Summary
The recent reports of artemisinin (ART) resistance in the Thai-Cambodian border area raise a serious concern on the long-term efficacy of ARTs. To elucidate the resistance mechanisms, we performed in vitro selection with dihydroartemisinin (DHA) and obtained two parasite clones from Dd2 with more than 25-fold decrease in susceptibility to DHA. The DHA-resistant clones were more tolerant of stressful growth conditions and more resistant to several commonly used antimalarial drugs than Dd2. The result is worrisome since many of the drugs are currently used as ART partners in malaria control. This study showed that the DHA resistance is not limited to ring stage, but also occurred in trophozoites and schizonts. Microarray and biochemical analyses revealed pfmdr1 amplification, elevation of the antioxidant defense network, and increased expression of many chaperones in the DHA-resistant parasites. Without drug pressure, the DHA resistant parasites reverted to sensitive in approximately eight weeks, accompanied by de-amplification of pfmdr1 and reduced antioxidant activities. The parallel decrease and increase in pfmdr1 copy number and antioxidant activity and the up and down of DHA sensitivity strongly suggest that pfmdr1 and antioxidant defense play a role in in vitro resistance to DHA, providing potential molecular markers for ART resistance.
Keywords: artemisinin resistance, drug selection, mechanism, pfmdr1, antioxidant defense
Introduction
Artemisinin (ART)-based combination therapy (ACT) has been endorsed by World Health Organization (WHO) and adopted by most malaria endemic countries for treating malaria (Bosman & Mendis, 2007). However, the recent report of delayed clearance of parasites after ART treatment in western Cambodia threatens to derail the current malaria control initiatives (Noedl et al., 2008; 2010; Dondorp et al., 2009). To address this urgent issue, WHO is coordinating large-scale risk evaluation and containment efforts to mitigate the spread of resistance (WHO, 2009, 2011). Yet, close surveillance for ART resistance at sentinel sites, an essential step in resistance management, is hindered by the lack of a clear understanding of the molecular mechanism of resistance and molecular markers.
Although the reports of potential ART resistance have generated interests in surveillance and containment of the resistance, the molecular mechanisms of ART action and resistance in Plasmodium falciparum remain largely unknown (Cui & Su, 2009; Ding et al., 2011). In addition, there is a lack of clarity regarding the definition of artemisinin resistance, which makes genetic associations difficult. Further complication arises when prolonged parasite clearance kinetics is used as a basis of defining resistance, since such a phenotype depends on host as well as parasite variables. Some reports suggested potential association of P. falciparum multiple drug resistance 1 (pfmdr1) and the sarco/endoplasmic reticulum calcium-dependent ATPase (SERCA) homologue PfATP6 with ART resistance (Cui & Su, 2009; Ding et al., 2011). Point mutations as well as copy number variations in pfmdr1 have been implicated in altered sensitivity to multiple structurally unrelated antimalarials including ART (Price et al., 1999; Duraisingh et al., 2000; Reed et al., 2000; Pickard et al., 2003; Sidhu et al., 2005). Whereas elevated pfmdr1 copy number is associated with increased risk for therapy failures of artesunate (ATS)/mefloquine (MQ) in Thailand and Cambodia and is also found in longitudinal studies of children treated with artemether (ATM)/lumefantrine (LUM) (Alker et al., 2007; Shah et al., 2008; Wongsrichanalai & Meshnick, 2008; Lim et al., 2009; Gadalla et al., 2011), recent clinical trials could not confirm a role of pfmdr1 in ART resistance (Dondorp et al., 2009; Imwong et al., 2010; Noedl et al., 2010). PfATP6 has been postulated to be an ART target, because ART could inhibit PfATP6 activity in a heterologous system, and a single amino acid change (L263E) in PfATP6 could abolish the inhibition (Eckstein-Ludwig et al., 2003; Uhlemann et al., 2005). A number of polymorphisms have been documented in this gene (Dahlstrom et al., 2008; Tanabe et al., 2011), but they do not seem to correlate with altered sensitivity to ART (Dondorp et al., 2009; Imwong et al., 2010). Only one mutation S769N has been linked to reduced ATM sensitivity in P. falciparum field isolates from French Guiana (Jambou et al., 2005), but this mutation has not been found in parasites from regions where highest levels of ART selection are expected (Zhang et al., 2008; Imwong et al., 2010). In addition, allelic exchange study did not yield conclusive evidence supporting the involvement of PfATP6 in ART responsiveness (Valderramos et al., 2010; Cui et al., 2012). In addition, analysis of the progeny of the HB3 × Dd2 genetic cross has identified two additional loci that are associated with the likelihood of acquiring genetically stable, high-level ART resistance (Beez et al., 2011).
In vitro selection for parasites resistant to a drug followed by investigations of phenotypic and genomic changes is an effective laboratory approach to identifying mutations associated with drug resistance. Several studies have attempted to use this approach to select resistance to ART and its derivatives in P. falciparum in vitro (Inselburg, 1985; Chavchich et al., 2010; Witkowski et al., 2010; Beez et al., 2011). Results from these studies varied, probably reflecting a multigenetic nature of ART resistance. Ring stage has been postulated to be able to endure high concentrations of ART through a quiescence mechanism (Teuscher et al., 2010; Witkowski et al., 2010), which may partially explain the ART resistance phenotype of delayed parasite clearance occurring in west Cambodia (Nosten, 2010). To further investigate the molecular mechanisms of ART action and resistance, we selected Dd2 parasite with DHA and obtained two parasite clones that were highly resistant to DHA. We show that the response to DHA was associated with increased pfmdr1 copy number and elevated antioxidant activities, providing potential molecular markers for monitoring the emergence of ART resistance.
Results
Selection for DHA resistance depends on parasite genetic backgrounds
It has been shown that malaria parasites with different genetic backgrounds acquire resistance to novel antimalarials at different frequencies, and certain parasite strains display a phenotype of accelerated resistance to multiple drugs (ARMD) (Rathod et al., 1997). To investigate the possibility of ARMD phenotype to DHA, we tested five laboratory parasite strains from different geographical origins for in vitro sensitivities to DHA (Table 1). All parasite strains had 50% inhibitory concentrations (IC50 values) for DHA in a close range (3.2 – 7.6 nM), although Dd2 and 7G8 had ~two-fold higher IC50 values than 3D7, D10, and HB3. To generate parasites resistant to DHA, we cultured parasites under constant DHA pressure with an initial concentration corresponding to 2×IC50 value of each strain. Parasites surviving this DHA selection were obtained from Dd2, 7G8, HB3, and D10 (except 3D7) in 37 – 70 days. When the selected parasites were subsequently selected using DHA concentrations at 4×IC50 values of the respective strains, only D10 and Dd2 yielded tolerant parasites. Further selection of these two parasite strains under DHA at 8×IC50 values only generated DHA-tolerant parasites from Dd2 (Table 1). These data suggest that field parasite populations may be highly heterogeneous in DHA sensitivity and propensity to develop resistance to DHA, consistent with the presence of an ARMD phenotype.
Table 1.
Selection of different parasite strains for reduced sensitivity to DHA.
| Strains | Origin | IC50 to DHA (nM)* | Days needed to reach 1% parasitemia#
|
||
|---|---|---|---|---|---|
| 2 × IC50 | 4 × IC50 | 8 × IC50 | |||
| 3D7 | Africa | 3.2±1.1 | – | – | – |
| D10 | PNG | 3.5±0.4 | 37 | 52 | – |
| HB3 | Honduras | 3.9±1.4 | 53 | – | – |
| Dd2 | Indochina | 7.6±2.1 | 43 | 60 | 72 |
| 7G8 | Brazil | 6.2±0.8 | 70 | – | – |
IC50 values of the parasite lines to DHA (mean ± standard deviation).
Parasite strains were subject to sequential DHA selection at a DHA concentration of 2 ×, 4 ×, and 8 × IC50 values of the respective parasite strains and days needed for the parasites to reach 1% parasitemia under the specific drug concentrations were recorded. –, parasite growth was not detected.
Using a similar scheme of step-wise selection we subjected Dd2 to DHA selection and obtained parasites that were able to tolerate constant exposure to 320 nM of DHA (Fig. S1A). This population of DHA-resistant parasites was subsequently cloned in the absence of DHA selection, and 26 clones were obtained after culturing for ~35 days. These parasite clones showed dramatically different susceptibilities to DHA, with IC50 values ranging from 3 to 243 nM (Fig. S1B), suggesting that the DHA-resistant parasites consisted of a heterogeneous parasite population and multiple genes might contribute to the resistance. Two clones (DHA1 and DHA2) selected for further characterization showed ~32- and ~25-fold increase in IC50 value (243 and 196 nM, respectively) compared with the wild type Dd2 strain. It is noteworthy that these lines, compared with the original DHA-resistant population which tolerated 320 nM of DHA, had a substantial reduction in resistance after the cloning procedure without DHA selection pressure.
DHA-resistant lines have a different growth phenotype
When cultures were initiated at a 0.5% ring-stage parasitemia in a drug-free medium with medium changed every other day, all three parasite lines had similar growth curves before reaching peak parasitemias of 8.3, 7.5 and 7.4%, respectively, on day 5 (Fig. 1A). Subsequently, Dd2 parasitemia dropped to 5.5% on day 6, possibly due to increased oxidative stress in the culture (Becker et al., 2004). In comparison, the parasitemias of DHA1 and DHA2 on day 6 and 7 were significantly higher than the respective parasitemias of Dd2 (unpaired t-test, P<0.017), suggesting that the two resistant lines were more tolerant of oxidative stress. We further determined the intraerythrocytic developmental cycle (IDC) durations of the three parasite lines by monitoring parasite growth every 2 h for 96 h (Fig. S2A). Compared with Dd2 with an IDC duration of 44.5 h (Reilly et al., 2007), DHA1 and DHA2 had significantly longer IDC durations of 47 and 47.5 h, respectively (unpaired t-test, P<0.017), largely due to more extended schizont development (Fig. S2A, C). The results also suggested that drug selection might have led to some deleterious changes in parasites that survived drug selection.
Figure 1. Growth phenotypes.
