Abstract
The transduction of sound by the receptor or hair cells of the cochlea leads to the activation of ion channels found in the basal and lateral regions of these cells. Thus, the processing of these transduced signals to the central nervous system is tied to the regulation of baso-lateral ion channels. The large conductance calcium-activated potassium or BK channel was revealed to interact with the small GTPase, Rab11b, which is one of many Rabs found in various endosomal pathways. Immunoelectron microscopy showed the colocalization of these two proteins in receptor cells and auditory neurons. Using Chinese hamster ovary cells as a heterologous expression system, Rab11b increased or decreased BK expression, depending on the overexpression or RNAi knockdown of Rab, respectively. Additional mutation analyses, using a yeast two-hybrid assay, suggested that this GTPase moderately interacts within a region of BK exclusive of the N- or C-terminal tails. These data suggest that this small GTPase regulates BK in a slow recycling process through the endocytic compartment and to the plasmalemma.
Keywords: ion channels, BK channel, Rab family, protein-protein interactions, cochlea, hair cell, ganglion cell
Introduction
The large conductance, calcium-activated potassium channel or BK, underlies tuning in non-mammalian hair cells and plays a role in hearing sensitivity in mammalian hair cells. Thus, the regulation of BK channel numbers will affect its role in mediating auditory signals in the cochlea. The purpose of this study was to discover the function of one of these potential regulators. Previously, we used a high-throughput screening method that used coimmunoprecipitation and LC-MS/MS to determine putative partners that interact with the BK channel [1]. These studies suggested a potential partner in the small GTPase, Rab11b.
Rabs are master regulators of endosomal trafficking, involved in protein transport to the cell membrane (early endosome), recycling from and to the membrane (recycling endosomes), and protein transport for degradation in the lysosome (late endosomes). To date, one of the oft-cited Rab/ion channel associations is with the cystic fibrosis transmembrane conductance regulator (CFTR), a cAMP-activated Cl− channel expressed on the apical side of a wide variety of epithelial cells [see 2 for review]. Rabs 4, 5, 7, 9, 11, and 27a are implicated in interactions with CFTR by either up or down regulating the expression of this protein via different intracellular pathways. Previous studies suggest that among the 60 plus Rab proteins of the Ras superfamily, Rab11b is involved primarily with recycling endosomes [3]. Ion channels regulated by this small GTPase include hERG, TRPV5 and V6, and the L-type Ca2+ channel, Cav1.2 [4–6].
In the present study, we investigated the possibility of a direct interaction between Rab11b in relation to BK in the mouse cochlea and what this interaction may entail in functional terms. Previous evidence suggests that the knockout of BK may decrease the susceptibility of the cochlea to damage caused by acoustic trauma [7]. Thus, insights into the regulatory mechanisms that control the expression of this channel at the plasmalemma will likely control the process of noise-induced hearing loss. To the best of our knowledge, this is the first study to show a relationship between the BK channel and the small GTPase Rab11b.
