Abstract
The development of suitable scaffolds for bone tissue engineering requires an in-depth understanding of the interactions between osteoblasts and scaffolding biomaterials. Although there have been a large amount of knowledge accumulated on the cell-material interactions on two-dimensional (2D) planar substrates, our understanding of how osteoblasts respond to a biomimetic nano-structured three-dimensional (3D) scaffold is very limited. In this work, we developed an approach to use confocal microscopy as an effective tool for visualizing, analyzing and quantifying osteoblast-matrix interactions and bone tissue formation on 3D nanofibrous gelatin scaffolds (3D-NF-GS). Integrin β1, phosphor-paxillin, and vinculin were used to detect osteoblasts responses to the nanofibrous architecture of 3D-NF-GS. Unlike osteoblasts cultured on 2D substrates, osteoblasts seeded on 3D-NF-GS showed less focal adhesions for phospho-paxillin and vinculin and the integrin β1 was difficult to detect after the first 5 days. Bone sialoprotein (BSP) expression on the 3D-NF-GS was present mainly in the cell cytoplasm at 5 days and inside secretory vesicles at 2 weeks, whereas most of the BSP on the 2D gelatin substrates was concentrated either in cell interface towards the periphery or at focal adhesion sites. Confocal images showed that osteoblasts were able to migrate throughout the 3D matrix within 5 days. By 14 days, osteoblasts were organized as nodular aggregations inside the scaffold pores and a large amount of collagen and other cell secretions covered and remodeled the surfaces of the 3D-NF-GS. These nodules were mineralized and were uniformly distributed inside the entire 3D-NF-GS after being cultured for 2 weeks. Taken together, these results give insight into osteoblast-matrix interactions in biomimetic nanofibrous 3D scaffolds and will guide the development of optimal scaffolds for bone tissue engineering.
Keywords: gelatin, scaffold, cell-material interactions, biomimetic, nanofibers, osteoblasts, confocal
INTRODUCTION
Understanding cell-matrix interactions is pivotal for the development of suitable three dimensional (3D) scaffolds for tissue regeneration.1–3 Cell adhesion, migration, proliferation and signaling on two dimensional (2D) planar substrates have been extensively studied for over three decades.4–6 By using the 2D models, considerable knowledge has been gained to help understand how cells interact with biomaterials.7–12 In the body; however, cells are surrounded by and interact with the extracellular matrix (ECM), which is a nano-structured 3D network. It is generally recognized that the 2D flat substrates that are widely used for cell culture are not representative of the cellular environment found in the body.13–16 Therefore, the knowledge of cell-material interactions from the artificial 2D models may not be applicable to predict cell behavior in physiological 3D environment. In fact, recent studies have shown that cell-matrix interactions in the body are significantly different from that on 2D substrates.4,17
In view of the importance of 3D architecture to mimic the in vivo microenvironment, hydrogels such as collagen, matrigel, and poly(ethylene glycol) (PEG), have been used to study cell-matrix interactions in vitro.18–21 However, most of these studies have been focused on the response of fibroblasts or carcinoma cells inside the hydrogels18–21 and the results may be considerably different from the interactions of normal osteoblasts with the ECM in vivo. Furthermore, while hydrogels have been widely studied for soft tissue regeneration, they may not be good candidates as bone scaffolding materials due to their inherent low mechanical properties.22
A variety of non-hydrogel scaffolds have been developed for bone regeneration in recent years.23–27 However, those non-transparent scaffolds do not have well-defined inter-connected open pore structure. Therefore, light cannot be transmitted through those scaffolds and light microscopy (the most widely used tool in biology) cannot be used to visualize cell-matrix interactions. In addition, those scaffolds cannot mimic the nano-structured architecture of the natural ECM, which is important for cell behavior, including adhesion, proliferation, differentiation, and tissue formation.5,28,29 Therefore, those non-hydrogel scaffolds are not good candidates to study osteoblast-matrix interactions. To date, little is known about how osteoblasts interact with biomimetic nano-structured 3D scaffolds in vitro.