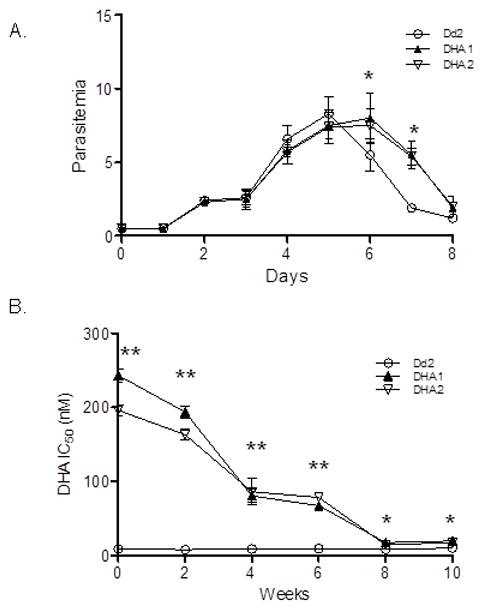
A. Growth curves of parasite clones DHA1, DHA2 and Dd2. Synchronized ring-stage parasites were seeded into 24-well plate, and parasitemias of each line were counted daily for four IDCs. Medium was changed every 48 h. The asterisk (*) denotes significant difference in parasitemia between Dd2 and DHA selected parasite lines DHA1 and DHA2 (unpaired t-test, P < 0.003). B. IC50 values of Dd2, DHA1 and DHA2 to DHA during in vitro culture without drug. Asterisk (*) and asterisks (**) indicate significant difference between Dd2 and either DHA1 or DHA2 at P=0.017 and P=0.003 level (unpaired t-test).
To determine the stability of the DHA resistance phenotype, DHA1 and DHA2 were cultured without DHA and their sensitivity to DHA was measured at a bi-weekly interval. Without drug pressure, the DHA1 and DHA2 lines gradually lost their resistance, and IC50 values decreased to 20 and 18 nM, respectively, in eight weeks (Fig. 1B). Continuous monitoring of the two parasite lines for two more weeks did not detect a further decrease of DHA sensitivity, although their IC50 values were still significantly higher than that of Dd2 (unpaired t-test, P<0.003). The loss of DHA resistance was also correlated with the restoration of the IDC duration to levels that were not significantly different from that of Dd2 (Fig. S2B, C). To determine how these parasite lines respond to further DHA selection, we re-applied the step-wise DHA selection on these revertant parasites. Remarkably, parasites having high DHA IC50 values (244 and 221 nM, respectively) were obtained in ten weeks, as compared to 15 months required for the initial selection of the DHA1 and DHA2 lines. The results suggested that the DHA1 and DHA2 had gained some stable genetic changes that can confer low-level DHA resistance, and these changes are required for acquiring high-level resistance.
Selection for DHA resistance results in reduced susceptibility to other antimalarial drugs
To examine whether selection for DHA resistance also affected sensitivities to other antimalarial drugs, we determined the IC50 valuesof DHA1 and DHA2 to some commonly used antimalarial drugs (Table 2). Compared with Dd2, DHA1 and DHA2 had significant increases in IC50 values to most drugs tested (unpaired t-test, P<0.0083). Specifically, DHA1 and DHA2 had >16-fold increase in IC50 values to ART, but only showed ~5- and ~10-fold increase in IC50 values to the two ART derivatives, ATM and ATS. Also noteworthy, DHA1 and DHA2 were ~10 times more resistant to LUM than Dd2. However, the changes in IC50 values to chloroquine (CQ), amodiaquine (AQ), piperaquine (PQ), MQ and quinine (QN) were modest (2 – 3 folds). In comparison, the DHA-selected lines DHA1 and DHA2 did not show significant changes in sensitivity to atovaquone (ATQ) (unpaired t-test, P>0.0083), a drug that acts on the mitochondria (Table 2). Remarkably, in vitro sensitivities of Dd2, DHA1, and DHA2 to most of these drugs except for ATQ were significantly correlated (Table S1), suggesting selection of a common resistance mechanism.
Table 2.
In vitro responses of Dd2, two DHA-selected strains (DHA1 and DHA2), their respective revertant lines DHA1-R and DHA2-R, and reselected lines by DHA (DHA1-R-DHA and DHA2-R-DHA) and MQ (DHA1-R-MQ and DHA2-R-MQ) to different antimalarial drugs (nM)*
| Strains | DHA | ATM | ATS | ART | CQ | AQ | PQ | MQ | LUM | QN | ATQ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dd2 | 7.6±2.1 | 7.0±1.8 | 9.8±2.2 | 5.5±1.4 | 161.1±27.5 | 25.7±3.4 | 14.8±1.1 | 40.2±8.6 | 37.6±6.3 | 307.2±54.7 | 1.4±0.2 |
| DHA1 | 197.9±27.5* | 38.3±10.0* | 86.5±17.7* | 93.4±12.6* | 307.5±97.2* | 65.3±5.6* | 37.0±6.5* | 102.6±17.8* | 398.3±87.0* | 467.6±120.8 | 1.2±0.4 |
| DHA2 | 167.5±14.5* | 30.1±7.7* | 102.0±24.2* | 91.5±26.0* | 270.3±79.0* | 40.2±12.3* | 40.7±10.5* | 146.7±24.3* | 487.9±78.8* | 578.2±54.3* | 1.6±0.5 |
| DHA1-R | 19.8±3.7* | 11.1±3.7 | 12.9±2.2 | 15.1±1.7* | 231.3±30.4* | 43.5±11.7* | 15.8±5.1 | 54.5±3.8* | 46.5±7.8 | 464.5±87.4* | 2.2±0.3 |
| DHA2-R | 17.6±4.1* | 12.8±4.0* | 17.6±4.2* | 18.2±4.3* | 290.6±16.2* | 39.4±7.8* | 19.0±3.8 | 60.2±6.4* | 58.3±8.0* | 449.8±95.1* | 1.7±0.1 |
| DHA1-R-DHA | 244.6±19.4* | 26.4±3.6* | 99.6±22.8* | 120.5±23.0* | 409.7±107.3* | 49.6±14.1* | 45.2±13.0* | 140.2±25.0* | 214.5±54.5* | 420.9±104.7 | 1.5±0.3 |
| DHA2-R-DHA | 221.4±45.2* | 58.3±23.6* | 79.9±7.6* | 116.7±24.7* | 307.5±97.2* | 45.3±3.4* | 35.1±2.2* | 122.5±12.1* | 236.3±37.3* | 567.2±52.5* | 1.4±0.5 |
| DHA1-R-MQ | 52.4±3.6* | 9.6±1.3 | 16.5±3.0* | 37.5±8.8* | 148.5±20.6 | 56.3±12.5* | 26.7±6.9* | 156.2±24.3* | 435.1±76.6* | 372.5±62.7 | 2.2±0.7 |
| DHA2-R-MQ | 65.0±3.2* | 11.4±2.9* | 10.3±2.2 | 29.7±11.2* | 156.3±13.6 | 61.1±10.5* | 30.3±1.8* | 147.0±8.8* | 411.3±52.8* | 422.9±38.3* | 1.3±0.2 |
Numbers represent the IC50 values (means ± standard deviations) in nM from at least six independent determinations. For each drug, significant differences between Dd2 and each of the re-selected strains were marked with * indicate significance at P = 0.0083 (unpaired t-test), respectively.
DHA, dihydroartemisinin; ATM, artemether; ATS, artesunate; ART, artemisinin; CQ, chloroquine; AQ, amodiaquine; PQ, piperaquine; MQ, mefloquine; LUM, lumefantrine; QN, quinine; ATQ, atovaquone.
The increased resistance to quinoline drugs prompted us to further characterize the resistance phenotype and potential mechanisms. CQ resistance mechanism is by far the best understood; mutations in the P. falciparum CQ resistance transporter (PfCRT) are the major determinant of CQ resistance (Fidock et al., 2000). A hallmark of PfCRT-dependent CQ resistance is chemosensitization by verapamil (VP), a Ca2+ channel blocker (Martin et al., 1987; Patel et al., 2010). To determine the VP-reversibility of CQ resistance of the two DHA-selected lines, we determined their CQ IC50 values in combination with VP. We found VP could re-sensitize the CQ resistance in all three lines, but the IC50 values in DHA1 and DHA2 had 2.3- and 1.8-fold decreases, from 308 and 270 to 133 and 156 nM, respectively, as compared to a 5.6-fold decrease (from 161 to 28.75 nM ) in Dd2 (Fig. S3). This result implies the resulted increase in CQ resistance in DHA1 and DHA2 was mostly not reversible by VP.
The drug sensitivity profiles for DHA-selected parasites revealed that they not only showed significant decreases in sensitivities to all ART related drugs, but also displayed reduced sensitivities to a number of currently used ACT partner drugs. Here we compared the IC50 values of Dd2, DHA1 and DHA2 to two commonly used ACTs: DHA/PQ and ATM/LUM. Compared with Dd2, both DHA1 and DHA2 showed significantly reduced sensitivities to these two ACTs (unpaired t-test, P<0.003) (Fig. S4). Specifically, DHA-selected lines had ~5-fold and >2-fold higher IC50 values than Dd2 to DHA/PQ and ATM/LUM, respectively.
DHA resistance is mediated by, but not limited to, ring stage dormancy mechanism
Previous selection for ART resistance using an intermittent drug selection scheme showed that tolerance to ART was mediated by a quiescence mechanism occurring only at the ring stage (Witkowski et al., 2010). This ART-induced temporary growth arrest at ring stage occurs readily in laboratory strains, and the recovery rates depend on genetic backgrounds of the strains (Teuscher et al., 2010). To determine whether DHA resistance in DHA1 and DHA2 also involves the ring-stage quiescence mechanism, we treated the ring-stage cultures with 1 or 3 μM of DHA for 48 h and monitored parasite recovery from drug treatment. After treatment with 1 μM of DHA, DHA1 and DHA2 recovered and reached 10% parasitemia in seven days, as compared to 16 days for Dd2 (Fig. 2A). When treated with 3 μM of DHA, DHA1 and DHA2 reached 10% parasitemia in 10 and 12 days, respectively, whereas it took 21 days for Dd2 to do so. These results showed that DHA1 and DHA2 could survive and quickly recover from high doses of DHA treatment, possibly reflecting the recrudescence frequently associated with ART monotherapy.
Figure 2. Survivorship and growth recovery of Dd2, DHA1 and DHA2 after DHA treatment.
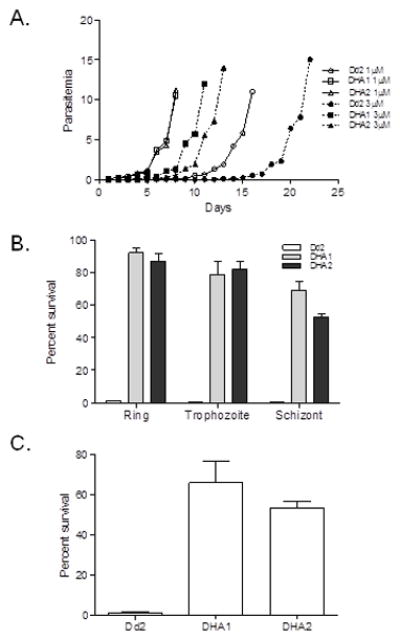
A. Recovery of growth in Dd2, DHA1 and DHA2 after DHA treatment. Synchronized parasites of DHA1, DHA2, and Dd2 were treated at ring stage with 1 or 3 μM DHA for 48 h. After treatment, drug-free medium was changed daily, and parasitemias were counted daily. B. Survival of parasites after DHA treatment. Synchronized parasites at ring, trophozoite and schizont stages were treated with 1 μM of DHA for 2 h and percentages of survived parasite were determined 6 h later by flow cytometry as the rhodamine 123/To-Pro III ratios. C. Unsynchronized parasites were treated with 100 nM of DHA for 48 h and survivorship of the parasites was determined by flow cytometry. All results from DHA1 and DHA2 in B and C are significantly different from those with Dd2 (unpaired t-test, P <0.003)
To determine whether this recrudescence mechanism also plays a role in other developmental stages, synchronized parasites at ring, trophozoite and schizont stages were exposed to 1 μM of DHA for 2 h and parasites that survived the treatment were counted 6 h later using flow cytometry (Fig. 2B). Significantly more DHA1 and DHA2 parasites were recovered after DHA treatment from all three stages than Dd2 (unpaired t-test, P < 0.003). Similar results were obtained when unsynchronized parasites were exposed to 100 nM of DHA for 48 h (Fig. 2C). These results showed that parasite resistance to ART was not limited to ring stage, but also occurred in trophozoite and schizont stages.