Materials and methods
Immunoelectron Microscopy
Tympanic bullae were removed rapidly from 30 day-old CBA/J mice and fixed by opening the round window and creating a small hole in the apex to expose the tissue to 4% paraformaldehyde/0.1% glutaraldehyde (Ted Pella) in 0.1 M phosphate buffer (pH 7.4) for 2 h at RT. Tissues were then rinsed in PBS followed by partial or complete removal of the bony walls of the cochlea and decalcification for 3 days at 4°C in 5 mM EDTA/0.1 M phosphate buffer. Tissues were washed over the course of 2 h at 15 min intervals and then dehydrated as follows: 1 × 15 min in 50% EtOH, 2 × 30 min in 70% EtOH and 1 × 30 min in 85% EtOH. Infiltration with LR White medium grade resin (Ted Pella) involved placing tissues for 1 h in 2:1 resin/70% EtOH, followed by 2 × 1 h and then overnight (O/N) in fresh 100% resin. The next day, tissues were placed in fresh 100% resin for 1 h, embedded in 00 gelatin capsules and cured at 50°C for 48 h. Sections of gold-interference thickness were collected on Formvar-coated 100 mesh hexagonal nickel grids (EMS) using a Sorvall Porter-Blum ultramicrotome and dried O/N. Immunostaining was accomplished using the post-embedding technique. Sections were rehydrated 3 × 5 min in TBS (pH 8.2) and free aldehyde groups were blocked for 30 min in 50 mM glycine/TBS. Blocking solution consisted of 5% normal goat serum/1% BSA/0.1% cold-water fish gelatin/0.1% Tween 20/TBS. After 30 min, sections were rinsed briefly in 0.1% BSA-c (Ted Pella) followed by O/N incubation at RT in anti-BK polyclonal antibody (1:25, Abcam, epitope human aa 945–961) and monoclonal anti-Rab11b (1:10, Sigma) diluted in 0.1% BSA-c/0.1% Tween 20/TBS. The next day, sections were washed 3 × 5 min in TBS (pH 8.2) and incubated in goat anti-rabbit IgG, conjugated with Au particles of 10 nm diameter, and goat anti-mouse IgG, conjugated with Au particles of 5 nm diameter (BBI, Ted Pella). These secondary antibodies were diluted 1:10 in 0.1% BSA-c/0.1% Tween 20/TBS (pH 8.2) for 90 min. After several washes in TBS followed by H2O, sections were osmicated (2%) for 15 min, rinsed and then stained with 1% KMnO4 (aq) for 15 s followed by a rinse. Sections were placed in 0.025% citric acid for 45 s, rinsed and then stained with 1% aqueous uranyl acetate for 30 min and finally rinsed 3 × 5 min in H2O. Sections were permitted to dry O/N. Tissues were imaged at 60 kV using an FEI Morgagni Model 268D TEM equipped with an AMT XR16 ActiveVu 16 MP camera.
Cloning and hemagglutinin (HA)-tagging of a BK Splice Variant from Mouse Cochlea
A BK-DEC splice variant (GenBank Acc. No. FJ872117) was cloned from total RNA extracted from adult mouse cochlea by RT-PCR, using a forward primer with a BglII site, 5′-GGAAGATCTCCCAAGATGGATGCGCTCATCA-3′ and a reverse primer, with a SalI site, 5′-ACGCGTCGACAGTTCTGGTCTCCTGGGAGT-3′. The PCR product was purified (QIA quick PCR Purification Kit, Qiagen) and cut at restriction sites BglII and SalI at 37°C O/N. The insert was gel-purified (StrataPrep DNA Gel Extraction Kit, Stratagene) and ligated into pcDNA3.1(+) (Invitrogen) at restriction sites BamHI (5′) and XhoI (3′). Tandem HA-tagged BK vector was generated using the pcDNA3.1-BK-DEC variant as a PCR template. The forward primer, which included a HindIII (5′) site and a tandem HA-tag sequence for linkage to the N-terminus of BK-DEC, consisted of 5′-CCCAAGCTTACCATGGGATACCCTTACGACGTTCCTGA TTACGCTTACCCTTACGACGTTCCTGATTACGCTATGGATGCGCTCATCATA CCGGTGA-3′. The reverse primer was set for the BK HindIII site and consisted of 5′-CCCAAGCTTCACAAAACACAGCTCACAAACAGTAGGGA-3′. The fragment was gel-purified and ligated into pcDNA3.1/BK-DEC. All primers were acquired from Integrated DNA Technologies.