In our previous study, we developed a technology to prepare 3D nanofibrous gelatin scaffolds (3D-NF-GS), which mimic both the nanoscale architecture and the chemical composition of natural collagen (a major component of many tissues).30 The 3D-NF-GS possess high surface area (>32 m2/g), high porosities (>96%), good mechanical properties, and nanofibrous pore wall structures, which pose them as attractive scaffolds for tissue regeneration. Meanwhile, the well-defined inter-connective pore structure of the 3D-NF-GS allows the use of light microscopy (e.g. confocal microscopy) to visualize cell behaviors with the scaffold. Our recent publication further indicated that the 3D-NF-GS are excellent scaffolds to support bone tissue formation.31 Therefore, the ECM-like 3D-NF-GS may serve as a great platform to study osteoblast-matrix interactions in 3D.
In this work, we used confocal microscopy as well as other tools to study the behavior of osteoblastic cells on the 3D-NF-GS in vitro. The osteoblasts adhesion, migration, proliferation, differentiation, and mineralization on the 3D-NF-GS were visualized and analyzed using confocal microscopy. An image processing method was further developed to quantify the osteoblast-matrix interactions.
Materials and Methods
Materials
Gelatin (type B, from bovine skin, approx 225 Bloom) was purchased from Sigma Chemical Company (St. Louis MO). N-hydroxy-succinimide (97%) (NHS) and (2-(Nmorpholino) ethanesulfonic acid) hydrate (MES) were purchased from Aldrich Chemical (Milwaukee, WI). 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide HCl (EDC) was purchased from Pierce Biotechnology (Rockford, IL). Ethanol, hexane, cyclohexane and 1,4-dioxane were purchased from Fisher Scientific (Fair Lawn, NJ).
Preparation of 3D nanofibrous gelatin scaffolds (3D-NF-GS)
3D-NF-GS were fabricated by combining a thermally induced phase separation (TIPS) technique and a porogen-leaching process as reported previously.30 Briefly, paraffin spheres (0.40g, 250–420 µm) were pretreated in teflon molds at 37°C for 20 minutes. Gelatin (2.0g) was dissolved in water (10 mL) and ethanol (10 mL) solvent mixture at 45°C and this solution was cast onto the paraffin sphere assembly. The gelatin solution in the paraffin assembly was then phase separated at −76°C for at least 4 h. After leaching paraffin, solvent exchange, the obtained scaffolds were crosslinked using 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) as reported previously in a 90/10 (acetone/water) mixture.30,31 The scaffolds were freeze-dried and cut to required size (6.0 mm in diameter and 1.0 mm in height) for later use. The resulting 3D-NF-GS had a surface area of 36.9 m2/g, a porosity of 96.45±0.19%, average fiber diameter of 159±49 nm, a average fiber length of 497±62 nm, and compressive modulus of 801±108 kPa.30 Meanwhile, the macropore size of the 3D-NF-GS ranges from 250 to 420 µm with the average size of about 320 µm. In comparison, the commercial available gelatin foam — Gelfoam® (Pharmacia & Upjohn Company, Kalamazoo, MI) with a similar porosity and pore sizes ranged from 100 to 400 µm, had a surface area of 0.046 m2/g and compressive modulus of 80±8 kPa.30
Preparation of TRITC-labeled gelatin
Gelatin (0.1 g) was dissolved in 4 ml distilled water at 37°C and the pH of the solution was adjusted to 10.0. Tetramethyl Rhodamine Iso-Thiocyanate (TRITC) (2.0 mg) was dissolved in 0.2 ml anhydrous N,N-dimethylformamide (DMF). This TRITC solution was then mixed with the gelatin solution and was incubated in the dark at 37°C for 24 hours. The reacted mixture was then extensively dialyzed against distilled water. The dialyzing water was changed every 12 hours continuing the dialysis for 4 days until no free TRITC was detected in the dialyzing water. This TRITC-conjugated gelatin was then used for making 3D-NF-GS.
