Abstract
Previous studies have demonstrated that alcohol use disorders (AUDs) are regulated by multiple mechanisms such as neurotransmitters and enzymes. The neurotransmitter, serotonin (5-hydroxytryptamine, 5-HT) may contribute to alcohol effects and serotonin receptors, including 5-HT3, play an important role in AUDs. Recent studies have also implicated histone deacetylases (HDACs) and acetyltransferases (HATS) in regulation of drug addiction, and HDAC inhibitors (HDACi) have been reported as transcriptional modulators of monoaminergic neurotransmission. Therefore, we hypothesize that HDACs may play a role in ethanol induced serotonergic modulation. The effects of ethanol on serotonin and 5-HT3, and the role HDACs, HDAC activity and the HDACi, trichostatin A (TSA), play in alcohol- induced serotonergic effects were studied. Human SK-N-MC and neurons, were treated with ethanol (0.05, 0.1 and 0.2%), and/or TSA (50 nM), and 5-HT3 levels were assessed at 24-72 hrs. Gene expression was evaluated by qRT-PCR and protein by western blot and flow cytometry. Serotonin release was assessed by ELISA and HDAC activity by fluorometric assay. Our results show an increase in 5-HT3 gene after ethanol treatment. Further, ethanol significantly increased HDACs 1 and 3 genes accompanied by an increased in HDAC activity while TSA significantly inhibited HDACs. Studies with TSA show a significant up-regulation of ethanol effects on 5-HT3, while surprisingly TSA inhibited ethanol-induced serotonin production. These results suggest that ethanol affects 5-HT3 and serotonin through mechanisms involving HDACs and HATs. In summary, our studies demonstrate some of the novel properties of HDAC inhibitors and contribute to the understanding of the mechanisms involve in alcohol-serotonergic modulation in the CNS.
Keywords: Alcohol, histone deacetylases, serotonin, 5-HT, SK-N-MC, neurons
Introduction
Alcohol dependence (AD) is a complex addiction regulated by multiple mechanisms including neurotransmitters and enzymes. One of the neurotransmitters that have been linked to alcohol's effects on the CNS and immune systems and to alcohol abuse is serotonin. In our previous studies, we have reported that alcohol upregulates the serotonin transporter (SERT) and the enzyme monoamine oxidase-A (MAO-A) in human dendritic cells (Babu et al., 2009). Others, have reported genetic studies that implicate several serotonin (5-Hydroxytryptamine, 5-HT) genes with multiple drug and alcohol addictions (Crabbe et al., 2006; Lovinger, 1997; Tabakoff et al., 2009). For instance, some findings have shown that 5-HT3 receptor overexpression decreases ethanol self administration in transgenic mice (Engel et al., 1998); and other serotonin receptor and transporter studies have been shown to regulate ethanol self-administration in animal models (Rodd et al., 2010) and humans (Johnson et al., 2011). Clinical and preclinical findings have shown that antagonism of the 5-HT3 receptor reduces alcohol consumption and some of its subjective effects. (Engleman et al., 2008). Overall, from previous literature, it is clear that ethanol modulation of 5-HT3 receptor-mediated responses may have important consequences in the intoxicating and addictive properties of ethanol (Feinberg-Zadek and Davies, 2010). However, such conflicting results regarding the effects of alcohol on these receptors and the lack of knowledge on the mechanisms of action lead us to the search for novel research studies involving ethanol, HDACs, and serotonin modulation in CNS cells.
Some of the mechanisms of interest are acetylation and deacetylation. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are enzymes that add and remove acetyl groups to and from target lysine residues within histones, respectively (Min-Hao and Allis, 1998; Peterson and Laniel, 2004); http://www3.interscience.wiley.com/cgi-bin/fulltext/119399306/main.html,ftx_abs- b28 resulting in regulation of gene expression, where HATs tend to be transcriptional activators whereas HDACs tend to be repressors. The class I HDAC family members include HDACs 1, 2, 3 and 8. Recent reports suggest that class I HDACs may function together in multiprotein complexes that bind and deacetylate transcription factors (Hildmann et al., 2007). Overall, epigenetic mechanisms that alter the chromatin structure at specific gene promoters have been demonstrated to play a major role in drug dependence through their ability to regulate genes that induce drug reinforcement such as in the case of cocaine addiction (Wang et al., 2009) . In terms of alcohol addiction, ethanol has been shown to increase the acetylation of histone H3 in primary culture of rat hepatocytes (Park et al., 2003). Further studies have indicated that ethanol exposure induces global protein hyperacetylation which likely leads to major physiological consequences that contribute to alcohol-induced hepatotoxicity (Shepard and Tuma, 2009). Such histone modifications have been known to underlie the mechanisms involved in ethanol-induced cellular injury (Shukla and Aroor, 2006). Although chromatin modifications including alterations in histones and DNA, as well as epigenetic effects have been shown to be altered by alcohol (Mandrekar., 2011); future studies are still needed to understand how the deacetylation and acetylation activities are specifically coordinated leading to changes in gene expression.
HDAC inhibitors (HDACi) have recently been known to modulate genes involved in drug addiction such as the µ opioid receptor gene (Lin et al., 2008), and decrease cocaine self administration in rats (Romieu et al., 2008). However, very little is known about the involvement of HDACi in alcohol use disorders, particularly their ability to modulate serotonin and serotonin genes associated with alcohol addiction such as 5-HT3. Our previous reports have indicated the ability of alcohol to induce enzymes and transporters involved in serotonin neurotransmission such as MAO and SERT (Babu et al., 2009). Since alcohol is known to affect the serotonin neurotransmitters that convert chemical signals produced by serotonin into functional changes in the cells and drugs that act on these neurotransmitters may affect alcohol intake in humans and animals (Lovinger, 1997); in the present study, we will further analyze the effects of ethanol treatment on 5-HT3 and serotonin release, and for the first time we will study the modulatory effect of the HDACi, TSA, on the expression of 5-HT3 and serotonin release in the human neuroblastoma cell line, SK-N-MC and fetal brain derived primary neurons. Based on previous findings, we hypothesize that alcohol modulates serotonin release and serotonin genes such as 5-HT3 while TSA will counteract the effects of ethanol on serotonin release and serotonin gene (5-HT3) expression. Our research studies will analyze for the first time the epigenetic mechanisms of ethanol-induced modulation of serotonin, specifically the ability of TSA to modulate ethanol-effects on serotonin release and 5-HT3 gene expression, leading to new insights on the promising role of HDACi on alcohol related effects.