Coimmunoprecipitation
A total of 1.0 μg of polyclonal anti-Rab11b antibody (Cell Signaling Technology) was complexed to a 50 μl volume of protein G Sepharose beads (Invitrogen) over the course of 2 h, after which octyl-β-glucoside-solubilized/cleared cochlear preparation was added for O/N incubation [8]. The next day, immunocomplexed beads were washed 5X in lysis buffer and eluted by boiling in sample buffer (Sigma). The same procedure was employed for the reciprocal coimmunoprecipitation by using 6 μg of polyclonal anti-BK antibody (Millipore). Samples were fractionated on a 4–15% SDS-PAGE gradient gel (Bio-Rad) and transferred to a nitrocellulose membrane (Protran BA; Schleicher & Schuell). BK blots were blocked in TBST (50 mM Tris-HCl (pH 8.0), 120 mM NaCl, 0.05% Tween 20), containing 4% milk for 30 min, then probed with anti-rabbit BKα polyclonal antibody at 1:150 (Chemicon), followed by donkey anti-rabbit HRP-conjugated secondary antibody at 1:5000 (Amersham). For Rab11b, blots were blocked as before and probed with anti-Rab11b polyclonal antibody at 1:750 (Cell Signaling Technology) followed by the same secondary as before. Controls consisted of incubating samples with uncomplexed beads, and immunoprecipitating BK using anti-BK antibody and Rab11b using anti-Rab11b antibody. Immunoreactive bands were developed using ECL (Amersham). Magic Mark XP (Invitrogen) was used as the protein standard to estimate relative mobilities.
Cloning Rab11b
Mouse Rab11b was amplified by RT-PCR using total RNA extracted from postnatal day 14 mouse cochleae using a total RNA extraction kit (Qiagen). Cloning primers were designed from the sequence of accession number: NM_008997 (Genbank, NCBI). Forward, 5′-CGCGGATCCAGGACAATGGGGAC CCGGGACGACGAGT-3′ and reverse, 5′-TGCTCTAGAAGTCACAGGCTCTGGC AGCACTGC-3′ primers had BamHI and XbaI sites, respectively, at the 5′ end for insertion into pcDNA3.1. The RT reaction at 50°C for 30 min, was followed by PCR at the following parameters: 94°C for 2 min followed by 35 cycles of denaturing at 94°C for 15 s, annealing at 55°C for 30 s, extending at 68°C for 1 min, and then a final extension at 72°C for 8 min.
Rab11b RNAi and overexpression
CHO cells were cultured in 60 mm dishes and maintained in Minimal Essential Medium (α-MEM; Invitrogen) supplemented with 10% FBS. Exogenous siRNAs to mouse Rab11b was used along with scrambled RNAs (scRNA) (Stealth Select RNAi; Invitrogen) that served as negative controls for low-, medium-, and high-GC content RNAi. Transfections were performed using 10 μl of Lipofectamine2000 (Invitrogen), 8 μg of HA-tagged BK-DEC/pcDNA3.1, and 300 pmoles pooled siRNA, along with equal concentrations of scRNA for the controls. siRNAs were mixed in a 1:1:1 ratio using the following sense strands (5′–3′) for Rab11b: CCGGGACGACGAGUACGAUUACCUA, ACGACGAGUACGAUUACCUAUUC AA, and CGGGAUCAUGCAGAUAGCAACAUUG. Transfected cells were maintained for 48 hrs after which lysates were prepared by sonication, solubilization in 0.1% ASB-14 for 10 min and cleared by spinning at 14,000 xg at 4°C. Protein concentrations were determined by RC-DC Protein assay (Bio-Rad) per manufacturer’s instructions. 1.5 μg of protein was fractionated on a 7.5% SDS-PAGE gel and transferred to a nitrocellulose membrane. Primary antibody consisted of monoclonal anti-HA tag (1:5000; Sigma). Immunoreactive bands were developed using chemiluminescence with Magic Mark XP (Invitrogen) as the protein standard to estimate relative mobilities.
CHO cells were maintained as described previously for overexpression studies [8]. Transfections were performed using 4 μg of HA-tagged BK-DEC/pcDNA3.1 and 4 μg of Rab11b/pcDNA3.1. The control consisted of equal quantities of HA-tagged BK and empty plasmid. Immunoreactive bands were developed as before.