Cell culture and seeding
Cell culture
Osteoblasts were isolated from new born mice calvaria after sacrificing as described by Ecarotcharrier et.al.32 The osteoblasts were cultured in alpha minimum enriched medium (Alpha-MEM, GIBCO™ Invitrogen Corporation, Carlsbad, CA), 10% FBS, and 1% antimycotic/antibiotic (10,000 IU/ml penicillin, 10,000 µg/ml streptomycin, 25 µg/ml amphotericin B; Cellgro, Mediatech Inc., VA) and 100 µg/ml ascorbic acid (Sigma Aldrich, UK) in an incubator at 37°C with a humidified gas mixture (5% CO2 and 95% air) and the medium was changed every 48 h. Primary culture of osteoblasts was allowed to grow for a week before passaging. Cells were then passaged at 1:3 ratio using a 0.25% trypsin solution (GIBCO™ Invitrogen, Corporation, Carlsbad, CA) and plated in new tissue culture dishes (FALCON™ BD Biosciences Discovery Labware, Bedford, MA). Passages 2–5 were used in all of the further experiments.
Seeding the cells on 3D-NF-GS
The 3D-NF-GS were sterilized with 70% ethanol overnight and then washed twice with 1× PBS for 10 minutes each. The scaffolds were then washed with complete medium (Alpha MEM, 10% FBS, 10,000 IU/ml penicillin, 10,000 µg/ml streptomycin, 25 µg/ml amphotericin B, 100 µg/ml ascorbic acid) twice for 2 hours each time on an orbital shaker (3520, Lab-Line Instruments, INC). The osteoblasts, which were cultured for at least 2 generations in tissue culture dishes were centrifuged and pelleted. They were resuspended in 20 µl of medium and seeded on rehydrated 3D-NF-GS. Each scaffold was seeded with 1 ×105 cells and cultured in an incubator at 37°C with 5% CO2. The medium volume was increased to 10 ml after 2 hours. Initially, the medium was changed after 24 hours followed by changing it every 48 hours for further culture period. These seeded 3D-NF-GS were cultured for either for 5 days (short term culture, no or little mineral deposition of cultured osteoblasts at this time point) or 14 days (long term culture, abundant mineral deposition of osteoblasts at this time point) and fixed with 4% paraformaldehyde (PFA) or used unfixed for mRNA experiments. Three 3D-NF-GS were seeded per group per experiment and every experiment was repeated three times.
Cell culture on 2D gelatin surface
The osteoblasts obtained after culture for at least 2 passages were plated on eight well Labtek chamber slides (the slides were pre-coated with a layer of gelatin on the surface using 5 mg/ml gelatin solution and then were crosslinked using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide HCl (EDC), see ref33,34) at a density of 50–100 cells per chamber and cultured for 5 days in Alpha MEM with 10% FBS, 10,000 IU/ml penicillin, 10,000 µg/ml streptomycin, 25 µg/ml amphotericin B and 100 µg/ml ascorbic acid in an incubator at 37°C with 5% CO2. Medium was changed every 48 hours. They were then fixed with 4% PFA for use in the following experiments.
Immunofluorescence staining
Immunofluorescence staining was performed on osteoblasts cultured on Labtek chamber slides as well as osteoblasts seeded 3D-NF-GS. Samples were fixed for 30 minutes with 4% PFA and permeabilized with 0.3% Triton X-100 for 10 minutes. Blocking was performed with 10% goat serum for 30 minutes. Samples were incubated with the indicated primary antibodies (Table 1) at room temperature for 60 minutes or at 4°C overnight, followed by detection with Alexa Flour secondary antibodies (Invitrogen Corporation, Carlsbad, CA) (1:500) for 2 hours at room temperature. The filamentous actin (F-actin) was labeled using Alexa Flour phalloidin 488 or 546 (Invitrogen Corporation, Carlsbad, CA) at a dilution of 1:50. The nuclei were labeled using Hoechst 33342 (Invitrogen Corporation, Carlsbad, CA). Stained samples were mounted in Slowfade anti-fade Gold reagent (Invitrogen Corporation, Carlsbad, CA).