Materials and methods
Culture of SK-N-MC and primary human fetal brain-derived neurons
The neuroblastoma cell line, SK-N-MC was purchased from ATCC (catalog # HTB-10, Manassas, VA) and cultured in eagle's minimum essential medium (MEM) (catalog # 30-2003) supplemented with fetal bovine serum to a final concentration of 10% (catalog # 30-2020) and 1% antibiotic/antimycotic solution (Sigma-Aldrich, St. Louis, MO). For all the experiments, SKN-MC were cultured at a concentration of 0.5 × 106 cells/ml in 6 well plates overnight to allow them to reach at least 60% confluency before any further treatment. Primary human fetal brain-derived neurons were purchased (catalog # 1520-5), supplemented with neuronal media, and cultured as per manufacture's recommendations (ScienCell, Carlsbad, CA). The major differences between both cell types are that the primary neurons are mixed fetal-brain derived cells, while the SK-N-MC cells are derived from a supra-orbital brain tumor.
Treatments
SK-N-MC and primary human neurons were treated with 0.05% (~10 mM), 0.1% (~20 mM), or 0.2% (~40 mM) ethanol. For the TSA experiments, after reaching confluency, the cells were pre-treated for two hours with 50 nM TSA, then treated with 20 mM ethanol for 24 to 72 hrs. This concentration of TSA has been previously tested to be effective and shown to inhibit HDAC 2 (Agudelo et al., 2011). For the ondansetron studies, the cells were pre-incubated with ondansetron (1-100 uM) for two hours and then treated with TSA (50 nM). Ethanol (catalog # E7023), TSA (catalog # T1952), and ondansetron (catalog # O3639) were obtained from Sigma-Aldrich. The medium was changed and reagents were replenished every 24 hrs. At these concentrations, ethanol, TSA, and ondansetron did not affect the viability of the cells as tested by MTT assay or trypan blue staining (data not shown).
RNA extraction and quantitative real-time PCR (qRT-PCR)
SK-N-MC and primary neurons were harvested at different time points (0- 72 hrs), and the RNA was extracted from the cell pellets using the RNAeasy mini kit from Qiagen (Valencia, CA). The RNA was eluted and stored at -80 °C. Equal quantities of RNA (1ug) from all the samples were reversed transcribed using the high-capacity cDNA reverse transcription kit from Applied Biosystems (Foster City, CA) to perform qRT-PCR using Taqman gene expression assays (Applied Biosystems) for the expression of 5-HT3A (Assay ID Hs00356082_m1), HDAC 1 (Assay ID Hs02621185_s1), HDAC 2 (Assay ID Hs00231032_m1), HDAC 3 (Assay ID Hs00187320_m1), and HDAC 8 (Assay ID Hs00218503_m1) genes. GAPDH (Hs99999905_m1) and 18S rRNA (catalog # 4333760F) were used as internal controls. Relative abundance of each mRNA species was assessed as previously described by us (Agudelo et al., 2011). Relative mRNA species expression was quantitated and expressed as transcript accumulation index (TAI=2-ΔΔCT), calculated using the comparative CT method. All data were controlled for quantity of RNA input by performing measurements on two endogenous reference genes GAPDH and 18S rRNA. The 18S rRNA control was used to rule out any modulatory effects caused by ethanol and TSA treatments on GAPDH. Results obtained from calculations normalized to 18S rRNA were similar to results from calculations normalized to GAPDH. In addition, results on RNA from treated samples were normalized to results obtained on RNA from control, untreated samples. Although the 5-HT3 receptor is comprised of five subunits (Boess et al., 1995), 5HT3A is the subunit of interest due to its association with alcohol drinking (Hodge et al., 2004).
HDAC activity assay
For the HDAC activity assay, SK-N-MC cells were harvested at different time points (24-72 hrs) and lysed followed by nuclear and cytoplasmic extraction using the nuclear extraction kit from Active Motif (Carlsbad, CA). After obtaining the specific lysates, the HDAC enzyme activity was measured with the fluorometric HDAC assay kit following the manufacture's protocol (Active Motif) and as previously described in other HDAC activity assays using nuclear and cytoplasmic lysates (Watson and Riccio, 2009). The fluorescent HDAC assay kit utilizes a short peptide substrate that contains an acetylated lysine residue that can be deacetylated by class I, II and IV HDAC Enzymes. HDAC enzyme activity in pmoles/min/mg were calculated based on standard curve. Then, the amounts of HDAC activity in pmoles/min/mg for each treated group were normalized to the respective control groups from each time point. The fluorescence was measured in a BioTek microplate reader with excitation wavelength at 360 nm and emission wavelength at 460 nm. Data are representative of two individual experiments ran in duplicates.
Intracellular 5-HT3 receptor analysis by flow cytometry
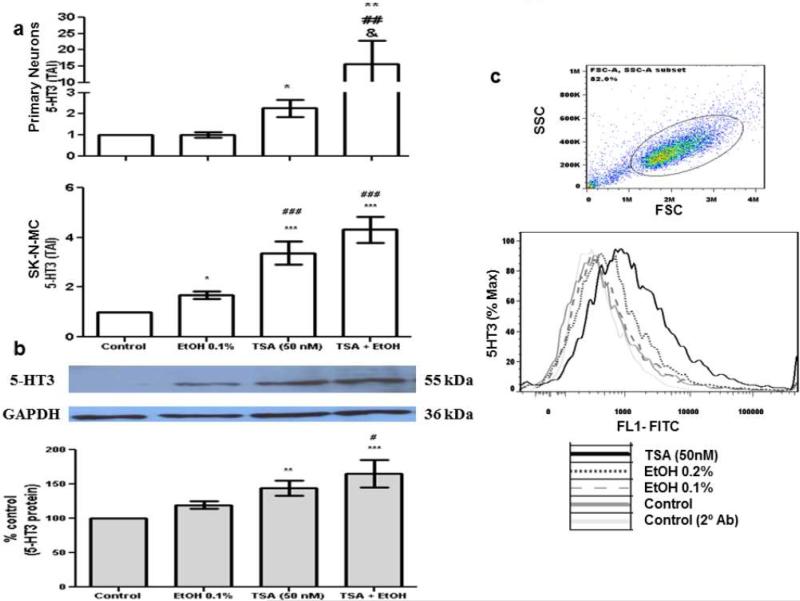
To assess the levels of 5-HT3 protein in SK-N-MC, the cells were treated with 10-40 mM ethanol, or 50 nM TSA. At 72 hrs, SK-N-MC (1 × 106 ) cells were harvested and aliquoted in 12 × 75 mm polystyrene falcon tubes, catalog # 352058 (BD Biosciences, San Jose, CA), blocked with human serum and normal goat serum (Chemicon International, Temecula, CA), fixed, and permeabilized with cytofix/cytoperm solution (BD Bioscience). The 5-HT3 protein was detected with an unlabeled primary polyclonal antibody, rabbit anti-serotonin receptor 3A (Millipore, Bedford, MA) and secondary antibody, goat anti-rabbit IgG fluorescein (FITC)-conjugated (Millipore). Cells were incubated for 20 min. on ice with both antibodies, washed, and resuspended in PBS containing 2% fetal calf serum. Cells were acquired on an Accuri C6 flow cytometer (BD Accuri; Ann Arbor, MI) and analyzed with FlowJo software (Tree Star, Inc.; Ashland, OR). A total of 10,000 events were collected for each sample. Cells were gated based on unlabeled and secondary antibody controls. Cells positive for specific protein are shown in the histograms with shifted mean fluorescence intensity compared to controls. (figure 4c).