DHA resistance does not involve known mutations associated with drug resistance
Reduced susceptibility to ART has been reported to associate with mutations in multiple genes, including pfmdr1 (Sanchez et al., 2008), pfcrt, pftctp (Bhisutthibhan et al., 1998), pfubcth (Hunt et al., 2007), putative ABC transporters G7 and G49 (Mu et al., 2003; Anderson et al., 2005), pfmrp (Raj et al., 2009), and PfATP6 (Jambou et al., 2005). Sequencing of the coding regions of all these genes in DHA1 and DHA2 did not detect any differences from those in Dd2. These data also verified that DHA1 and DHA2 were derived from Dd2.
Comparison of global gene expression in Dd2 and DHA1
Microarray analysis was performed to determine differences in gene expression at three stages between Dd2 and DHA1 and in response to DHA treatments. Microarray data from the two biological replicates for each treatment were highly correlated (R2>0.96, P<0.001). Based on IDC progression pattern in Dd2 and DHA1 (Fig. S2A), we selected three matching time points to compare differences in gene expression in ring, trophozoite and schizont stages between Dd2 and DHA1. Comparison of the microarray results of the parasite lines Dd2 and DHA1 with the predetermined 48 h expression profile of Dd2 (Llinas et al., 2006) revealed that gene expression profiles at three time points from both lines had the best fit with Dd2 gene expression profiles at 14, 27 and 38 h, respectively (Fig. S5, R2>0.93, P<0.001). These results strongly indicated that differences between the Dd2 and DHA1 very likely reflected their true biological differences. We first compared differences in gene expression between Dd2 and DHA1 at three stages during the IDC and detected 519 and 213 genes with ≥2-fold up- and down-regulation in DHA1, respectively (Fig. 3A, Table S2). More genes with ≥2-fold changes in expression were detected in trophozoite and schizont stages than in ring stage (Fig. 3A). Based on functional annotations of parasite genes in PlasmoDB, we classified the 5333 genes in P. falciparum genome into 12 categories. Our results showed that gene ontology (GO) categories of “catalytic/metabolic activity” and “chaperone” were significantly enriched in DHA1 in both up- and down-regulated gene pools (Z-test, P<0.01) (Fig. 3B). Strikingly, seven genes annotated as heat shock proteins (HSPs) in the “chaperone” category and at least 22 genes out of 72 in the genome involved in antioxidant defense in the “catalytic/metabolic activity” category were among the up-regulated genes in DHA1 (Fig. 3C). In addition to these two categories, at least 14 genes annotated as “transporters” were also detected in both up- and down-regulated gene pools (Table S2), including >6-fold up-regulation of pfmdr1. Several genes potentially involved in transcription such as AP2 domain proteins and chromatin modifiers were also up-regulated in DHA1, which might be responsible for significantly greater number of genes up-regulated in this line. Despite that a large portion of parasite genes in DHA1 showed ≥2-fold changes in transcript abundance, only 16 up-regulated and three down-regulated genes were shared among the three stages (Fig. 3A, Table 2). Noticeably, these shared up-regulated genes include pfmdr1, five genes associated with antioxidant defense (PFF1130c, PF14_0164, PFI0925w, PFI1250w and PFL0725w), and hsp70. Up-regulation of hsp70 is consistent with a recent report of positive selection around the hsp70 locus in relation to the ATS-treatment delayed parasite clearance phenotype seen in Cambodia (Cheeseman et al., 2012).
Figure 3. Microarray comparison of gene expression at three developmental stages.
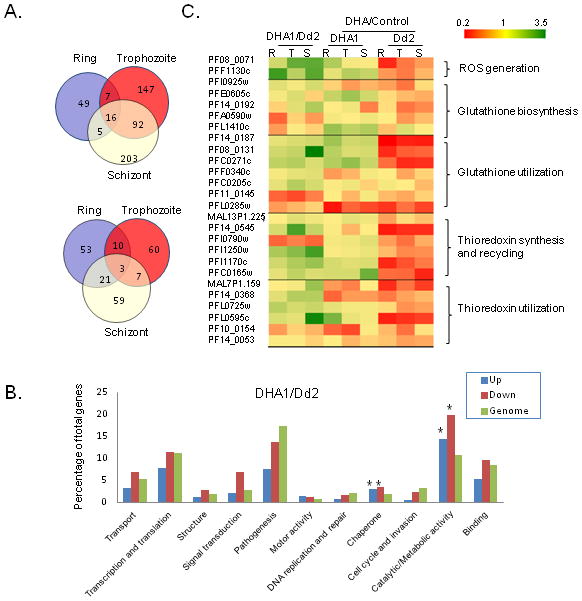
A.Venn diagrams show the overlap of genes with ≥2-fold up-regulation (upper panel) and down-regulation (lower panel) in DHA1 versus Dd2 at three stages. B. Representations (percentages) of the number of genes from different functional categories in the gene pools with ≥2-fold up-(blue) and down-regulation (red) are compared to the overall representations of the functional categories in the genome (green). Genes encoding hypothetical proteins are not shown. Asterisk (*) denotes categories significantly enriched in up- or down-regulated gene pools as compared to genomic representations (Z-test, P < 0.05). C. Expression changes of genes involved in antioxidant defense in Dd2 and selected DHA-resistant clone DHA1 with or without DHA treatment. R, T, and S denote ring, trophozoite, and schizont stages, respectively.
Treatment of Dd2 with DHA at 1 μM for 2 h resulted in significantly more genes with ≥2-fold reduction than those with ≥2-fold increase in transcript levels (Fig. S6A, Z-test, P<0.01). Interestingly, DHA treatment of Dd2 caused substantial disturbance in gene expression in most GO categories, as compared to only two GO categories in DHA1 (Fig. S6B, Z-test, P<0.05). This divergent response to DHA treatment is further illustrated from comparisons showing insignificant overlaps in up- or down-regulated genes between the two parasite lines (Fig. S7). It is noteworthy that in the “catalytic/metabolic activity” category at least 22 genes involved in antioxidant defense were further up-regulated in DHA1 after DHA treatment (Table S3). In sharp contrast, only two genes involved in antioxidant defense were up-regulated in Dd2 as compared to eight such genes down-regulated after DHA treatment (Table S4). In addition, a tryptophan-rich gene (PF10_0026) implicated in tolerance to ATS was also up-regulated in DHA1 in ring stage (Natalang et al., 2008; Deplaine et al., 2011), and upon stimulation with DHA, it had more than 2-fold up-regulation in ring and schizont stages (Table S2–4). Again, this DHA-induced up-regulation of PF10_0026 was not observed in pre-selected Dd2 parasites.
Pfmdr1 gene amplification is associated with DHA resistance
Real-time RT-PCR analysis confirmed over-expression of pfmdr1 in both DHA1 and DHA2 (Fig. 4A). To elucidate the mechanism of increased pfmdr1 expression, we quantified the copy numbers of pfmdr1 in the three parasite lines. Compared with ~4 copies of pfmdr1 in Dd2, DHA1 and DHA2 had 8.5 and 9.3 copies, respectively (Fig. 4B). Mapping of the amplified region in DHA1 using real-time PCR confirmed further amplification of the same amplicon as in Dd2 (Fig. S8A), which was mapped to the ~880 – 970 k position on chromosome 5 (Gonzales et al., 2008). Microarray data showed that all the 14 genes in this region were up-regulated, but only pfmdr1 had >6-fold over-expression (Fig. S8B), suggesting that it might be a major contributor to DHA resistance in our selected parasites. To further incriminate pfmdr1 amplification as a key factor of DHA resistance, we cultured DHA1 and DHA2 in drug-free medium for 10 weeks and monitored the dynamics of pfmdr1 copy number. Without DHA, pfmdr1 copy number gradually decreased and reached 4.1 and 4.2 copies by eight weeks, a level similar to that of Dd2 (Fig. 4B, unpaired t-test, P>0.017). Concomitantly, IC50 values to DHA also gradually declined to 20 and 18 nM, respectively (Fig. 1B). Finally, the revertant parasites had higher IC50 values than Dd2 to all drugs tested except ATQ, and for DHA, ART, CQ, MQ, and QN the differences between the revertant populations and Dd2 remained significant (unpaired t-test, P<0.0083) (Table 2). These results suggested that pfmdr1 copy number variation plays a direct and major role in DHA resistance in the selected parasites, but other mechanisms may be responsible for the residual low-level resistance to DHA and other drugs in the revertant parasites.
Figure 4. Pfmdr1 amplification and correlation with DHA resistance in DHA1 and DHA2.
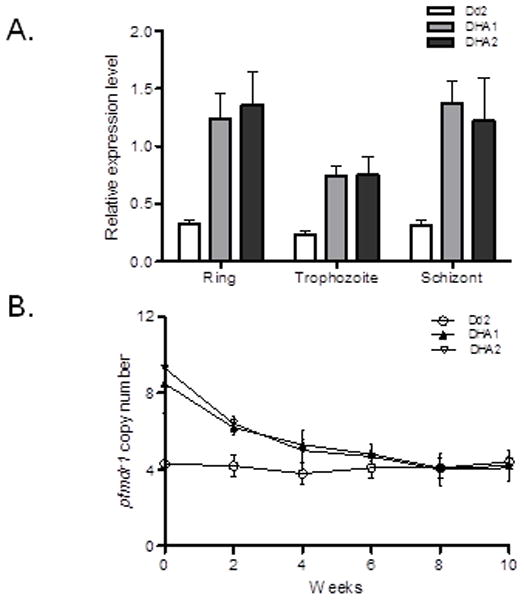
A. Confirmation of elevated pfmdr1 expression in Dd2 and two DHA-selected lines DHA1 and DHA2. Pfmdr1mRNA levels in rings (14 h), trophozoites (27 h) and schizonts (38 h) were determined by real-time RT-PCR with the constitutively expressed seryl-tRNA synthetase as an internal reference gene. B. Pfmdr1 copy numbers in Dd2, DHA1 and DHA2. Pfmdr1 copy numbers were determined biweekly by real-time PCR in the three parasite lines during in vitro culture without drug.