Table 1.
Primary Antibody Information
| Antibody | Company | Dilution |
|---|---|---|
| Monoclonal anti- Vinculin antibody (mouse) | Sigma Aldrich (V9131) | 1:250 |
| Rabbit polyclonal to Collagen I | Abcam Inc.(ab59435) | 1:250 |
| Integrin β1 antibody (rabbit) | Abcam Inc. (ab52971) | 1:200 |
| Phospho-Paxillin (Tyr118) antibody (rabbit) | Cell-Signaling technologies (2541S) | 1:50 |
| Bone sialoprotein antibody (mouse) | Dr. Chunlin Qin’s laboratory [31, 32] | 1:500 |
Laser scanning confocal microscopy
Immunofluorescence microscopy images were obtained using a Leica TCS SP5-II upright microscope equipped with confocal fixed stage system (Leica DM 6000 CFS). Images were taken using an A-plan apochromat 63× objective (0.9 N.A.). The collagen staining on TRITC labeled scaffolds was visualized using apochromat 2.5× air (0.07 NA), apochromat 10× air (0.4 NA) and apochromat 20× air (0.7 NA) objective lenses. The 405 Diode, 488nm Argon (25% power), 561nm DPSS (40% power), and 633nm HeNe (50% power) lasers were used to excite Hoechst 33342 (absorption 350 to 440), Alexa Fluor 488 (absorption 450 to 520), 546 (absorption 500 to 580) and 633 (absorption 540 to 640) dyes respectively. The pinholes for each laser line were aligned for optimal confocality. Various z stacks were acquired for all samples. The maximum depth for z stacks was approximately 400 µm from each side of the 1 mm thick 3D-NF-GS. The z range for most of the z stacks was from 10 microns to 150 microns depending upon different experiments. Each figure shown is representative of a combination of projected z stacks into a two dimensional (2D) image. The experiments were repeated and analyzed a minimum of three times for each set of experiments.
Image processing for pixel intensity
Images were processed using Leica Application Suite - Advanced Fluorescence (LASAF)- Quantify software. Z stacks acquired for 3D-NF-GS were maximally projected to create 2D images. Data from these images were quantified by comparing the number of pixels highlighted in the total frame in desired laser wavelength channels, thus calculating the pixel intensity for the desired channel. These calculations were used to quantify the collagen matrix production by osteoblasts on TRITC labeled 3D-NF-GS, collagen secretion by osteoblasts on unlabeled 3D-NF-GS, calculation of the length of focal/fibrillar adhesions and calculating the alizarin red expression to quantify mineralization. Collagen I detection on TRITC labeled 3D-NF-GS and alizarin red staining experiments were repeated five times (n=5) and collagen I detection experiments on unlabeled scaffolds were repeated three times and 10 different regions of interest (ROIs) for each experiment were analyzed (n=30).
Quantification of cell morphology and cell-migration assessment
Phalloidin (F-actin) and Hoechst 33342 (nuclei) labeled cells were subjected to confocal imaging on 2D surfaces and 3D-NF-GS. Several confocal z stacks were acquired to encompass the entire cell outlines and were maximally projected to create a 2D image. LASAF- Quantify software was used to delineate cell outlines and calculate individual cell spread area of various osteoblasts in each of the 2D and 3D matrix environments. The cell numbers were calculated by counting number of nuclei per 40,000 µm2 area of each ROI in each projected image from 3 different experiments. In each experiment, 15 ROIs were analyzed (total n=45). The focal adhesion structures were manually counted per 10,000 µm2 area of ROI per projected confocal z stack image from 3 different sets of experiments for each adhesion molecule. 10 different ROIs were analyzed per experiment (total n=30 for each).