Fig. 4.
TSA significantly enhances ethanol effects on 5-HT3. After reaching 60% confluency, SK-N-MC and human primary neurons were pre-incubated with TSA for 2 hours, then treated with EtOH (0.1%) for 48 and 72 hrs. Gene expression (48 hrs) was performed in both primary neurons and SK-N-MC (figure 4a) as described above. Data are expressed as mean ± SEM of 5-HT3 gene expression of six independent experiments. Protein levels (72 hrs) were measured in SK-N-MC by western blot and flow cytometry. For the western blot experiments (figure 4b), 20 μg of protein from SK-N-MC whole cell lysates were stained with primary anti-5-HT3 and secondary anti-IgG-HRP antibodies. GAPDH was used as a loading control. Data presented show a representative blot. Protein quantification is expressed as % control ± SEM of 5-HT3 protein of six independent experiments. * represents significance compared to control (*** p<0.0001, ** p<0.005, * p<0.05). # represents significance compared to ethanol treatment (### p<0.0001, ## p<0.005, #p<0.05). & represents significance compared to TSA (& p<0.05). Comparisons between groups were performed using one-way ANOVA and Dunnett's Multiple Comparison Test. For the flow cytometry experiments, 1 × 106 SK-N-MC cells were fixed and permeabilized with cytofix/cytoperm solution prior to intracellular staining with primary anti-5HT3 and secondary anti-IgG-FITC antibody. Figure 4c show a representative histogram overlay of the gated cells stained with anti-5HT3. Gate is shown on the scatter graph. The light gray and gray solid line histograms represent the secondary antibody and the untreated controls respectively; the gray dashed line histogram represents the ethanol (0.1%) treated cells, the gray dotted line histogram represent the ethanol 0.2 % and the black solid line histogram represents the TSA (50 nM) treated cells. Data are representative of three independent experiments.
Western blot analysis
After 72 hours of alcohol treatment, the SK-N-MC cells were harvested, cell lysates were prepared in protein extraction reagent (Pierce Biotechnology, Rockford, IL) containing protease inhibitor (Pierce Biotechnology), shaken gently for 10 minutes, and centrifuged at 14,000 g for 10 minutes to get rid of cell debris. Protein levels were quantified using Bradford protein assay reagent (Bio-Rad laboratories, Hercules, CA). Equal quantities of protein (20 ug) were subjected to SDS-PAGE and transferred into a nitrocellulose membrane (Bio-Rad laboratories), blocked with 10% nonfat dry milk, washed with tris-buffered saline-tween 20 (TBST), and incubated overnight with primary antibody, rabbit anti-serotonin receptor 3A (Millipore) or rabbit anti-GAPDH as loading control (sigma). After overnight incubation, the membranes were washed and incubated for 1 hr. with secondary antibody, goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated (Millipore). The blots were developed using the super signal west pico chemiluminescent substrate (Pierce Biotechnology).
Serotonin detection
SK-N-MC cells were harvested and the cell culture supernatants were collected at 72 hrs. Serotonin levels in the supernatants were detected as per manufacture's recommendations with a competitive ELISA kit obtained from GenWay Biotech (San Diego, CA). Amounts of serotonin (nmol/L) were calculated based on the standard curve created with known standards provided by the manufacture. The optical density was measured in a BioTek microplate reader (Winooski, VT) at 405 nm within 60 minutes after adding the stop solution. Data are representative of three individual experiments performed in duplicates.
Statistics
Experiments were performed at least three times in duplicates unless otherwise indicated in the figure legend. The values obtained were averaged and data are represented as the mean ±SE. All the data were analyzed using GraphPad Prism software. Comparisons between groups were performed using one-way ANOVA and Dunnett's Multiple Comparison post test. Differences were considered significant at p ≤ 0.05.
Results
Ethanol modulates 5-HT3 gene
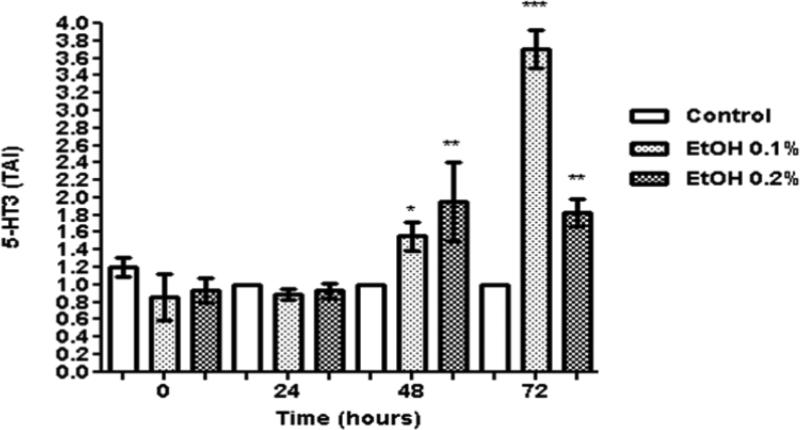
Data presented on figure 1 show the dose response and kinetic effects of 0.1-0.2% (~20 mM - 40 mM) ethanol on 5-HT3 gene expression in SK-N-MC cells. There is no significant effect on 5-HT3 gene expression at 24 hrs, but the expression is significantly up-regulated by 48 and 72 hrs, specially the 0.1 % ethanol treatment dose. Based on the dose-response and kinetic results, 0.1% and the 48 hr. time point were selected to perform subsequent gene expression studies. 0.1% is also a physiological significant concentration because it is close to the legal blood alcohol concentration of 0.08%. The ethanol concentrations used are non-toxic and do not affect cell viability as demonstrated by MTT cell viability assay and trypan blue cell count (data not shown).
Fig. 1.