Y2H studies
BK and Rab11b interactions were studied by cotransforming yeast cells with wild type Rab11b plus one of four different BK constructs cloned from the BK-DEC variant [9]. Each of the four components was inserted into a pBD-GAL4 plasmid and consisted of: the first 127 amino acids (BK1–127), which includes the N-terminus, transmembrane segment S0, and the linker region; the first 350 aa (BK1–350), which includes the N-terminus and transmembrane segments S0–S6; aa 122–625 (BK122–625), which includes transmembrane segments S1–6 and the RCK1 domain (S7, S8); and residues 616–1175 (BK616–1175), which includes a portion of the RCK1 domain, the RCK2 domain, the Ca2+ bowl, and the remaining C-terminus. Competent AH109 yeast cells (Clontech) were transformed using the lithium acetate/SS-DNA/PEG method of [10] Briefly, cells were suspended in a lithium acetate solution for transformation with pAD-GAL4-Rab11b plasmid and one of the BK constructs inserted in pBD-GAL4. Cells were plated under lowly to highly stringent conditions as defined by SD medium lacking two, three, or four of the following amino acids, Histidine, (-H), Leucine (-L), Adenine (-A), or Tryptophan (-T). Once colonies grew to 2 – 3 mm in diameter a colony lift filter assay was performed using either –HLT or –HLAT colonies, to determine lacZ reporter gene expression through β-galactosidase activity. This assay involved transferring the colonies to filter paper, fracturing them in LN2 and exposing them to 4-chloro-3-indolyl-b-D-galactopyranoside (X-gal; Agilent Technologies).
Results and discussion
Rab11b and BK interact and colocalize in the cochlea
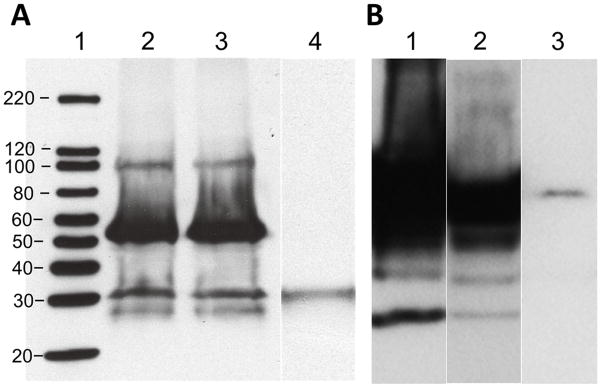
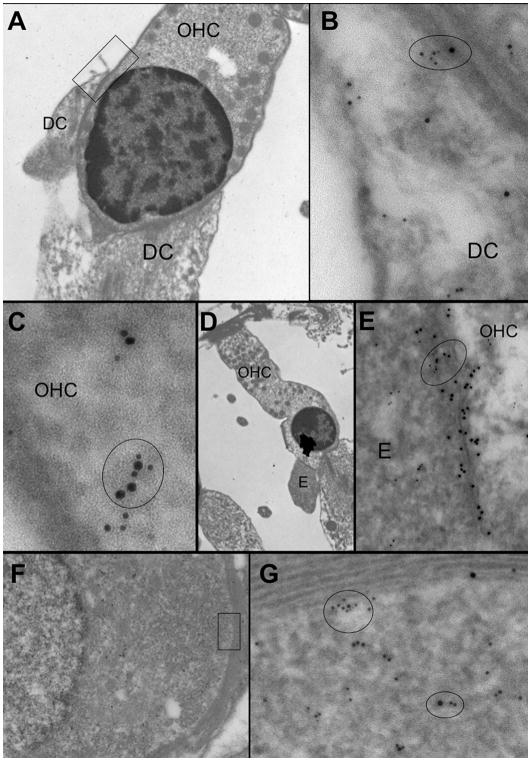
Reciprocal coimmunoprecipitation was used to verify the association of the Rab11b and BK interaction using mouse protein lysates obtained from cochlear sensory epithelium and antibodies specific to both proteins (Fig. 1). Products of immuno-precipitation using the anti-BK antibody were analyzed for coprecipitation of Rab11b. A protein species at 25 kDa was observed, which is equivalent to the molecular weight of Rab11b, as shown when compared to the immunoprecipitation of Rab11b alone. The reciprocal coimmunoprecipitation was accomplished using an antibody to Rab11b. The result shows that Rab11b coprecipitates a protein at ~100–110 kDa, which is equivalent to the immunoprecipitation of BK alone. To further verify potential interactions, we used immunogold labeling to determine if BK and Rab11b colocalize in hair, supporting, and ganglion cells of the mouse cochlea (Fig. 2). The results show that BK and Rab11b, immunolabeled with colloidal gold particles of 10 and 5 nm, respectively, were colocalized in various cell types of the cochlea. These types included outer hair cells, Deiter’s cells and ganglion cells. In OHCs, BK and Rab11b were colocalized at efferent synapses, while both proteins also colocalized in the cytoplasm of Deiter’s cells and near the membrane of adjacent OHCs. Similarly, both proteins were colocalized in the cytoplasm and near the membrane of spiral ganglion cell bodies surrounded by myelin. These results suggested the potential for a direct physical interaction between these proteins. To date, little is known about the regulation of ion channels in the cochlea. Other systems, however, show that channels such as hERG, TRP and voltage-gated Ca2+ channels are regulated by Rab11 proteins. TRPV5 and TRPV6 expression increases at the plasmalemma, for example, by direct contact with locked GDP-bound Rab11 [4], whereas plasmalemma expression of the hERG channel increases through the GTPase, Rab11b [6]. In contrast, Rab11b is part of a degradative pathway for the L-type Ca2+ channel, Cav1.2 [5].