A second component is required for high-level ART resistance
To further dissect the genetic components contributing to DHA resistance, we applied a similar step-wise selection pressure of DHA and MQ to the two revertant parasite populations. Pfmdr1 amplification has been associated with MQ resistance and can be selected by in vitro MQ treatment (Wilson et al., 1989; Cowman et al., 1994). Under DHA and MQ selection, pfmdr1 copy numbers in the revertant DHA1 and DHA2 populations were doubled in 8–10 weeks (Fig. S9). DHA re-selection fully restored the DHA-resistant phenotype in the DHA1 and DHA2 populations with DHA IC50 values of 245 and 221 nM, respectively (Table 2). However, whereas MQ re-selection fully restored the reduced susceptibility to AQ and LUM, it only partially restored DHA resistance with DHA IC50 values of 52 and 65 nM, respectively (Table 2). In addition, the increase in pfmdr1 copy number from MQ selection did not affect the sensitivity to CQ. These results suggested that some specific components acting in concert with higher-level pfmdr1 expression are needed in order to confer high-level resistance to DHA, which could not be fully selected by MQ.
Elevated antioxidant activity is correlated with DHA resistance
Microarray and RT-PCR studies detected that the expression of many genes involved in antioxidant pathways was elevated under DHA pressure (Fig. 3C and S10), suggesting that antioxidant defense system might be another component contributing to DHA resistance in DHA1 and DHA2. In malaria parasites, the antioxidant networks include superoxide dismutases (SODs), glutathione (GSH) and thioredoxin (Trx) systems (Muller, 2004; Becker et al., 2005). Both SOD-1 (cytosolic, Fe-dependent) and SOD-2 (mitochondrial) were up-regulated in DHA-selected DHA1 (Fig. 3C). In addition, several components of GSH and Trx metabolic pathways were up-regulated after a brief exposure to DHA (Fig. 3C, S10). In agreement, the antioxidant GSH and Trx levels, and intracellular SOD and glutathione-S-transferase (GST) activities were significantly higher in DHA1 and DHA2 than in Dd2 (Fig. 5). The increase in the antioxidant defense system is likely due to DHA selection, since the baseline SOD and GST activities between the different parasite strains before DHA selection were not significantly different (Fig. S11). Similar to the pfmdr1 copy number variation, the DHA1 and DHA2 parasites’ intracellular GSH and Trx levels, and SOD and GST enzyme activities gradually declined without DHA pressure; and at week 10, the Trx levels in DHA1 and DHA2 were similar to that in Dd2 (Fig. 5B, unpaired t-test, P>0.017). However, GSH level, SOD and GST activities at ten weeks were nonetheless significantly higher than those in Dd2 (Fig. 5A, C, and D, unpaired t-test, P<0.017). The significantly higher SOD and GST activities in the revertant lines than in Dd2 may account for the residual, yet significantly higher DHA and ART IC50 values detected in these revertant lines at week 10 (Table 2). These components were likely the stable changes that allowed parasites to quickly gain high-level resistance to DHA. To corroborate this conclusion, we monitored the SOD and GST activities during re-selection of the revertant parasites by DHA and MQ. We found that DHA re-selection resulted in continuous increases in SOD and GST activities of selected parasites, which reached similar levels of DHA1 and DHA2 at the end of selection. However, MQ re-selection did not result in noticeable increases in SOD and GST activities (unpaired t-test, P>0.017) (Fig. 5E and F).
Figure 5. Cellular levels or activities of major components of the antioxidant defense system.
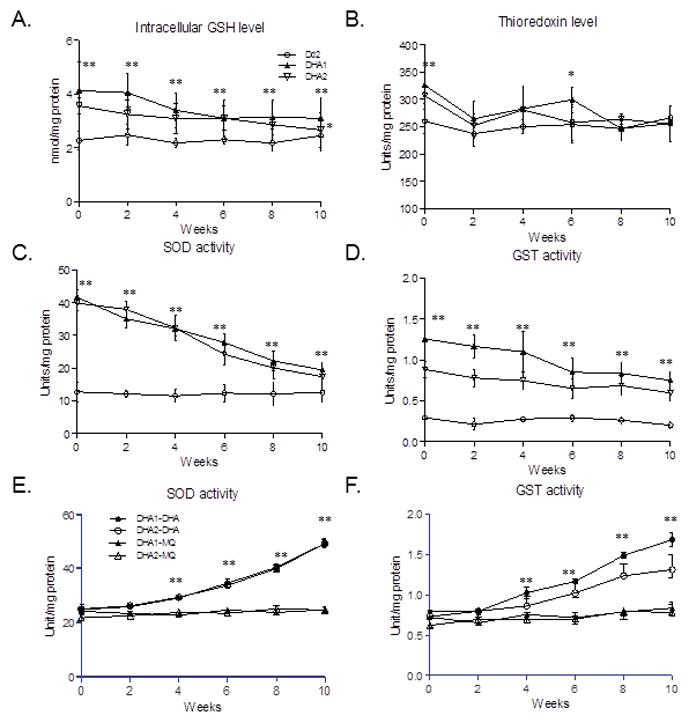
A. Glutathione (GSH). B. Thioredoxins. C. Superoxide dismutase (SOD). D. Glutathione S-transferase (GST). E. SOD activity in the DHA resistance-reverted DHA1 and DHA2 parasites during re-selection with DHA (DHA1-DHA and DHA2-DHA) and MQ (DHA1-MQ and DHA2-MQ). F. GST activity in the DHA resistance-reverted DHA1 and DHA2 parasites during re-selection with DHA (DHA1-DHA and DHA2-DHA) and MQ (DHA1-MQ and DHA2-MQ). Cellular levels of GSH and thioredoxins, enzymatic activities of GST and SOD were measured at a bi-weekly interval during 10 weeks of culture of the DHA-resistant clones DHA1 and DHA2 without DHA (A–D), and during DHA and MQ re-selection (E–F). Statistical analysis was performed at each time point by comparing the differences (unpaired t-test). Asterisk (*) and asterisks (**) indicate significant difference at P=0.017 and P=0.003, respectively, between Dd2 and either DHA1 or DHA2 (A–D), and between DHA re-selected and MQ re-selected parasites (E–F).
We noticed that a brief DHA treatment further raised transcript levels of some antioxidant defense genes including SODs and GST only in DHA1 but not in Dd2 (Fig. 3C). Likewise, both SOD and GST activities in DHA1 and DHA2 were significantly increased by DHA treatment (Fig. 6A and B, unpaired t-test, P<0.003). On the contrary, DHA treatment led to a significant reduction in activity of these two enzymes in Dd2 (Fig. 6A and B, P<0.017). The overall increase in the antioxidant network in DHA1 and DHA2 suggests that they are more capable of dealing with intracellular reactive oxygen species (ROS). To validate this speculation, we treated the parasites with curcumin and tert-butyl peroxide (TBP) at their respective IC50 values to induce oxidative stress in the parasites (Cui et al., 2007a). Whereas the baseline levels of ROS in Dd2, DHA1 and DHA2 were not significantly different (P>0.017), treatment with 50 μM of curcumin or 200 μM of TBP for 4 h resulted in a significant increase of intracellular ROS level in Dd2 (Fig. 6C). However, ROS levels in DHA1 and DHA2 remained relative constant during treatment with these two compounds (unpaired t-test, P>0.017). This was also reflected from the significantly lower TBP IC50 value in Dd2 (85.3±12.5 μM) than in DHA1 (182.6±25.7 μM) and DHA2 (194.6±12.7 μM). These results suggested that some permanent changes in the ROS response pathways have occurred in the DHA1 and DHA2 parasites.
Figure 6. Antioxidant activities in Dd2, DHA1 and DHA2 after treatment with DHA, curcumin, or tert-butyl peroxide (TBP).
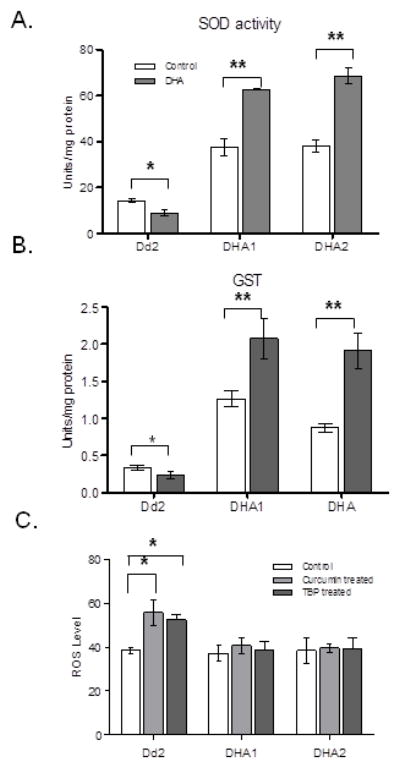
A. SOD levels in parasites treated with or without (control) 100 nM of DHA for 2 h. B. GST levels in parasites treated with or without 100 nM DHA for 2 h. The result represents Mean ± SD from three replications. C. ROS generation in Dd2, DHA1 and DHA2. The parasites Dd2, DHA1 and DHA2 at trophozoite stage were treated with 50 μM of curcumin or 200 μM of TBP for 4 h and intracellular ROS levels were determined. Asterisk (*) and asterisks (**) indicate significant difference between drug-treated and control parasites at P=0.05 and P=0.003, respectively (unpaired t-test).
Discussion
We successfully selected two P. falciparum parasite lines that were able to grow under continuous exposure to 320 nM of DHA, the highest fold increase in IC50 value obtained so far from in vitro induced ART resistance. We have shown that the resistance to ART is not limited to ring-stage dormancy, but also occurs in trophozoites and schizonts. Using molecular and biochemical approaches, we have provided strong evidence showing that sensitivity to DHA is dynamically correlated with pfmdr1 copy number and the antioxidant activity. To our knowledge, this study, for the first time, integrates pfmdr1 gene amplification and enhanced antioxidant defense as the mechanism of ART resistance. This finding has significant implications for development of ART resistance in Southeast Asia.