Ethanol modulates 5-HT3 gene expression. SK-N-MC (0.5 × 106 cells/ml) were incubated in 6-well plates overnight. After reaching 60% confluency, the cells were treated with ethanol (0.1 and 0.2%) for 24-72 hours. The cells were harvested, RNA was extracted and reverse transcribed followed by qRT- PCR for 5-HT3 and endogenous GAPDH and 18S rRNA gene expression. Data are expressed as mean ± SE of TAI values of three independent experiments. Comparisons between groups were performed using one-way ANOVA and Dunnett's Multiple Comparison Test. Comparison between groups, “Control vs. ethanol 0.1%” and “Control vs. ethanol 0.2%” revealed significant differences among the groups at 48 and 72 hrs, *p<0.05; ** p<0.005; ***p<0.0001.
Ethanol induces HDACs 1 and 3 genes
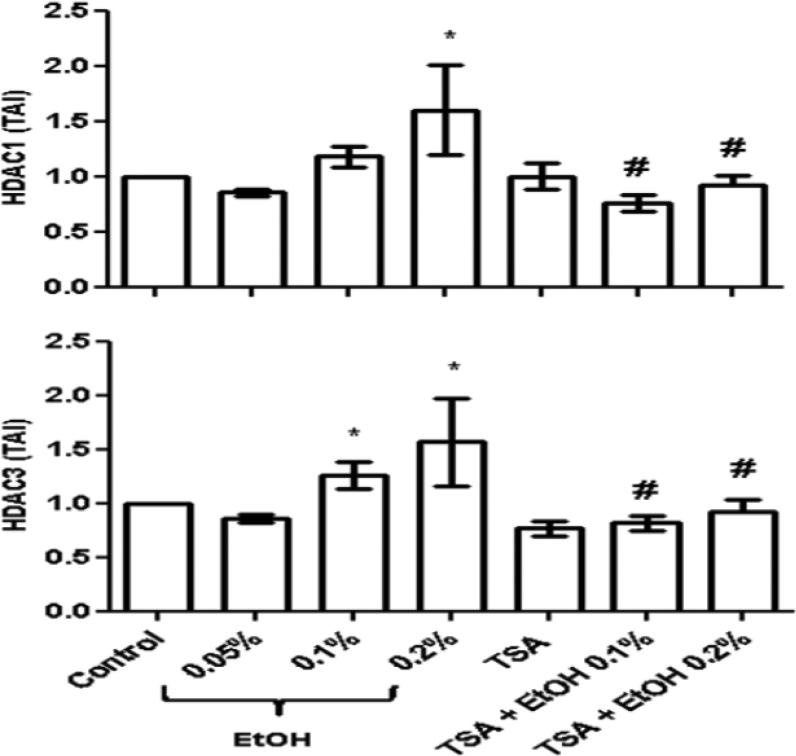
Recent studies have demonstrated that HDACs play a major role in drug dependence through their ability to regulate other genes. Our lab has demonstrated that ethanol significantly upregulates HDAC2 and reactive oxygen species (ROS) production while the HDACi, TSA, has neuroprotective effects and significantly reduces ROS (Agudelo et al., 2011). Further, HDACi have recently been known to modulate genes involved in drug addiction such as the μ opioid receptor gene (Lin et al., 2008). In particular, TSA is a widely use HDACi known to inhibit most of the class I HDACs; therefore, in this set of experiments, we tested the ability of TSA to inhibit genes from the class I HDAC family of enzymes, and subsequently measured the effects of the inhibition of HDACs 1, 2, 3 and 8 on the expression of 5-HT3.
Data on figure 2 show a significant upregulation of HDACs 1 and 3 genes in SK-N-MC cells treated with ethanol (~20 mM). Further, treatment with TSA alone or TSA (50 nM) prior to ethanol (~20 mM) resulted in a significant inhibition of HDACs 1 and 3 as expected. HDAC2 upregulation after ethanol treatment and inhibition after TSA treatment was reported previously by us (Agudelo et al., 2011). HDAC8 was tested in this study; however, there was no significant upregulation of HDAC8 after ethanol treatment and TSA did not inhibited HDAC8 (data not shown). TSA is a nonspecific inhibitor and the mechanisms of action in terms of specificity of inhibition of the class I HDACs 1, 2, 3, and 8 are not fully understood.
Fig. 2.
Ethanol induces HDACs 1 and 3 genes. After reaching 60% confluency, SK-N-MC were pre-treated with TSA (50 nM) for 2 hours, then treated with ethanol (0.1%) for 48 hours. After incubation, cells were harvested, RNA was extracted and reverse transcribed followed by qRTPCR for HDACs 1 and 3 and housekeeping GAPDH and 18S rRNA gene expression. Ethanol significantly enhanced HDACs 1 and 3, while TSA + ethanol treatments significantly inhibited HDACs 1 and 3 genes. Data are expressed as mean ± SE of TAI values of four independent experiments. Comparisons between groups were performed using one-way ANOVA and Dunnett's Multiple Comparison Test. Comparison between groups revealed significant differences, “Control vs. ethanol ” (*p<0.05) and “ethanol vs. TSA + ethanol” (#p<0.05).
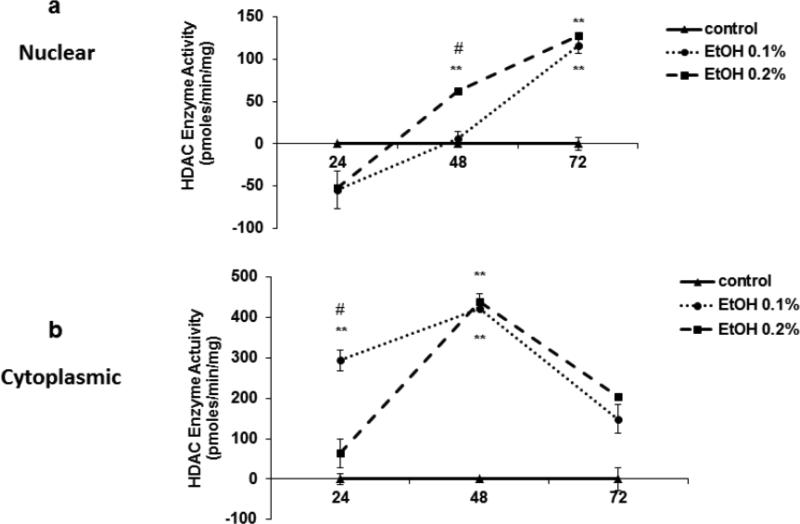
Ethanol induces HDAC activity
From our results above, we know that alcohol induces HDACs 1 and 3 genes and that TSA is significantly inhibiting HDACs 1 and 3 by 48 hrs. Therefore, in this set of experiments, we measured HDAC activity in nuclear and cytoplasmic extracts from SK-N-MC at different time points, 24-72 hrs, (Figure 3). Overall, the nuclear activity levels are lower than the cytoplasmic levels. Interestingly, the nuclear HDAC activity of the ethanol treated samples is lower than the control at 24 hrs, but increases significantly over time (48 and 72 hrs). While at early time points (24 and 48 hrs), the HDAC cytoplasmic activity is significantly higher compared to the controls, it peaks by 48 hrs and goes down by 72 hrs. Overall, the nuclear and cytoplasmic HDAC activities seem to be inversely proportional at early time points (24 and 48 hrs), but normalized to similar levels by 72 hrs.