Fig. 1.
Reciprocal coimmunoprecipitation of BK and Rab11b in cochlea. (A) Rab11b immunoprecipitates the BK channel, which has a molecular weight of ~100 kDa (lane 2), when compared to the control experiment showing immunoprecipitation of BK alone (lane 3). Lane 1 is the molecular weight standard and lane 4 is a bead control. (B) BK coimmunoprecipitates the protein Rab11b, which has a molecular weight of ~25 kDa (lane 1), when compared to the immunoprecipitation of Rab11b alone (lane 3). Lane 3 is a bead control.
Fig. 2.
Colocalization of BK and Rab11b in cochlear cells as seen using immunoelectron microscopy. Rab11b and BK were immunolabeled with colloidal gold particles of 5 and 10 nm, respectively. (A) Colocalization of BK and Rab11b in a Deiter’s cell (DC) and outer hair cell (OHC), in the region depicted in the square at low magnification, is revealed (B, C) at a higher magnification (ovals). (D, E) Colocalization of BK and Rab11b (oval) is observed at an efferent (E) synapse on an OHC, shown at low and high magnifications. (F, G) Colocalization of both proteins, in a region (square) near the myelin-coated membrane and cytoplasm of a spiral ganglion cell at low magnification, is revealed at a higher magnification (ovals).
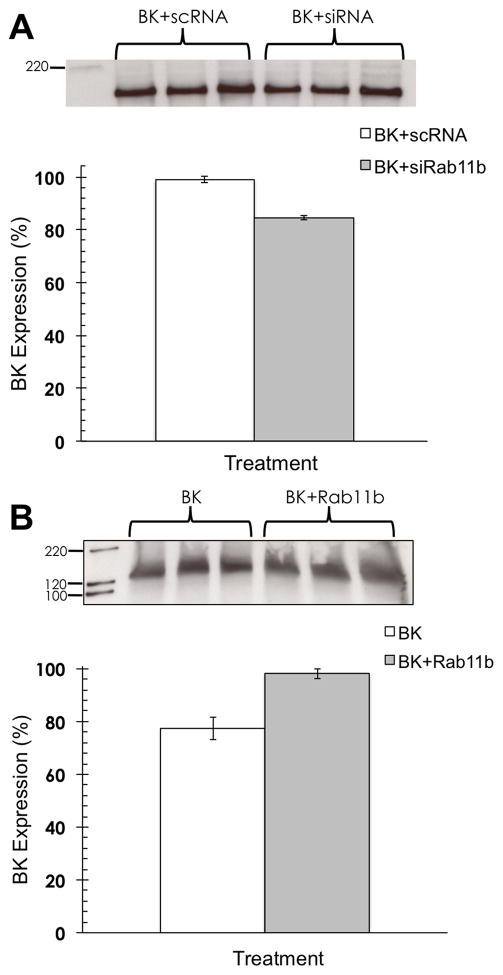
Rab11b increases BK through a moderate interaction
CHO cells doubly transfected with BK and Rab11b were used to determine the effect on BK expression in the presence of Rab (Fig. 3). Knockdown of Rab resulted in a decrease in BK expression of ~15%, whereas overexpression of Rab resulted in an increase in BK of ~20%. These results suggest a mechanism similar to that of TRP and hERG channels in that Rab11 increases the BK expression at the plasmalemma. Moreover, this regulation may occur via the slow recycling pathway as opposed to the degradative pathway exemplified by the Rab11b association with the Ca2+ channel. Previous studies suggest that slow recycling via Rab11 occurs through the endocytic recycling compartment, whereas Rab4 and Rab35 are required for rapid recycling [3].