In vitro selection for antimalarial drug resistance is an effective approach to studying drug resistance mechanisms. It is noteworthy that the DHA selection protocol used here differs from the way that the parasites are exposed to ACTs in patients, which may explain the differences between in vitro selected parasites and parasites isolated from clinical resistance manifested as delayed parasite clearance. Nevertheless, the in vitro selected DHA resistance revealed several important points. First, although ART, ART derivatives and synthetic trioxanes have been proposed to act by a similar mechanism, DHA selection resulted in strikingly distinctive responses to ART family drugs, revealing surprising structural specificity to the selection of resistance phenotype. The selected DHA-resistant lines displayed a >25 and >16 fold decrease in sensitivity to DHA and ART, respectively, but the increase in resistance to other ART derivatives was modest (~5 and ~10 fold reduction in ATM and ATS sensitivity, respectively). In parallel, the revertant parasites still maintained residual, yet significant resistance to DHA and ART. On one hand, since DHA is less stable under aqueous conditions in vitro (Haynes et al., 2007), the selected phenotype may reflect the increased ability of the parasites to decompose DHA (and ART) compared with other more stable derivatives. On the other hand, this result may imply that a specific resistance mechanism selected by DHA (and potentially ART) is yet to be discovered. Therefore, this structure-related finding and especially the distinction of ATM underscores that future studies should be directed to address the structure-specific resistance mechanisms. Second, DHA selection resulted in general elevation of resistance to a number of commonly used antimalarials, suggesting of common potential resistance mechanisms. Together with the results from MQ re-selection, this study pointed out that pfmdr1 amplification largely explains the increased resistance to MQ, LUM and QN. However, the divergent responses between MQ and LUM suggest the involvement of different components in addition to pfmdr1. Third, the results also present a troubling clinical outlook since many of these tested drugs are ACT partner drugs. This further emphasizes that selection of ART partner drugs needs to be carefully evaluated to avoid cross resistance. In this study, DHA selection did not lead to noticeable changes in sensitivity to ATQ, a drug targeting the mitochondria. This may suggest that drugs targeting parasite’s mitochondria and apicoplast deserve future considerations in ACTs.
ART monotherapy of P. falciparum malaria is often associated with recrudescence (Li et al., 1984), and there are clear indications that the recrudescence rates associated with ART treatments have increased in regions of Southeast Asia where ART and derivatives have been used extensively (WHO, 2010). The recently confirmed ART resistance in the Thai-Cambodian border area is manifested as delayed parasite clearance (Noedl et al., 2008; Dondorp et al., 2009). A ring-stage dormancy mechanism has been invoked to explain this phenomenon (Teuscher et al., 2010; Witkowski et al., 2010). In cultured parasite strains, ART-induced temporary arrest of growth can occur readily at the ring stage (Teuscher et al., 2010), and after selection, ring stage parasites can tolerate high-dose ART pressure (Witkowski et al., 2010). Ring-stage tolerance to ART is likely associated with low-level hemoglobin digestion at this stage, since hemoglobin digestion is required for the action of ARTs (Klonis et al., 2011). However, our study showed that increased tolerance to DHA is not limited to ring stage. Throughout the IDC, the DHA-resistant parasites displayed much faster recovery rates and drastically higher survivorship after exposure to high doses of DHA. A recent genome-wide association study has identified a selective sweep on chromosome 13 that shows strong association with reduced parasite clearance rates (Cheeseman et al., 2012). Interestingly, hsp70 gene is located in the region under positive selection and shows increased expression, which is consistent with our in vitro DHA selection result. Furthermore, among genes up-regulated in DHA1, many encode chaperones including HSPs and HSP-like proteins. Although the roles of HSPs in drug resistance in P. falciparum are mostly unexplored, an analogy drawn from other eukaryotic systems suggests that up-regulation of HSPs may contribute to the overall increase in resistance to multiple antimalarial drugs.
Pfmdr1 mutations have been associated with altered susceptibilities to multiple antimalarial drugs (Reed et al., 2000; Duraisingh & Cowman, 2005) and many more non-antimalarial drugs (Yuan et al., 2009; 2011). In particular, pfmdr1 gene amplification is associated with increased resistance to ARTs in both field parasite isolates and in vitro selected ART-resistant parasites (Wilson et al., 1993; Price et al., 2004; Chavchich et al., 2010; Rodrigues et al., 2010; Borges et al., 2011). By following the dynamics of pfmdr1 copy number and corresponding DHA IC50 values during in vitro culture and selection, our study conclusively showed that DHA resistance is dynamically associated with the pfmdr1 copy number. Furthermore, we have shown that the pfmdr1 up-regulation in selected parasite DHA1 is unique among the 14 genes in the amplified fragment, and further pfmdr1 up-regulation can be induced by DHA treatment. Since pfmdr1 amplification has been shown to affect the parasite’s fitness (Hayward et al., 2005), parasites reverted to the original pfmdr1 copy number in the absence of drug selection, which was accompanied by a gradual loss of the pfmdr1 amplification. This observation might explain the findings that parasites obtained from areas where low-grade ART resistance has been found from clinical studies do not exhibit such resistance when they are tested in vitro. The low-grade resistance could have been reverted during in vitro culture without drug selection pressure.
Our study has pinpointed elevated antioxidant defense in the parasite as another major mechanism of ART resistance. Although increased antioxidant defense activity and up-regulation of some antioxidant genes in Plasmodium in response to ARTs and several antimalarial drugs such as CQ have been reported (Ginsburg & Golenser, 1999; Ginsburg et al., 1999; Natalang et al., 2008; Radfar et al., 2008; Nogueira et al., 2010; Chandra et al., 2011), our study showed that up-regulation of many genes of the antioxidant network was critical for high-level resistance to DHA. Not only are these genes highly enriched among up-regulated genes in DHA1, they were further up-regulated in response to DHA treatment. In contrast, up-regulation of antioxidant defense genes was not apparent in Dd2. In agreement, the intracellular antioxidants GSH and Trx, as well as two important enzymes involved in redox metabolism SOD and GST, were markedly augmented in the selected parasites. Moreover, by following the dynamics of GSH, Trx, SOD and GST during culture of the parasite without drug pressure, we conclusively showed that gradual decreases of the antioxidant levels and enzymatic activities were highly correlated with gradual reduction of the DHA IC50 values. Also noteworthy is that re-selection of the revertant parasite lines with DHA fully restored the SOD and GST activities, but MQ selection had only negligible effect on the activities of these enzymes (Fig. 5E and F). These data further underline the significant contribution of elevated antioxidant defense in DHA resistance. These findings are consistent with the proposed mode of action of ART family drugs because these drugs were shown to increase oxidative stress and reduce the levels of GSH in the parasites (Krungkrai & Yuthavong, 1987; Meshnick, 2002; Hartwig et al., 2009). Enhanced antioxidant defense in selected parasites is well reflected in their increased capability of tolerating stressful culturing conditions under high parasitemia and dealing with induced ROS. The enhanced antioxidant activities in DHA-resistant parasites DHA1 and DHA2 may partially explain the overall decrease in sensitivity to many antimalarial drugs. MQ selection normally leads to pfmdr1 amplification and increased CQ sensitivity, although it does not fully revert the parasites to wild-type CQ sensitivity. DHA selection, however, resulted in pfmdr1 amplification but increased CQ resistance, which suggests a direct link of the DHA-selected CQ resistance with elevated antioxidant defense (Ginsburg & Golenser, 2003). Collectively, this study indicates that antioxidant defense system in the malaria parasites might be another important component for induced ART resistance.
Although many factors such as pfmdr1 amplification, increased antioxidant activity, and mutations in PfATP6 and other genes have been implicated in P. falciparum response to ART (Cui & Su, 2009; Ding et al., 2011), our carefully designed experiments and extensive data allow us to dissect relationship of these factors and to propose a potential DHA resistance mechanism for the first time (Fig. 7). It is clear that increased pfmdr1 copy number and elevated expression of genes in many antioxidant pathways are required for high-level resistance to DHA. Selection of DHA will lead to some complex stable changes in SOD and GST pathways, which may take times, and reversible changes in the Trx pathway. These changes in antioxidant pathways only lead to low-grade DHA resistance (also possibly to many other antimalarials). Selection with MQ leads to increased pfmdr1 copy number and medium-level resistance to DHA, consistent with previous reports of shared components in resistance to MQ and DHA (Sidhu et al., 2005; Sidhu et al., 2006). In addition, enhanced production of antioxidants will facilitate the removal of free radicals, ROS, as well as heme, thus increasing parasite’s capacity to tolerate oxidative stress from drugs. Upon the changes in antioxidant pathways, increased copy number of pfmdr1 can result in high-level resistance to DHA. Based on a current view suggesting that PfMDR1 likely imports drugs into rather than pumps drugs out of the food vacuole (Sanchez et al., 2011), increased pfmdr1 expression potentially enhances transport of DHA into the food vacuole, which may sequester the drug away from its sites of activity. Yet, a recent study suggested that the food vacuole might be an initial site of antimalarial activity of endoperoxides (del Pilar Crespo et al., 2008). Therefore, the mechanism by which pfmdr1 amplification leads to increased resistance to DHA requires further investigations.
Figure 7. A proposed mechanism for the in vitro DHA resistance in P. falciparum.
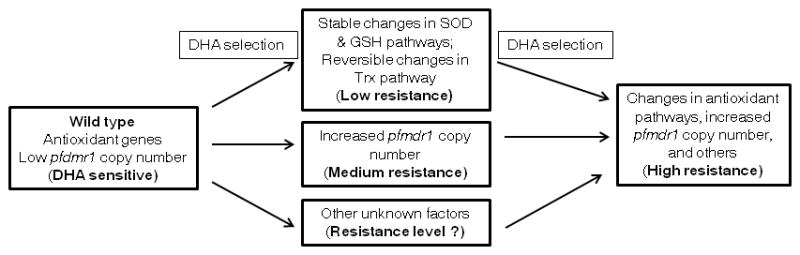
DHA selection can lead to stable changes in some antioxidant pathways (such as the SOD and GSH pathways) and reversible changes in the Trx pathway, which enable the parasites to more effectively remove heme and neutralize free radicals generated from the action of the drug, resulting in low-level resistance to DHA. DHA selection can also lead to pfmdr1 amplification, resulting in medium-level resistance to DHA. In parasites with the background of elevated antioxidant defense, selection of pfmdr1 amplification results in high-level resistance to DHA. Other unknown factors may contribute to additional resistance to DHA.