Fig. 3.
Ethanol induces HDAC activity. SK-N-MC were treated with ethanol (0.1%). The cells were harvested at 24, 48 and 72 hrs, lysed followed by nuclear and cytoplasmic extraction. HDAC enzyme activity was measured in both nuclear and cytoplasmic lysates with a fluorometric HDAC assay kit. The fluorescence was measured in a BioTek microplate reader with excitation wavelength at 360 nm and emission wavelength at 460 nm. Data are expressed as mean ± SE of HDAC enzyme activity (pmoles/min/mg) of EtOH treated cells normalized to controls from two independent experiments ran in duplicates. Comparisons between groups were performed using one-way ANOVA and Dunnett's Multiple Comparison Test. * represents significance compare to control (* p<0.05; **p<0.005). # represents significance between ethanol treatments (# p<0.05).
TSA significantly enhances ethanol effects on 5-HT3
After confirming an increase in HDAC gene expression and activity after ethanol treatment, and the inhibition of HDACs 1 and 3 genes with TSA, we proceeded to measure the expression of the 5-HT3 gene, and surprisingly there was a significant increase in 5-HT3 expression in TSA and TSA + ethanol treated cells (figure 4a). Since alcohol and TSA are known to have neuromodulatory properties, we wanted to compare their effect on cells with neurogenic origin.
Therefore, primary fetal brain-derived neurons and the cell line SK-N-MC were used in our studies. Alcohol and TSA are having similar effects on 5-HT3 gene in both cell models with TSA having a more robust and significant effect compared to ethanol, suggesting a role of TSA in neuromodulation of both types of neurons. For all subsequent studies, SK-N-MC cells were used.
Besides gene expression studies, we also determined protein expression by western blot and flow cytometry. SK-N-MC were treated with ethanol (20 mM), TSA (50 nM), or pretreated for 2 hrs with TSA and treated with ethanol for 72 hrs. Then, the cell lysates or the cell pellets were analyzed by immunoblotting and immunofluorescence, respectively. Data in figure 4b show an increase in protein levels of 5HT3 after treatment with ethanol compared to the control, and this effect is significantly enhanced after TSA treatment as shown by western blot and flow cytometry (figures 4b and 4c). These results are consistent with the gene expression results presented in figure 4a.
The effects of TSA on 5-HT3 are blocked by the 5-HT3 antagonist, ondansetron
Although ondansetron works on the ion channel protein (5-HT3), the interaction of ondansetron and TSA may be resulting in activation of other receptors, which can have effects on the regulation of gene expression such as in the case of serotonin activation of G-protein coupled receptors (GPCRs). It is well established that GPCRs interact with G-proteins to activate many intracellular signaling pathways and modulate ion channel function (Bertil, 1994). Serotonin activation of GPCRs will result in the induction of second-messengers, which can affect ion channels or act on the nucleus to alter gene expression (Lovinger, 1997). In our studies, when the cells were treated with the 5-HT3 antagonist, ondansetron, the increase in 5-HT3 gene caused by TSA was significantly inhibited (figure 5a). Therefore, 5-HT3 is somehow being affected by the interaction of TSA and ondansetron, or possibly by the signaling cascades triggered by the activation of other receptors. To validate the observed results regarding gene expression, we performed western blot, and the 5-HT3 antagonist, ondansetron, is also blocking TSA effects on 5-HT3 protein (figure 5b).
Fig. 5.
The effects of TSA on 5-HT3 are blocked by the antagonist, ondansetron. The cells were pre-incubated with the 5-HT3 antagonist, ondansetron (10 uM), for 2 hrs, and treated with TSA (50nM) for 48-72 hrs, or treated with TSA or ondansetron alone. Comparisons between groups were performed using one-way ANOVA and Dunnett's Multiple Comparison Test. TSA significantly induced 5-HT3 levels compared to the untreated control; while treatment with ondansetron or the combination of ondansetron and TSA significantly inhibited the TSA-induction of 5-HT3. * represents significance compared to control (* p<0.05). # represents significance compared to TSA treatment (# p<0.05). Data (5a) are representative of four independent experiments, western blot (5b) is representative of two independent experiments performed in duplicates.
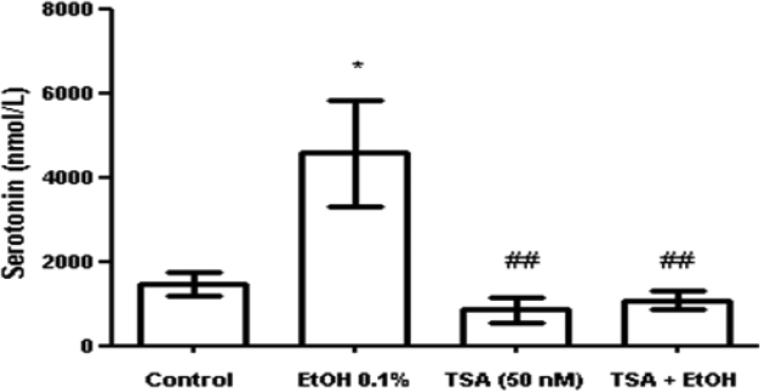
Ethanol induces serotonin secretion while TSA inhibits ethanol effects
There is evidence that alcoholics and experimental animals that consume large quantities of alcohol have differences in brain serotonin levels compared to non-alcoholics (Lovinger, 1997). Overall, it is known that serotonin and other neurotransmitters may contribute to alcohol's intoxicating and rewarding effects (Lovinger, 1997). Since serotonin's actions have been associated with alcohol's effects on the brain and alcohol abuse, we tested the effects of alcohol on serotonin release in SK-N-MC and the ability of TSA to modulate the action of ethanol on serotonin release. As expected ethanol increased serotonin release; however, when we tested the effects of TSA on serotonin, surprisingly, there was a significant decreased in serotonin levels in both TSA treated cells and TSA plus alcohol treated cells (figure 6). TSA is somehow blocking serotonin release and inhibiting the action of alcohol. Amazingly, the effects of TSA on 5-HT3 have no action upon serotonin levels, suggesting that serotonin regulation may involve other serotonin receptors or complex mechanisms that are worth of future exploration.
Fig. 6.