Fig. 3.
Rab11b alters BK expression. (A) CHO cells cotransfected with Rab11b siRNAs and BK show a decrease in BK expression compared to CHO cells transfected with BK and scrambled (sc) RNA. Transfections were done in triplicate for each treatment (BK+scRNA vs. BK+siRNA) and densitometry measurements were normalized to the highest densitometry value (normalized to 100%) within a given set of 6 lanes. Statistical significance was determined using an unpaired, two-tailed t-test to obtain p < 0.05. Error bars represent the standard deviation. (B) CHO cells cotransfected with BK and Rab11b cDNAs showed an increase in BK expression compared to cells cotransfected with BK and empty plasmid. Transfections were done in triplicates for each treatment (BK+plasmid vs. BK+Rab11b) and analyzed as before.
In order to determine the strength as well as a putative BK region for Rab interaction, AH109 yeast cells were cotransformed with Rab11b and four different components of the BK channel, and then grown on three different dropout media. Comparisons were made to a positive control consisting of the interaction between Kv4.2 and pentraxin [11]. Results revealed that Rab11b has a moderate association with a region of the BK channel that includes aa 127–616 (Fig. 4), since these cultures were positive on –HLT dropout medium. These residues encompass essentially the core of BK that includes transmembrane segment S2 through the RCK1 domain, but exclude the rest of the C-terminus including RCK2 and the Ca2+ bowl (S8–S10). Thus far, there are no data delineating regions of ion channel interaction other that Rab11a, which interacts with the carboxy terminus of TRP channels [4].
Fig. 4.
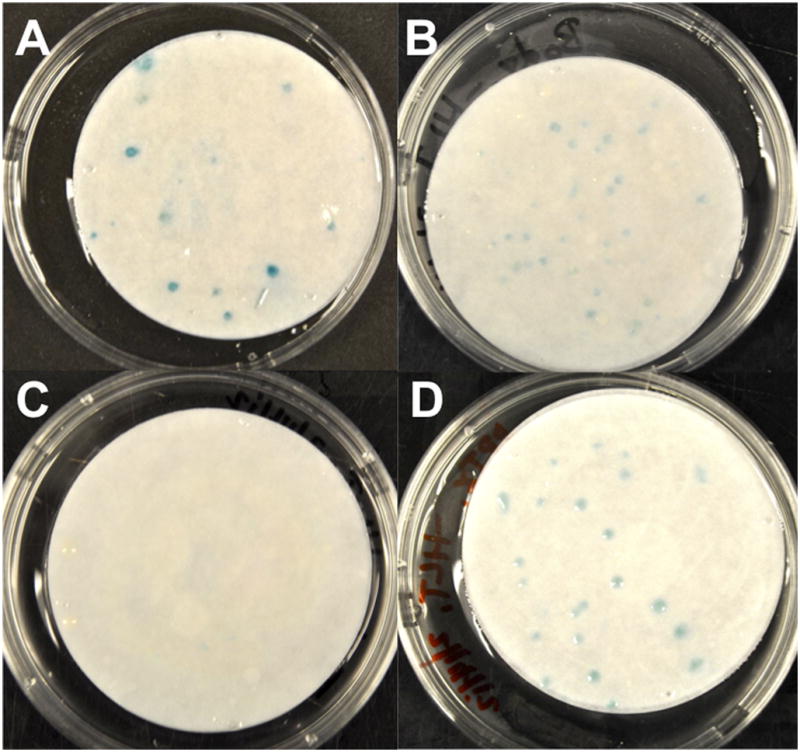
Rab11b interacts within core regions of BK as determined by a yeast two-hybrid assay. Colonies grown on –HLT dropout medium were positive for lacZ reporter gene expression, when grown with specific components of BK. Cotransformation of AH109 cells with (A) Rab11b and BK1–350 or Rab11b and BK122–625 produced positive lacZ colonies. LacZ expression was absent when yeast cells were cotransformed with (C) Rab11b and BK1–127 or Rab and BK616–1175 (data not shown). (D) LacZ expression in a positive control, resulting from the interaction between Kv4.1 and pentraxin.