Our observations have significant implications for the emerging ART resistance in the Thai-Cambodian area of the Greater Mekong Subregion (GMS). Our results indicate that selection for resistance to ART is dependent on genetic changes in the parasite genome, particularly those of antioxidant pathways. Among the five laboratory strains tested, Dd2 displayed a much higher propensity to develop resistance to DHA, which is reminiscent of the “mutator” ARMD phenotype showing that certain parasite isolates are predisposed to develop resistance to multiple antimalarial drugs (Rathod et al., 1997). This ARMD phenotype has been verified in the MQ-selected Dd2 line and its parental strain W2 from Indochina (Oduola et al., 1988; Rathod et al., 1997). While this phenotype may be attributed to defective DNA mismatch repair in these parasites (Trotta et al., 2004; Castellini et al., 2011), we also noticed that among the five strains tested only Dd2 has more than one copy of pfmdr1, suggesting that the pfmdr1 amplification background may favor development of resistance to ART family drugs. In Thailand and Cambodia, MQ alone or in combination with ATS has been used extensively to treat uncomplicated falciparum malaria, and MQ resistance was evident (Mockenhaupt, 1995). Increased pfmdr1 gene copy number, the main cause of MQ resistance, has been found prevalent in this region (Price et al., 2004). Although pfmdr1 amplification was not associated with the clinical ART-resistant isolates (Imwong et al., 2010; Mok et al., 2011), it is possible that the resistance phenotype is unstable and resistant parasites could rapidly revert to ART sensitivity. However, we showed that parasites with some specific genetic backgrounds (mutations or elevated expression in antioxidant pathways) could regain ART resistance at much accelerated rates. This highlights the significance of extensive surveillance at sentinel sites in this area of the GMS. Furthermore, we speculate that parasites originated from the GMS might also have elevated antioxidant levels, which again favor the development of drug resistance. In the GMS, glucose-6-phosphate dehydrogenase (G6PD) deficiency, which confers resistance against malaria, is very common (Cappellini & Fiorelli, 2008). In G6PD-deficient red cells, defense against oxidative damage is reduced. Malaria parasite growing in G6PD-deficient erythrocytes displays enhanced, coordinated expression of antioxidant enzymes and HSPs (Akide-Ndunge et al., 2009), which may inevitably reduce the parasite’s sensitivity to antimalarial drugs. Taken together, our study has unified two potential pathways for the development of ART resistance in malaria parasite. The genetic backgrounds of the parasite in the GMS with high-frequency pfmdr1 amplification and elevated antioxidant defense shaped by drug history and host genetic factor, respectively, underline the high likelihood of emergence of ART resistance in this region. Further studies are necessary to dissect the molecular events in the antioxidant pathways leading to stable increased resistance to DHA.
Experimental procedures
Parasite culture, drug selection and assays
Laboratory P. falciparum strains (3D7, Dd2, 7G8, HB3, and D10) were obtained from MR4. Parasite culture and synchronization were done as described previously (Miao et al., 2010). To investigate the possibility of ARMD phenotype to DHA, we subjected five parasite strains (3D7, Dd2, 7G8, HB3, and D10) with different genetic backgrounds to DHA selection. Ten milliliters of cultures of parasites were maintained in 25 cm2 flask at 5% hematocrit. For each strain, 5 ×108 parasites at 5% parasitemia were continuously exposed to a stepwise increase of DHA concentrations starting at two-fold IC50 values of the respective parasite strains. The time when parasites reappeared in culture and reached 1% parasitemia was recorded for each parasite strain. Afterwards, selection was continued with 2-fold increase of the DHA concentrations, and the highest DHA concentrations used were 8-fold IC50 values of the respective parasite strains. Meanwhile, DHA selection was performed on Dd2 with increasing DHA pressure in stepwise two-fold increments from 10 to 320 nM. Each time when the parasite appeared and reached 2% parasitemia, a two-fold increase of the drug pressure was re-applied. Once the parasites could grow steadily under 320 nM of DHA, they were cloned by limiting dilution (Rosario, 1981). Two parasites clones DHA1 and DHA2 with highest IC50 values of DHA were routinely cultured under 100 nM of DHA.
For drug assays, parasites were synchronized and zero time point of the IDC was set at 1 h post invasion. IC50 values of antimalarials were determined by using the SYBR-Green I method (Smilkstein et al., 2004). A 2-fold serial dilution was used with a starting concentration of 1000 nM for DHA, ATM, ATS, ART, AQ, PQ and MQ, 2000 nM for CQ, 20 nM for ATQ, and 3000 nM for LUM and QN. For the two ACTs, the stock concentrations of DHA/PQ and ATM/LUM were 500 nM/1100 nM and 500 nM/1500 nM, respectively, according to the commercial formulations of Duocotexcin® and Coartem®, respectively. To determine VP-reversibility of CQ resistance in DHA1 and DHA2, IC50 values of CQ were measured together with VP at a starting concentration of 2/25 μM of the combination (Patel et al., 2010). Each drug or drug combination was tested in three technical replicates and three biological replicates. IC50 values were calculated using the program GraphPad Prism V5 (La Jolla, CA, USA) by constructing a dose-response curve.
Phenotype analysis
To measure parasite proliferation, cultures of synchronized Dd2, DHA1, and DHA2 parasites were initiated in 24-well plates with 0.5% rings and 5% hematocrit. Medium was changed every 48 h and parasitemia was monitored daily for eight days. Cell cycle duration of each parasite line was precisely measured using Giemsa-stained smears every 2 h for two cycles and determined as the interval between the peak trophozoite parasitemias of two cycles (Reilly et al., 2007).
To determine the responses of the parasites to brief or more extended treatment of ARTs, we measured the survivorship of the parasites after DHA treatment. For brief treatments, synchronized parasites at 12, 24 and 36 h were treated with 1 μM of DHA for 2 h. After removal of the drug, parasites were let to recover in a drug-free medium for 6 h, and viability of the parasites was determined by flow cytometry as described (Witkowski et al., 2010). To determine the potential involvement of ring-specific dormancy in ART resistance, synchronized ring-stage parasites were treated with 1 or 3 μM DHA for 48 h. Subsequently, cultures were maintained in drug-free medium and examined daily using microscopy to determine parasitemias. Medium was changed daily and the amount of time needed for the each parasite culture to reach 10% parasitemia was recorded.
Microarray analysis
To determine genome-wide transcriptional changes in the parasites, microarray analysis was performed for Dd2 and DHA1. A custom-designed expression microarray was developed by Roche NimbleGen (Madison, WI) based on the P. falciparum 3D7 genomic sequence. A total of 35809 45–60-mer probes were replicated and arranged on the array slides using ArrayScribe array design software (NimbleGen). For transcripts of <299 bp, 300–3999 bp and >4000 bp in length, 3, 6 and 11 probes per transcript were selected, respectively. No 3′ bias was used during probe selection and the melting temperatures of the probes are distributed between 60.9 and 77.8. For microarray analysis, 3×109 tightly synchronized Dd2 and DHA1 parasites were treated for 2 h with or without 1 μM DHA at approximately equivalent developmental stages (14, 27 and 38 h for Dd2; 15, 29, and 41 h for DHA1). Total RNA was isolated from untreated control or treated parasites using Trizol Reagent, and treated with RNase-free DNase I to remove contamination of genomic DNA. The quality of the RNA samples was assessed using RNA Nano Chips on the Agilent Bioanalyzer (Agilent, Santa Clara, CA). RNA labeling and array hybridization followed the manufacturer’s instructions (Nimblegen). Slides were scanned using Axon GenePix 4000B. Signals were normalized using the RMA (Robust Microarray Analysis) method. Signal values for each chip were adjusted using RMA background correction and data across all chips were normalized using the quantile method to adjust the probe intensity values to achieve a uniform distribution. Finally, data was summarized into expression level signal values using RMA’s median polish summarization. The transformed microarray data (log2 values) were generated using the ArrayStar 4 software (DNASTAR, Madison, WI). Data were further filtered to remove spots with standard deviations of ≥1 from two independent experiments. The correlations of the data between Dd2 and DHA1 and between our data and the 48-h gene expression profiles in Dd2 (Llinas et al., 2006) were compared to ensure the selected parasites were from similar developmental time points. The data between untreated parasites of Dd2 and DHA1 and between drug treated and untreated control of same line were further compared. The genes with two-fold changes were used for gene ontology (GO) analysis based on data from the PlasmoDB (plasmodb.org).
Analysis of candidate genes associated with drug resistance
Genes that are associated with drug resistance in P. falciparum including pfmdr1 (PFE1150w), pfatp6 (PFA0310c), pftctp (PFE0545c), pfubcth (PFA0220w), pfcrt (MAL7P1.27), G7 (PF13_0271), G49 (PF08_0078), and ABC transporter pfmrp (PFA0590w) were analyzed for variation in copy number and sequence. Copy number was determined by real-time PCR analysis as previously described using the single-copy gene seryl-tRNA synthetase (PF07_0073) as an internal reference (Cui et al., 2007b). The coding regions of these candidate genes were amplified from the genomic DNA, sequenced, and compared with Dd2 sequences to identify mutations.
Quantitative analysis of redox metabolism
To measure ROS levels in the parasites, cultures were maintained in drug-free medium for three cycles, and late trophozoites were labeled with 2′,7′-dichlorfluorescein-diacetate A (50 mM) at 37°C for 15 min and treated with 50 μM curcumin or 200 μM TBP for 4 h. ROS levels, represented by the fluorescence intensity, were measured using flow cytometry (Cui et al., 2007a). To determine the cellular levels of major antioxidants GSH and Trx, and the activities of GST and SOD, 3×109 parasites were harvested from Dd2, DHA1 and DHA2, and at a biweekly interval during the ten-week culture period of these lines in the absence of selective drug pressure. Cell lysates were prepared after sonication and centrifugation, and protein concentrations were determined using a protein assay kit (Bio-Rad, Hercules, CA). GSH level, GST and SOD enzyme activities in protein extract were determined using respective assay kits (Cayman Chemical, Ann Arbor, MI). The unit activity of GST was defined as the amount of 1-chloro-2,4-dinitrobenzene (mM) reduced GSH/min/mg protein. The SOD unit activity is defined as the amount of SOD needed for 50% dismutation of the superoxide radical. The levels of Trxs were determined using the Thioredoxin Assay kit (Redoxica, Little Rock, AR).
Statistical analysis
The differences of IC50 values among parasite lines were compared by unpaired t-tests as previously described (Cui et al., 2012). We used the Bonferroni correction to control for multiple tests; differences were considered significant when P≤0.05/n (n=test number). Cross resistance to different drugs was evaluated using Pearson’s correlation coefficient. Differences between percent representations of functional categories from microarray experiments were compared using the Z-test; differences were considered significant at P<0.05 and P<0.01.
Supplementary Material
Table 3.