Ethanol induces serotonin secretion while TSA inhibits ethanol effects. SK-N-MC culture supernatants were collected at 72 hrs. Serotonin levels in the cell culture supernatants were detected by ELISA. The optical density was measured in a BioTek microplate reader at 405 nm. Ethanol enhanced serotonin levels compared to the untreated control while TSA significantly reduced serotonin levels compared to the ethanol treated supernatants. Comparisons between groups were performed using one-way ANOVA and Dunnett's Multiple Comparison Test. * represents significance compared to control (* p<0.05) and # represents significance compared to ethanol treatment (## p<0.007). Data are representative of three independent experiments.
Discussion
Alcohol acts through numerous pathways to affect the brain leading to the development of neurodegeneration. Many mechanisms, including neurotransmitters and enzymes, act together to reflect the actions of alcohol; therefore, in the current study we explore the novel role of the HDACi, TSA in alcohol's serotonergic mechanisms. This study is aimed to clarify the link between TSA and serotonin and their role in alcohol effects on serotonin release and 5-HT3 in neuronal cells. Alcohol is known to increase serotonin release in the CNS and modulation of neurotransmitters such as serotonin and serotonergic receptors has a great influence on addictive behaviors. In particular, 5-HT3 is known to regulate alcohol intake and contribute to alcohol's effects (Lovinger, 1997). Activation of certain serotonin receptors has been linked to neuronal survival and neuroprotection in other neurodegenerative disorders (Cho and Hu, 2007), and drugs that affect serotonergic signal transmission have been shown to alter alcohol consumption in animals (LeMarquand et al., 1994).
Recent studies have indicated that chronic ethanol exposure induces global protein hyperacetylation (Shepard and Tuma, 2009). These studies were done in alcohol fed animals and most of the hyperacetylated proteins were mitochondrial proteins that play a role in liver injury. Although we did not measure histone acetylation or hyperacetylation, it is well known that HDAC inhibition has been associated with histone acetylation and hyperacetylation of several proteins and gene promoters (Xiong et al., 2012). In our in vitro studies with SK-N-MC cell cultures, we observed a significant increase in HDAC expression and activity after ethanol treatment (figures 2 and 3); and we were able to block HDAC 1 and 3 expression with TSA (figure 2). HDACs are associated with a number of well characterized cellular oncogenes and tumor-suppressor genes that lead to an aberrant recruitment of HDAC activity, which in turn results in changes in gene expression (Cress and Seto, 2000). Interestingly, recent reports have demonstrated that inhibition of histone deacetylases with TSA results in transcriptional modulation of monoaminergic neurotransmission genes in neuroblastoma cells (Bence et al., 2011). Their studies showed that TSA significantly upregulated the expression of dopamine and serotonin transporters while transcript levels of monoamino oxidase A and catechol-omethyltransferase were significantly reduced. However, they did not measure the effects of TSA on serotonin production and 5-HT3 expression. Overall, to date no studies have been reported to elucidate the role of HDACi on alcohol-induced serotonergic effects. Therefore, in the current study, we are the first ones to elucidate the role TSA plays on alcohol modulatory effects on serotonin production and 5-HT3 expression.
It is evident from our kinetic studies that ethanol treatment is enhancing 5-HT3 overtime (figure 1). Further, our study is the first one to show a significant increase in 5-HT3 (figure 4) after inhibition of HDACs 1 and 3 with TSA (figure 2). We are also the first ones to report that the significant upregulation of 5-HT3 by TSA is being blocked by the 5-HT3 antagonist, ondansetron (figure 5). According to previous reports, TSA is a non-specific HDACi known to inhibit class I family of HDACs (1, 2, 3, and 8). Although in the current study we did not test other HDAC inhibitory effects of TSA related to HDAC activity or HDAC protein levels; TSA is known to inhibit HDAC2 gene and protein as previously published by us (Agudelo et al., 2011), and HDAC 1 and 3 genes as shown in figure 2. HDAC 8 was also tested; however, alcohol had no effect on this HDAC and TSA did not inhibit it (data not shown); which may be due to the specificity of TSA.
It is well established that when TSA is used to inhibit HDACs, HATs get activated and may initiate a cascade of posttranslational modifications resulting in hyperacetylation as shown in other systems (Ekwall et al., 1997; Park et al., 2002), and which may be accountable for the effects observed in our studies regarding the induction of 5-HT3 (figure 4) However, more studies will be necessary to elucidate the exact mechanisms of action of TSA and how HDACs and HATs are regulating serotonergic signals. There are reports supporting that TSA can stimulate gene expression via mechanisms involving transcription factors. In particular, the effects of HDACi on the promoter activities of various genes have been actively examined, and these inhibitors have been suggested to modulate gene transcription through transcription factors such as Sp1 and Sp3 (Her et al., 2010).
There is plenty of evidence suggesting a functional link between HATs and HDACs in regulating the balance of histone acetylation (Peserico and Simone, 2011). HAT-HDAC interplay has been shown to modulate global histone acetylation in gene-coding regions during stress (Johnsson et al., 2009). Further, HDACi are known to cause general and local histone hyperacetylation in yeast and mammalian cells (Ekwall et al., 1997; Park et al., 2002) For instance, TSA induces promoter activity and acetylation of transcription factors (Huang et al., 2005). Other reports have shown that HDACi such as valproic acid and TSA might restore HAT activity by inhibiting HDAC activity and by repressing HAT targeting proteins; implying an indirect induction of HAT activation (Fortson et al., 2011).
Moreover, our data (figure 3) indicate that HDAC activity is significantly changing through the gene activation process and alcohol-induced modulatory mechanisms. For instance, at early time points (24 hrs) during ethanol (0.1 and 0.2%) treatments, the nuclear HDAC activity (3a) levels were significantly lower compared to the respective controls increasing over time by 48-72 hrs. While the cytoplasmic activity (3b) shows a different pattern of activation. First, the HDAC cytoplasmic activity is overall higher than the nuclear activity. Further, the cytoplasmic activity in the ethanol treated cells significantly peaks by 48 hrs and then drops by 72 hrs to similar activity levels detected in the nuclear lysates. Overall, both nuclear and cytoplasmic HDAC activities seem to be inversely proportional during early time points (24-48 hrs) of ethanol treatments, which could be explained by the co-localization and transport of some of these HDACs. HDACs 1, 2, and 8 are found primarily in the nucleus, whereas HDAC3 is found in both the nucleus and the cytoplasm (de Ruijter et al., 2003). Further findings have reported that some HDACs are able to move in and out of the nucleus, depending on different signals http://en.wikipedia.org/wiki/Histone_deacetylase-cite_note-pmid12429021-5 (de Ruijter et al., 2003; Longworth and Laimins, 2006); which can also lead to changes in HDAC activity.
