this couple [17]. In this regard, this girl was conceived by simple mixture of sperms and oocytes, whereas most patients with Beckwith-Wiedermann syndrome and Angelman syndrome with loss of imprinting of maternally imprinted genes such as LIT1 (long QT intronic transcript 1) as well as a single SRS patient with hypomethylated H19-DMR have been conceived by ISCI [8–12, 17]. Such technical difference might underlie the partial hypermethylation in this patient.
It is uncertain whether the partial hypermethylation is relevant to the development of SRS in this patient. However, hypomethylation of the H19-DMR was excluded, and the partial hypermethylation might have affected the expression of PEG1/MEST, contributing to the SRS phenotype. In support of this, while no PEG1/MEST mutation has been identified in patients with SRS [15], Peg1/Mest knockout mice show pre- and post-natal growth failure when the mutant gene is transmitted from the father [18]. In addition, while it is unlikely that such abnormal methylation has occurred in multiple DMRs, as indicated by the normal methylation pattern of the SNRPN-DMR, it might be possible that such abnormal methylation is also present in other DMRs of the imprinted gene(s) for SRS, impairing the expression of the corresponding gene(s). In this regard, methylation pattern was not examined for the DMR region of GRB10, because GRB10 shows biparental expression in most tissues, except for the brain in which the paternally derived allele is preferentially expressed [19].
The results, though fragmentary, would provide a new insight into the genetic mechanism(s) for the development of LBW in individuals conceived by ART. The mUPD7 results in growth failure with and without SRS phenotype [20]. Furthermore, paternally expressed genes usually serve to promote fetal growth, and maternal disomy for several chromosomes are known to cause growth failure [20]. Thus, excessive methylation affecting the expression of paternally expressed gene(s) including PEG1/MEST may be involved in the high prevalence of LBW in individuals born after ART. In this context, while loss of imprinting of maternally imprinted genes and resultant overexpression of paternally expressed genes have been identified in most of the patients born after ART who have Beckwith-Wiedermann syndrome and Angelman syndrome associated with normal to relative excessive growth [17] as well as in ART-related large offspring syndrome in ruminants [21], hypermethylation of paternally expressed genes has not been studied in LBW patients conceived by ART. Thus, methylation pattern in paternally expressed genes would be worth analyzing in such patients.
In summary, we observed partial hypermethylation in an SRS girl conceived with use of ART. Further studies will serve to clarify the relevance of abnormal imprinting to the high prevalence of LBW with and without SRS phenotype in subjects born after ART.
Acknowledgements
This work was supported by a grant for Child Health and Development (17C-2) and a grant for Research on Children and Families from the Ministry of Health, Labor, and Welfare.
Footnotes
Partial hypermethylation was identified at the differentially methylated region of paternally expressed PEG1/MEST in a girl with Silver-Russell syndrome born after in vitro fertilization.
References
- 1.Hitchins MP, Stanier P, Preece MA, Moore GE. Silver-Russell syndrome: a dissection of the genetic aetiology and candidate chromosomal regions. J Med Genet. 2001;38:810–819. doi: 10.1136/jmg.38.12.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preece MA, Abu-Amero SN, Ali Z, Abu-Amero KK, Wakeling EL, Stanier P, Moore GE. An analysis of the distribution of hetero- and isodisomic regions of chromosome 7 in five mUPD7 Silver-Russell syndrome probands. J Med Genet. 1999;36:457–460. [PMC free article] [PubMed] [Google Scholar]
- 3.Monk D, Wakeling EL, Proud V, Hitchins M, Abu-Amero SN, Stanier P, Preece MA, Moore GE. Duplication of 7p11.2-p13, including GRB10, in Silver-Russell syndrome. Am J Hum Genet. 2000;66:36–46. doi: 10.1086/302717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannula K, Lipsanen-Nyman M, Kontiokari T, Kere J. A narrow segment of maternal uniparental disomy of chromosome 7q31-qter in Silver-Russell syndrome delimits a candidate gene region. Am J Hum Genet. 2001;68:247–253. doi: 10.1086/316937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, Barbu V, Danton F, Thibaud N, Le Merrer M, Burglen L, Bertrand AM, Netchine I, Le Bouc Y. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 6.Hansen M, Kurinczuk JJ, Bower C, Webb S. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. N Engl J Med. 2002;346:725–730. doi: 10.1056/NEJMoa010035. [DOI] [PubMed] [Google Scholar]
- 7.Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng G, Wilcox LS. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med. 2002;346:731–737. doi: 10.1056/NEJMoa010806. [DOI] [PubMed] [Google Scholar]
- 8.Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71:162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBaun MR, Niemitz E, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet. 2003;72:1338–1341. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, Macdonald F, Sampson JR, Barratt CL, Reik W, Hawkins MM. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART) J Med Genet. 2003;40:62–64. doi: 10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bliek J, Terhal P, Van Den Bogaard MJ, Maas S, Hamel B, Salieb-Beugelaar G, Simon M, Letteboer T, Van Der Smagt J, Kroes H, Mannens M. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am J Hum Genet. 2006;78:604–614. doi: 10.1086/502981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota T, Das S, Christian SL, Baylin SB, Herman JG, Ledbetter DH. Methylation-specific PCR simplifies imprinting analysis. Nat Genet. 1997;16:16–17. doi: 10.1038/ng0597-15. [DOI] [PubMed] [Google Scholar]
- 14.Kosaki K, Kosaki R, Robinson WP, Craigen WJ, Shaffer LG, Sato S, Matsuo N. Diagnosis of maternal uniparental disomy of chromosome 7 with a methylation specific PCR assay. J Med Genet. 2000;37:E19. doi: 10.1136/jmg.37.9.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi S, Uemura H, Kohda T, Nagai T, Chinen Y, Naritomi K, Kinoshita EI, Ohashi H, Imaizumi K, Tsukahara M, Sugio Y, Tonoki H, Kishino T, Tanaka T, Yamada M, Tsutsumi O, Niikawa N, Kaneko-Ishino T, Ishino F. No evidence of PEG1/MEST gene mutations in Silver-Russell syndrome patients. Am J Med Genet. 2001;104:225–231. doi: 10.1002/ajmg.10022. [DOI] [PubMed] [Google Scholar]
- 16.Kosaki K, McGinniss MJ, Veraksa AN, McGinnis WJ, Jones KL. Prader-Willi and Angelman syndromes: diagnosis with a bisulfite-treated methylation-specific PCR method. Am J Med Genet. 1997;73:308–313. doi: 10.1002/(SICI)1096-8628(19971219)73:3<308::AID-AJMG15>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 17.Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. Am J Hum Genet. 2004;74:599–609. doi: 10.1086/382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- 19.Blagitko N, Mergenthaler S, Schulz U, Wollmann HA, Craigen W, Eggermann T, Ropers HH, Kalscheuer VM. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue-and isoform-specific fashion. Hum Mol Genet. 2000;9:1587–1595. doi: 10.1093/hmg/9.11.1587. [DOI] [PubMed] [Google Scholar]
- 20.Hurst LD, McVean GT. Growth effects of uniparental disomies and the conflict theory of genomic imprinting. Trends Genet. 1997;13:436–443. doi: 10.1016/S0168-9525(97)01273-0. [DOI] [PubMed] [Google Scholar]
- 21.Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, Broadbent PJ, Robinson JJ, Wilmut I, Sinclair KD. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27:153–154. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]