Up and down regulated genes in selected DHA-resistant clone DHA1 during IDC.
| Gene ID | Gene description | Fold change (DHA1/Dd2)
|
||
|---|---|---|---|---|
| Ring | Trophozoite | Schizont | ||
| PF10_0336 | Conserved Plasmodium protein, unknown function | 9.2 | 6.4 | 5.0 |
| PF14_0164 | NADP-specific glutamate dehydrogenase | 3.1 | 3.0 | 2.4 |
| PF14_0463 | Chloroquine resistance marker protein | 3.0 | 2.8 | 3.4 |
| PFE1150w | Multidrug resistance protein 1 (pfmdr1) | 6.2 | 8.6 | 9.8 |
| PFF1130c | Superoxide dismutase | 4.8 | 2.5 | 2.6 |
| PFI0925w | Gamma-glutamylcysteine synthetase | 2.5 | 2.8 | 3.1 |
| PFI1250w | Thioredoxin-like protein 2 | 2.8 | 3.4 | 4.7 |
| PFL0725w | Thioredoxin peroxidase 2 | 2.5 | 2.0 | 2.2 |
| PF08_0054 | Heat shock protein 70 | 2.1 | 2.1 | 2.1 |
| PFI1780w | Plasmodium exported protein (PHISTc), | 66.0 | 21.4 | 5.1 |
| MAL7P1.58 | Pfmc-2TM | 8.9 | 62.0 | 13.0 |
| PFB0100c | Knob-associated histidine-rich protein | 8.7 | 3.1 | 2.2 |
| PFI1805w | Rifin | 15.7 | 3.0 | 2.0 |
| PFF0280w | Conserved Plasmodium protein, unknown function | 2.2 | 2.5 | 2.3 |
| PF13_0235 | Transcription factor with AP2 domain(s), putative | 3.5 | 4.2 | 2.8 |
| PFD0150w | Conserved Plasmodium protein, unknown function | 4.1 | 2.0 | 2.4 |
| PFD0295c | Apical sushi protein | −8.2 | −2.0 | −4.4 |
| PF10_0331a | Conserved Plasmodium protein, unknown function | −2.0 | −2.4 | −2.5 |
| PFC0345w | Conserved Plasmodium protein, unknown function | −7.7 | −4.1 | −9.4 |
Acknowledgments
We want to thank Hao Meng for technical assistance, Daniel Parker for Guodong Ning for helping with the statistical analysis, and Yongjun Liu for assistance with microarray data analysis. This work was supported by NIAID, NIH (1R21AI085518 and U19AI089672).
References
- Akide-Ndunge OB, Tambini E, Giribaldi G, McMillan PJ, Muller S, Arese P, Turrini F. Co-ordinated stage-dependent enhancement of Plasmodium falciparum antioxidant enzymes and heat shock protein expression in parasites growing in oxidatively stressed or G6PD-deficient red blood cells. Malar J. 2009;8:113. doi: 10.1186/1475-2875-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alker AP, Lim P, Sem R, Shah NK, Yi P, Bouth DM, Tsuyuoka R, Maguire JD, Fandeur T, Ariey F, Wongsrichanalai C, Meshnick SR. Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am J Trop Med Hyg. 2007;76:641–647. [PubMed] [Google Scholar]
- Anderson TJ, Nair S, Qin H, Singlam S, Brockman A, Paiphun L, Nosten F. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multidrug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob Agents Chemother. 2005;49:2180–2188. doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Koncarevic S, Hunt NH. Oxidative stress and antioxidant defence in malaria parasites. ASM Press; Washington: 2005. [Google Scholar]
- Becker K, Tilley L, Vennerstrom JL, Roberts D, Rogerson S, Ginsburg H. Oxidative stress in malaria parasite-infected erythrocytes: host-parasite interactions. Int J Parasitol. 2004;34:163–189. doi: 10.1016/j.ijpara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Beez D, Sanchez CP, Stein WD, Lanzer M. Genetic predisposition favors the acquisition of stable artemisinin resistance in malaria parasites. Antimicrob Agents Chemother. 2011;55:50–55. doi: 10.1128/AAC.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhisutthibhan J, Pan XQ, Hossler PA, Walker DJ, Yowell CA, Carlton J, Dame JB, Meshnick SR. The Plasmodium falciparum translationally controlled tumor protein homolog and its reaction with the antimalarial drug artemisinin. J Biol Chem. 1998;273:16192–16198. doi: 10.1074/jbc.273.26.16192. [DOI] [PubMed] [Google Scholar]
- Borges S, Cravo P, Creasey A, Fawcett R, Modrzynska K, Rodrigues L, Martinelli A, Hunt P. Genome-wide scan reveals amplification of mdr1 as a common denominator of resistance to mefloquine, lumefantrine and artemisinin in P. chabaudi malaria parasites. Antimicrob Agents Chemother. 2011;55:4858–4856. doi: 10.1128/AAC.01748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman A, Mendis KN. A major transition in malaria treatment: the adoption and deployment of artemisinin-based combination therapies. Am J Trop Med Hyg. 2007;77:193–197. [PubMed] [Google Scholar]
- Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74. doi: 10.1016/S0140-6736(08)60073-2. [DOI] [PubMed] [Google Scholar]
- Castellini MA, Buguliskis JS, Casta LJ, Butz CE, Clark AB, Kunkel TA, Taraschi TF. Malaria drug resistance is associated with defective DNA mismatch repair. Mol Biochem Parasitol. 2011;177:143–147. doi: 10.1016/j.molbiopara.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R, Tripathi LM, Saxena JK, Puri SK. Implication of intracellular glutathione and its related enzymes on resistance of malaria parasites to the antimalarial drug arteether. Parasitol Int. 2011;60:97–100. doi: 10.1016/j.parint.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Chavchich M, Gerena L, Peters J, Chen N, Cheng Q, Kyle DE. Role of pfmdr1 amplification and expression in induction of resistance to artemisinin derivatives in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54:2455–2464. doi: 10.1128/AAC.00947-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, McGready R, Ashley E, Imwong M, Stepniewska K, Yi P, Dondorp AM, Mayxay M, Newton PN, White NJ, Nosten F, Ferdig MT, Anderson TJ. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci U S A. 1994;91:1143–1147. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Miao J, Cui L. Cytotoxic effect of curcumin on malaria parasite Plasmodium falciparum: inhibition of histone acetylation and generation of reactive oxygen species. Antimicrob Agents Chemother. 2007a;51:488–494. doi: 10.1128/AAC.01238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Miao J, Furuya T, Li X, Su XZ, Cui L. PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot Cell. 2007b;6:1219–1227. doi: 10.1128/EC.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Su XZ. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Rev Anti Infect Ther. 2009;7:999–1013. doi: 10.1586/eri.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Wang Z, Jiang H, Parker D, Wang H, Su XZ, Cui L. Lack of association of the S769N mutation in Plasmodium falciparum SERCA (PfATP6) with resistance to artemisinins. Antimicrob Agents Chemother. 2012;56:2546–2552. doi: 10.1128/AAC.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom S, Veiga MI, Ferreira P, Martensson A, Kaneko A, Andersson B, Bjorkman A, Gil JP. Diversity of the sarco/endoplasmic reticulum Ca(2+)-ATPase orthologue of Plasmodium falciparum (PfATP6) Infect Genet Evol. 2008;8:340–345. doi: 10.1016/j.meegid.2008.02.002. [DOI] [PubMed] [Google Scholar]
- del Pilar Crespo M, Avery TD, Hanssen E, Fox E, Robinson TV, Valente P, Taylor DK, Tilley L. Artemisinin and a series of novel endoperoxide antimalarials exert early effects on digestive vacuole morphology. Antimicrob Agents Chemother. 2008;52:98–109. doi: 10.1128/AAC.00609-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplaine G, Lavazec C, Bischoff E, Natalang O, Perrot S, Guillotte-Blisnick M, Coppee JY, Pradines B, Mercereau-Puijalon O, David PH. Artesunate tolerance in transgenic Plasmodium falciparum parasites overexpressing a tryptophan-rich protein. Antimicrob Agents Chemother. 2011;55:2576–2584. doi: 10.1128/AAC.01409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding XC, Beck HP, Raso G. Plasmodium sensitivity to artemisinins: magic bullets hit elusive targets. Trends Parasitol. 2011;27:73–81. doi: 10.1016/j.pt.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- Eckstein-Ludwig U, Webb RJ, Van Goethem ID, East JM, Lee AG, Kimura M, O’Neill PM, Bray PG, Ward SA, Krishna S. Artemisinins target the SERCA of Plasmodium falciparum. Nature. 2003;424:957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla NB, Adam I, Elzaki SE, Bashir S, Mukhtar I, Oguike M, Gadalla A, Mansour F, Warhurst D, El-Sayed BB, Sutherland CJ. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrobial agents and chemotherapy. 2011;55:5408–5411. doi: 10.1128/AAC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg H, Golenser J. Redox metabolism in glucose-6-phosphate dehydrogenase deficient erythrocytes and its relation to antimalarial chemotherapy. Parassitologia. 1999;41:309–311. [PubMed] [Google Scholar]
- Ginsburg H, Golenser J. Glutathione is involved in the antimalarial action of chloroquine and its modulation affects drug sensitivity of human and murine species of Plasmodium. Redox Rep. 2003;8:276–279. doi: 10.1179/135100003225002907. [DOI] [PubMed] [Google Scholar]
- Ginsburg H, Ward SA, Bray PG. An integrated model of chloroquine action. Parasitol Today. 1999;15:357–360. doi: 10.1016/s0169-4758(99)01502-1. [DOI] [PubMed] [Google Scholar]
- Gonzales JM, Patel JJ, Ponmee N, Jiang L, Tan A, Maher SP, Wuchty S, Rathod PK, Ferdig MT. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS Biol. 2008;6:e238. doi: 10.1371/journal.pbio.0060238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig CL, Rosenthal AS, D’Angelo J, Griffin CE, Posner GH, Cooper RA. Accumulation of artemisinin trioxane derivatives within neutral lipids of Plasmodium falciparum malaria parasites is endoperoxide-dependent. Biochem Pharmacol. 2009;77:322–336. doi: 10.1016/j.bcp.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes RK, Chan HW, Lung CM, Ng NC, Wong HN, Shek LY, Williams ID, Cartwright A, Gomes MF. Artesunate and dihydroartemisinin (DHA): unusual decomposition products formed under mild conditions and comments on the fitness of DHA as an antimalarial drug. ChemMedChem. 2007;2:1448–1463. doi: 10.1002/cmdc.200700064. [DOI] [PubMed] [Google Scholar]
- Hayward R, Saliba KJ, Kirk K. pfmdr1 mutations associated with chloroquine resistance incur a fitness cost in Plasmodium falciparum. Mol Microbiol. 2005;55:1285–1295. doi: 10.1111/j.1365-2958.2004.04470.x. [DOI] [PubMed] [Google Scholar]
- Hunt P, Afonso A, Creasey A, Culleton R, Sidhu AB, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol. 2007;65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Dondorp AM, Nosten F, Yi P, Mungthin M, Hanchana S, Das D, Phyo AP, Lwin KM, Pukrittayakamee S, Lee SJ, Saisung S, Koecharoen K, Nguon C, Day NP, Socheat D, White NJ. Exploring the contribution of candidate genes to artemisinin resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54:2886–2892. doi: 10.1128/AAC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inselburg J. Induction and isolation of artemisinine-resistant mutants of Plasmodium falciparum. Am J Trop Med Hyg. 1985;34:417–418. doi: 10.4269/ajtmh.1985.34.417. [DOI] [PubMed] [Google Scholar]