When serotonin levels were measured, as expected, there was an upregulation of serotonin after ethanol treatment; and surprisingly, TSA significantly inhibited serotonin release. Overall, ethanol effects on 5-HT3 expression were mild compared to the effects observed after TSA and TSA + ethanol treatments, which significantly enhanced 5-HT3 gene and protein expression. These effects of ethanol on 5-HT3 modulation could be explained by two major findings. First, ethanol significantly induces HDACs (figure 2) and HDAC activity (figure 3), which can result in transcriptional repression; therefore, not allowing a more robust increase in 5-HT3 gene transcription. Or second, the increase in serotonin levels caused by ethanol (figure 6) might result in binding of free serotonin to the 5-HT3 receptors, blocking further 5-HT3 production as a regulatory mechanism. Another major point is that TSA is inhibiting HDACs (figure 2), and when HDACs are low, histone acetyltransferases (HATs) get activated causing an induction of histone hyperacetylation leading to transcriptional regulation (Ekwall et al., 1997; Johnsson et al., 2009; Park et al., 2002), which may explain the significant enhancement of 5-HT3 expression observed after TSA treatment (figure 4). In turn, an increase in 5-HT3 expression and function will probably cause excessive stimulation of neurons. As a result of this stimulation, a decrease in serotonin levels is triggered as observed in our studies with TSA treatment in SK-N-MC (figure 6).
Another possibility is that TSA may be activating serotonergic functioning by increasing 5-HT3 receptors and blocking serotonin release. After all, our findings are indicating that the TSA-induced increase in 5-HT3 has no effect upon serotonin levels, which may be explained by the involvement of other serotonin receptors, such as 5-HT1 or the serotonin reuptake transporter (SERT) that may also play a major role in alcohol-induced serotonergic modulation (Babu et al., 2009; Thompson et al., 2011). In addition, it is important to acknowledge that the release of other neurotransmitters that play key roles in alcohol intoxication may be increased since there is plenty of evidence to suggest that serotonin does not act alone and some of the serotonin-mediated responses to alcohol may be due to interactions between serotonin and other neurotransmitters (Lovinger, 1997) that may also be affected by alcohol and TSA; and which in turn can have effects upon serotonin levels. Two key neurotransmitters that have been extensively reviewed (Lovinger, 1997) and reported to interact with the serotonergic system are gamma-aminobutyric acid (GABA) (Yoo et al., 2010) and dopamine (Gonzalez-Burgos et al., 2008).
In summary, the results presented in this study show for the first time a promising role of the histone deacetylase inhibitor, TSA, in alcohol's serotonergic modulation; and demonstrate some of the novel properties of histone deacetylation, which may prove useful for future studies to develop treatments for alcohol use disorders or for other serotonin related disorders.
Acknowledgment
This research was supported by the Institute of NeuroImmune Pharmacology at Florida International University and in part by the National Institute of Health, grants R01MH085259 and R01DA021537.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest statement
All authors declare that there are no conflicts of interest.
References
- Agudelo M, Gandhi N, Saiyed Z, Pichili V, Thangavel S, Khatavkar P, Yndart-Arias A, Nair M. Effects of Alcohol on Histone Deacetylase 2 (HDAC2) and the Neuroprotective Role of Trichostatin A (TSA). Alcohol. Clin. Exp. Res. 2011;35:1550–1556. doi: 10.1111/j.1530-0277.2011.01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu DK, Diaz A, Samikkannu T, Rao KVK, Saiyed ZM, Rodriguez JW, Nair MPN. Upregulation of Serotonin Transporter by Alcohol in Human Dendritic Cells: Possible Implication in Neuroimmune Deregulation. Alcoholism: Clinical and Experimental Research. 2009;33:1731–1738. doi: 10.1111/j.1530-0277.2009.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence M, Koller J, Sasvari-Szekely M, Keszler G. Transcriptional modulation of monoaminergic neurotransmission genes by the histone deacetylase inhibitor trichostatin A in neuroblastoma cells. J. Neural Trans. 2011:1–8. doi: 10.1007/s00702-011-0688-4. [DOI] [PubMed] [Google Scholar]
- Bertil H. Modulation of ion-channel function by G-protein-coupled receptors. Trends Neurosci. 1994;17:531–536. doi: 10.1016/0166-2236(94)90157-0. [DOI] [PubMed] [Google Scholar]
- Boess F, Beroukhim R, Martin I. Ultrastructure of the 5-Hydroxytryptamine 3 receptor. J. Neurochem. 1995;64:1401–1405. doi: 10.1046/j.1471-4159.1995.64031401.x. [DOI] [PubMed] [Google Scholar]
- Cho S, Hu Y. Activation of 5-HT4 receptors inhibits secretion of [beta]-amyloid peptides and increases neuronal survival. Exp.l Neurol. 2007;203:274–278. doi: 10.1016/j.expneurol.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. REVIEW: Alcohol-related genes: contributions from studies with genetically engineered mice. Addiction Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient Inhibition of Histone Deacetylation Alters the Structural and Functional Imprint at Fission Yeast Centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Engel S, Lyons C, Allan A. 5-HT3 receptor over-expression decreases ethanol self administration in transgenic mice. Psychopharmacology. 1998;140:243–248. doi: 10.1007/s002130050763. [DOI] [PubMed] [Google Scholar]
- Engleman E, Rodd Z, Bell R, Murphy J. The role of 5-HT3 receptors in drug abuse and as a target for pharmacotherapy. CNS Neurol. Disord. Drug Targets. 2008;7:454–467. doi: 10.2174/187152708786927886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg-Zadek PL, Davies PA. Ethanol Stabilizes the Open State of Single 5-Hydroxytryptamine3A(QDA) Receptors. J. Pharmacol. Exp. Ther. 2010;333:896–902. doi: 10.1124/jpet.109.164863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortson WS, Kayarthodi S, Fujimura Y, Xu H, Matthews R, Grizzle WE, Rao VN, Bhat GK, Reddy ESP. Histone deacetylase inhibitors, valproic acid and trichostatin-A induce apoptosis and affect acetylation status of p53 in ERG-positive prostate cancer cells. International J. Oncology. 2011;39:1:111–119. doi: 10.3892/ijo.2011.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos I, Feria-Velasco A, Giuseppe Di Giovann V.D.M.a.E.E. Serotonin/dopamine interaction in memory formation. Prog. in Brain Res. 2008:603–623. doi: 10.1016/S0079-6123(08)00928-X. [DOI] [PubMed] [Google Scholar]