In summary, our data show an interaction between the small GTPase Rab11b and the BK channel, which suggests that BK is shuttled through a slow recycling endocytic pathway. While little is known with regard to sites of interaction between Rabs and ion channels, we show that Rab11b interacts within the core segment of BK that includes transmembrane regions and the S7 segment of the C-terminus. Changes in this regulatory pathway will likely alter signal processing in receptor and neuronal cells.
We show the interaction of Rab11b and the BK channel in cochlea
We reveal that BK and Rab11b colocalize in the cochlea
We demonstrate that BK expression increases with an increase in Rab11b
We demonstrate that BK expression decreases with Rab11b knockdown
We reveal that Rab11b interacts with the core region of the BK channel
Acknowledgments
This study was supported by NIH/NIDCD grant R01-DC004295 to B.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kathiresan T, Harvey M, Orchard S, Sakai Y, Sokolowski B. A protein interaction network for the large conductance Ca2+-activated K+ channel in the mouse cochlea. Mol Cell Proteomics. 2009;8:1972–1987. doi: 10.1074/mcp.M800495-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saxena SK, Kaur S. Regulation of epithelial ion channels by Rab GTPases. Biochem Biophys Res Comm. 2006;351:582–587. doi: 10.1016/j.bbrc.2006.10.087. [DOI] [PubMed] [Google Scholar]
- 3.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nature Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Graaf SF, Chang Q, Mensenkamp AR, Hoenderop JGJ, Bindels RJM. Direct interaction with Rab11a targets the epithelial Ca2+ channels TRPV5 and TRPV6 to the plasma membrane. Mol Cell Biol. 2006;26:303–312. doi: 10.1128/MCB.26.1.303-312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best JM, Foell JD, Buss CR, Delisle BP, Balijepalli RC, January CT, Kamp TJ. Small GTPase Rab11b regulates degradation of surface membrane L-type Cav1.2 channels. Am J Physiol Cell Physiol. 2011;300:C1023–C1033. doi: 10.1152/ajpcell.00288.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delisle BP, Underkofler HAS, Moungey BM, Slind JK, Kilby JA, Best JM, Foell JD, Balijepalli RC, Kamp TJ, January CT. Small GTPase determinants for the Golgi processing and plasmalemmal expression of human ether-a-go-go related (hERG) K+ channels. J Biol Chem. 2011;284:2844–2853. doi: 10.1074/jbc.M807289200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyott SJ, Meredith AL, Fodor AA, Vázquez AE, Yamoah EN, Aldrich RW. J Biol Chem. 2007;282:3312–3324. doi: 10.1074/jbc.M608726200. [DOI] [PubMed] [Google Scholar]
- 8.Sokolowski B, Orchard S, Harvey M, Sridhar S, Sakai Y. Conserved BK channel-protein interactions reveal signals relevant to cell death and survival. PLoS ONE. 2011;6(12):e28532. doi: 10.1371/journal.pone.0028532. Epub 2011 Dec 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakai Y, Harvey M, Sokolowski B. Identification and quantification of BK channel full-length variants in the developing mouse cochlea. J Neurosci Res. 2011;89:1747–1760. doi: 10.1002/jnr.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 11.Duzhyy D, Harvey M, Sokolowski B. A secretory-type protein, containing a pentraxin domain, interacts with an A-type K+ channel. J Biol Chem. 2005;280:15165–15172. doi: 10.1074/jbc.M500111200. [DOI] [PubMed] [Google Scholar]