- Jambou R, Legrand E, Niang M, Khim N, Lim P, Volney B, Ekala MT, Bouchier C, Esterre P, Fandeur T, Mercereau-Puijalon O. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- Klonis N, Crespo-Ortiz MP, Bottova I, Abu-Bakar N, Kenny S, Rosenthal PJ, Tilley L. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc Natl Acad Sci U S A. 2011;108:11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krungkrai SR, Yuthavong Y. The antimalarial action on Plasmodium falciparum of qinghaosu and artesunate in combination with agents which modulate oxidant stress. Trans R Soc Trop Med Hyg. 1987;81:710–714. doi: 10.1016/0035-9203(87)90003-4. [DOI] [PubMed] [Google Scholar]
- Li GQ, Arnold K, Guo XB, Jian HX, Fu LC. Randomised comparative study of mefloquine, qinghaosu, and pyrimethamine-sulfadoxine in patients with falciparum malaria. Lancet. 1984;2:1360–1361. doi: 10.1016/s0140-6736(84)92057-9. [DOI] [PubMed] [Google Scholar]
- Lim P, Alker AP, Khim N, Shah NK, Incardona S, Doung S, Yi P, Bouth DM, Bouchier C, Puijalon OM, Meshnick SR, Wongsrichanalai C, Fandeur T, Le Bras J, Ringwald P, Ariey F. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11. doi: 10.1186/1475-2875-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 2006;34:1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SK, Oduola AM, Milhous WK. Reversal of chloroquine resistance in Plasmodium falciparum by verapamil. Science. 1987;235:899–901. doi: 10.1126/science.3544220. [DOI] [PubMed] [Google Scholar]
- Meshnick SR. Artemisinin: mechanisms of action, resistance and toxicity. Int J Parasitol. 2002;32:1655–1660. doi: 10.1016/s0020-7519(02)00194-7. [DOI] [PubMed] [Google Scholar]
- Miao J, Fan Q, Cui L, Li X, Wang H, Ning G, Reese JC, Cui L. The MYST Family Histone Acetyltransferase Regulates Gene Expression and Cell Cycle in Malaria Parasite Plasmodium falciparum. Mol Microbiol. 2010;78:883–902. doi: 10.1111/j.1365-2958.2010.07371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockenhaupt FP. Mefloquine resistance in Plasmodium falciparum. Parasitol Today. 1995;11:248–253. doi: 10.1016/0169-4758(95)80201-0. [DOI] [PubMed] [Google Scholar]
- Mok S, Imwong M, Mackinnon MJ, Sim J, Ramadoss R, Yi P, Mayxay M, Chotivanich K, Liong KY, Russell B, Socheat D, Newton PN, Day NP, White NJ, Preiser PR, Nosten F, Dondorp AM, Bozdech Z. Artemisinin resistance in Plasmodium falciparum is associated with an altered temporal pattern of transcription. BMC Genomics. 2011;12:391. doi: 10.1186/1471-2164-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Ferdig MT, Feng X, Joy DA, Duan J, Furuya T, Subramanian G, Aravind L, Cooper RA, Wootton JC, Xiong M, Su XZ. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- Muller S. Redox and antioxidant systems of the malaria parasite Plasmodium falciparum. Mol Microbiol. 2004;53:1291–1305. doi: 10.1111/j.1365-2958.2004.04257.x. [DOI] [PubMed] [Google Scholar]
- Natalang O, Bischoff E, Deplaine G, Proux C, Dillies MA, Sismeiro O, Guigon G, Bonnefoy S, Patarapotikul J, Mercereau-Puijalon O, Coppee JY, David PH. Dynamic RNA profiling in Plasmodium falciparum synchronized blood stages exposed to lethal doses of artesunate. BMC Genomics. 2008;9:388. doi: 10.1186/1471-2164-9-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Noedl H, Se Y, Sriwichai S, Schaecher K, Teja-Isavadharm P, Smith B, Rutvisuttinunt W, Bethell D, Surasri S, Fukuda MM, Socheat D, Chan Thap L. Artemisinin resistance in Cambodia: a clinical trial designed to address an emerging problem in Southeast Asia. Clin Infect Dis. 2010;51:e82–89. doi: 10.1086/657120. [DOI] [PubMed] [Google Scholar]
- Nogueira F, Diez A, Radfar A, Perez-Benavente S, do Rosario VE, Puyet A, Bautista JM. Early transcriptional response to chloroquine of the Plasmodium falciparum antioxidant defence in sensitive and resistant clones. Acta Trop. 2010;114:109–115. doi: 10.1016/j.actatropica.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Nosten F. Waking the sleeping beauty. J Infect Dis. 2010;202:1300–1301. doi: 10.1086/656478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oduola AM, Milhous WK, Weatherly NF, Bowdre JH, Desjardins RE. Plasmodium falciparum: induction of resistance to mefloquine in cloned strains by continuous drug exposure in vitro. Exp Parasitol. 1988;67:354–360. doi: 10.1016/0014-4894(88)90082-3. [DOI] [PubMed] [Google Scholar]
- Patel JJ, Thacker D, Tan JC, Pleeter P, Checkley L, Gonzales JM, Deng B, Roepe PD, Cooper RA, Ferdig MT. Chloroquine susceptibility and reversibility in a Plasmodium falciparum genetic cross. Molecular microbiology. 2010;78:770–787. doi: 10.1111/j.1365-2958.2010.07366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, Kawamoto F, Miller RS, Meshnick SR. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Cassar C, Brockman A, Duraisingh M, van Vugt M, White NJ, Nosten F, Krishna S. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–2949. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radfar A, Diez A, Bautista JM. Chloroquine mediates specific proteome oxidative damage across the erythrocytic cycle of resistant Plasmodium falciparum. Free Radic Biol Med. 2008;44:2034–2042. doi: 10.1016/j.freeradbiomed.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Raj DK, Mu J, Jiang H, Kabat J, Singh S, Sullivan M, Fay MP, McCutchan TF, Su XZ. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J Biol Chem. 2009;284:7687–7696. doi: 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod PK, McErlean T, Lee PC. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A. 1997;94:9389–9393. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Reilly HB, Wang H, Steuter JA, Marx AM, Ferdig MT. Quantitative dissection of clone-specific growth rates in cultured malaria parasites. Int J Parasitol. 2007;37:1599–1607. doi: 10.1016/j.ijpara.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues LA, Henriques G, Borges ST, Hunt P, Sanchez CP, Martinelli A, Cravo P. Experimental evolution of resistance to artemisinin combination therapy results in amplification of the mdr1 gene in a rodent malaria parasite. PLoS One. 2010;5:e11593. doi: 10.1371/journal.pone.0011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario V. Cloning of naturally occurring mixed infections of malaria parasites. Science. 1981;212:1037–1038. doi: 10.1126/science.7015505. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, Mayer S, Nurhasanah A, Stein WD, Lanzer M. Genetic linkage analyses redefine the roles of PfCRT and PfMDR1 in drug accumulation and susceptibility in Plasmodium falciparum. Mol Microbiol. 2011;82:865–878. doi: 10.1111/j.1365-2958.2011.07855.x. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, Rotmann A, Stein WD, Lanzer M. Polymorphisms within PfMDR1 alter the substrate specificity for anti-malarial drugs in Plasmodium falciparum. Mol Microbiol. 2008;70:786–798. doi: 10.1111/j.1365-2958.2008.06413.x. [DOI] [PubMed] [Google Scholar]
- Shah NK, Alker AP, Sem R, Susanti AI, Muth S, Maguire JD, Duong S, Ariey F, Meshnick SR, Wongsrichanalai C. Molecular surveillance for multidrug-resistant Plasmodium falciparum, Cambodia. Emerg Infect Dis. 2008;14:1637–1640. doi: 10.3201/eid1410.080080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Zakeri S, Palacpac NM, Afsharpad M, Randrianarivelojosia M, Kaneko A, Marma AS, Horii T, Mita T. Spontaneous mutations in the Plasmodium falciparum sarcoplasmic/ endoplasmic reticulum Ca2+-ATPase (PfATP6) gene among geographically widespread parasite populations unexposed to artemisinin-based combination therapies. Antimicrob Agents Chemother. 2011;55:94–100. doi: 10.1128/AAC.01156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuscher F, Gatton ML, Chen N, Peters J, Kyle DE, Cheng Q. Artemisinin-induced dormancy in Plasmodium falciparum: duration, recovery rates, and implications in treatment failure. J Infect Dis. 2010;202:1362–1368. doi: 10.1086/656476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta RF, Brown ML, Terrell JC, Geyer JA. Defective DNA repair as a potential mechanism for the rapid development of drug resistance in Plasmodium falciparum. Biochemistry. 2004;43:4885–4891. doi: 10.1021/bi0499258. [DOI] [PubMed] [Google Scholar]
- Uhlemann AC, Cameron A, Eckstein-Ludwig U, Fischbarg J, Iserovich P, Zuniga FA, East M, Lee A, Brady L, Haynes RK, Krishna S. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat Struct Mol Biol. 2005;12:628–629. doi: 10.1038/nsmb947. [DOI] [PubMed] [Google Scholar]
- Valderramos SG, Scanfeld D, Uhlemann AC, Fidock DA, Krishna S. Investigations into the role of the Plasmodium falciparum SERCA (PfATP6) L263E mutation in artemisinin action and resistance. Antimicrob Agents Chemother. 2010;54:3842–3852. doi: 10.1128/AAC.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Development of a strategy towards elimination of Plasmodium falciparum parasites with altered responses to artemisinins. World Health Organization; 2009. pp. 1–52. [Google Scholar]
- WHO. Global report on antimalarial drug efficacy and drug resistance. 2010. p. 115. [Google Scholar]
- WHO. Global plan for artemisinin resistance containment (GPARC) 2011. p. 87. [Google Scholar]
- Wilson CM, Serrano AE, Wasley A, Bogenschutz MP, Shankar AH, Wirth DF. Amplification of a gene related to mammalian mdr genes in drug-resistant Plasmodium falciparum. Science. 1989;244:1184–1186. doi: 10.1126/science.2658061. [DOI] [PubMed] [Google Scholar]
- Wilson CM, Volkman SK, Thaithong S, Martin RK, Kyle DE, Milhous WK, Wirth DF. Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol. 1993;57:151–160. doi: 10.1016/0166-6851(93)90252-s. [DOI] [PubMed] [Google Scholar]
- Witkowski B, Lelievre J, Barragan MJ, Laurent V, Su XZ, Berry A, Benoit-Vical F. Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob Agents Chemother. 2010;54:1872–1877. doi: 10.1128/AAC.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsrichanalai C, Meshnick SR. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg Infect Dis. 2008;14:716–719. doi: 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Cheng KC, Johnson RL, Huang R, Pattaradilokrat S, Liu A, Guha R, Fidock DA, Inglese J, Wellems TE, Austin CP, Su XZ. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Johnson RL, Huang R, Wichterman J, Jiang H, Hayton K, Fidock DA, Wellems TE, Inglese J, Austin CP, Su XZ. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum. Nat Chem Biol. 2009;5:765–771. doi: 10.1038/nchembio.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Guan Y, Zheng B, Wu S, Tang L. No PfATPase6 S769N mutation found in Plasmodium falciparum isolates from China. Malar J. 2008;7:122. doi: 10.1186/1475-2875-7-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