- Her S, Lee M-S, Morita K. Trichostatin A Stimulates Steroid 5α-Reductase Gene Expression in Rat C6 Glioma Cells via a Mechanism Involving Sp1 and Sp3 Transcription Factors. J. Mol. Neurosci. 2010;41:252–262. doi: 10.1007/s12031-009-9284-6. [DOI] [PubMed] [Google Scholar]
- Hildmann C, Riester D, Schwienhorst A. Histone deacetylases—an important class of cellular regulators with a variety of functions. Appl. Microbiol. Biotechnol. 2007;75:487–497. doi: 10.1007/s00253-007-0911-2. [DOI] [PubMed] [Google Scholar]
- Hodge CW, Kelley SP, Bratt AM, Iller K, Schroeder JP, Besheer J. 5-HT3A Receptor Subunit is Required for 5-HT3 Antagonist-Induced Reductions in Alcohol Drinking. Neuropsychopharmacology. 2004;29:1807–1813. doi: 10.1038/sj.npp.1300498. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhao S, Ammanamanchi S, Brattain M, Venkatasubbarao K, Freeman JW. Trichostatin A Induces Transforming Growth Factor Î2 Type II Receptor Promoter Activity and Acetylation of Sp1 by Recruitment of PCAF/p300 to a Sp1·NF-Y Complex. J. Biol. Chem. 2005;280:10047–10054. doi: 10.1074/jbc.M408680200. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Ait-Daoud N, Seneviratne C, Roache JD, Javors MA, Wang X-Q, Liu L, Penberthy JK, DiClemente CC, Li MD. Pharmacogenetic Approach at the Serotonin Transporter Gene as a Method of Reducing the Severity of Alcohol Drinking. Am. J. Psychiatry. 2011;168:265–275. doi: 10.1176/appi.ajp.2010.10050755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson A, Durand-Dubief M, Xue-Franzen Y, Ronnerblad M, Ekwall K, Wright A. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 2009;10:1009–1014. doi: 10.1038/embor.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMarquand D, Pihl RO, Benkelfat C. Serotonin and alcohol intake, abuse, and dependence: Findings of animal studies. Biol. Psychiatry. 1994;36:395–421. doi: 10.1016/0006-3223(94)91215-7. [DOI] [PubMed] [Google Scholar]
- Lin Y-C, Flock KE, Cook RJ, Hunkele AJ, Loh HH, Ko JL. Effects of trichostatin A on neuronal mu-opioid receptor gene expression. Brain Res. 2008;1246:1–10. doi: 10.1016/j.brainres.2008.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Laimins LA. Histone deacetylase 3 localizes to the plasma membrane and is a substrate of Src. Oncogene. 2006;25:4495–4500. doi: 10.1038/sj.onc.1209473. [DOI] [PubMed] [Google Scholar]
- Lovinger D. Serotonin's Role in Alcohol's Effects on the Brain. Alcohol Health Res. World. 1997;21:114–120. [PMC free article] [PubMed] [Google Scholar]
- Mandrekar P. Epigenetic regulation in alcoholic liver disease. World J Gastroenterol. 2011;17:2456–2464. doi: 10.3748/wjg.v17.i20.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min-Hao K, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Park P-H, Miller R, Shukla SD. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem. Biophys. Res. Comm. 2003;306:501–504. doi: 10.1016/s0006-291x(03)01040-4. [DOI] [PubMed] [Google Scholar]
- Park SH, Lee SR, Kim BC, Cho EA, Patel SP, Kang H-B, Sausville EA, Nakanishi O, Trepel JB, Lee BI, et al. Transcriptional Regulation of the Transforming Growth Factor Î2 Type II Receptor Gene by Histone Acetyltransferase and Deacetylase Is Mediated by NF-Y in Human Breast Cancer Cells. J. Biol. Chem. 2002;277:5168–5174. doi: 10.1074/jbc.M106451200. [DOI] [PubMed] [Google Scholar]
- Peserico A, Simone C. Physical and Functional HAT/HDAC Interplay Regulates Protein Acetylation Balance. J. Biomed. Biotechnol. 2011 doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Laniel M-A. Histones and histone modifications. Current Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Oster SM, Toalston JE, Pommer TJ, McBride WJ, Murphy JM. Serotonin-3 receptors in the posterior ventral tegmental area regulate ethanol self-administration of alcohol-preferring (P) rats. Alcohol. 2010;44:245–255. doi: 10.1016/j.alcohol.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu P, Host L, Gobaille S, Sandner G, Aunis D, Zwiller J. Histone Deacetylase Inhibitors Decrease Cocaine But Not Sucrose Self-Administration in Rats. J Neurosci. 2008;28:9342–9348. doi: 10.1523/JNEUROSCI.0379-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard B, Tuma P. Alcohol-induced protein hyperacetylation: Mechanisms and consequences. World J. Gastroenterol. 2009;15:1219–1230. doi: 10.3748/wjg.15.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Aroor A. Epigenetic effects of ethanol on liver and gastrointestinal injury. World J. Gastroenterol. 2006;12:5265–5271. doi: 10.3748/wjg.v12.i33.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson H, Kechris K, Bell R, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biology. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Cruz DA, Olukotun DY, Delgado PL. Serotonin receptor, SERT mRNA and correlations with symptoms in males with alcohol dependence and suicide. Acta Psychiatrica Scandinavica. 2011 doi: 10.1111/j.1600-0447.2011.01816.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Wang L, Lv Z, Hu Z, Sheng J, Hui B, Sun J, Ma L. Chronic Cocaine-Induced H3 Acetylation and Transcriptional Activation of CaMKII[alpha] in the Nucleus Accumbens Is Critical for Motivation for Drug Reinforcement. Neuropsychopharmacology. 2009;35:913–928. doi: 10.1038/npp.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PMD, Riccio A. Nitric oxide and histone deacetylases: A new relationship between old molecules. Communicative Integrative Biology. 2009;2:11–13. doi: 10.4161/cib.2.1.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Du W, Zhang Y-J, Hong J, Su W-Y, Tang J-T, Wang Y-C, Lu R, Fang J-Y. Trichostatin A, a histone deacetylase inhibitor, suppresses JAK2/STAT3 signaling via inducing the promoter-associated histone acetylation of SOCS1 and SOCS3 in human colorectal cancer cells. Mol. Carcinogenesis. 2012;51:174–184. doi: 10.1002/mc.20777. [DOI] [PubMed] [Google Scholar]
- Yoo J-H, Lee H-K, Kim H-C, Lee S-Y, Jang C-G. GABAA receptors mediate the attenuating effects of a 5-HT3 receptor antagonist on methamphetamine-induced behavioral sensitization in mice. Synapse. 2010;64:274–279. doi: 10.1002/syn.20726. [DOI] [PubMed] [Google Scholar]