To assess migration of cells inside the 3D-NF-GS, phalloidin 488 and Hoechst 33342 labeled cells were visualized using Leica confocal SP5 microscope at superior, middle and inferior levels of the 1mm thick 3D-NF-GS. The superior level was imaged 30–50 µm below the 3D-NF-GS’s upper surface, middle level was imaged 300–400 µm inside the 3D-NF-GS and lower level was imaged 30–50 µm below the surface of the 3D-NF-GS after inverting it. At each level a 10–15 µm thick z stack was obtained and projected into a single optical image. Various areas inside one specimen at each level were assessed for uniformity of migration and the experiment was repeated three times for confirmation.
Scanning electron microscopy (SEM) observation
Samples were fixed for 30 minutes with 4% PFA and briefly washed with deionized water for complete removal of any salts. Then they were dehydrated through a series of alcohol concentrations (20, 30, 40, 50, 60, and 70%). Hexamethyldisilazane (Sigma, UK) was used for the final dehydration step and sublimed under vacuum. The samples were sputter coated with gold (30 sec, 20 mA) for analysis by SEM at an accelerating voltage of 15 keV.
RNA extraction and real time PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol after the 3D-NF-GS were mechanically homogenized with a tissue tearor (Biospec products, Bartlesville, OK, USA) and the cells from 2D surfaces were lysed using a cell scraper. RNA concentrations were measured using a Nanodrop spectrophotometer (Nanodrop Technologies, Montchanin, USA). RNA samples with an optical density ratio of absorbance at 260nm (RNA) over 280nm (protein) greater than 1.8 were used to make cDNA. For PCR analysis, 2 µg RNA was reverse transcribed to cDNA with High capacity RNA to cDNA kit (Invitrogen Corporation, Carlsbad, CA) according the manufacturer’s protocol. For amplification of target genes, 10µl PCR master mix (Promega, Madison, USA) was used with 2µl cDNA (0.05µg) and 1–2µl sequence specific primers according to the manufacturer’s protocol with a standard PCR program (denaturation: 30 s at 94°C; primer hybridization: 2 min at target specific temperatures; elongation: 2 min at 72°C). PCR products were analyzed by electrophoresis on a 1.0% agarose gel and visualized with 0.5 mg/ml ethidium bromide. The cDNA from the osteoblasts grown on 2D surface were used as positive controls.
Real time PCR was set up using iTaq SYBR Green with ROX (Bio-Rad, Richmond, CA, USA) for sequence specific primers (Table 2) with 3 minutes incubation at 94°C, a 10 second Taq activation at 94°C, a 30 second annealing at 56°C followed by an extension for 30 seconds minute at 72°C on Bio-Rad CFX96 Real-Time PCR System (Applied Biosystem, Bio-Rad, Richmond, CA). Target genes were normalized against GAPDH using relative standard curve.
Table 2.
PCR Primer Information
| Gene | Forward Primer | Reverse primer |
|---|---|---|
| Coll I | 5’ GAGCGGAGAGTACTGGATCG 3’ | 5’ GCTTCTTTTCCTTGGGGTTC 3’ |
| BSP | 5’ GAAGCAGGTGCAGAAGGAAC 3’ | 5’ ACTCAACGGTGCTTTTT 3’ |
| OCN | 5’ AAGCAGGAGGGCAATAAGGT 3’ | 5’ CCGTAGATGCGTTTGTAGGC 3’ |
| Runx2 | 5’GCCGGGAATGATGAGAACTA 3’ | 5’ GGACCGTCCACTGTCACTTT 3’ |
| GAPDH | 5’AACTTTGGCATTGTGGAAGG 3’ | 5’ ACACATTGGGGGTAGGAACA 3’ |
Alizarin red and von Kossa staining
Fixed 3D-NF-GS that were cultured with osteoblasts for 5 days and 14 days as well as unseeded 3D-NF-GS (controls) were stained with alizarin red. Since the scaffold surface is adsorbent, fluorescent signal detection of alizarin via the Leica SP5 confocal microscope was a better method of identifying mineralization over background. The fixed 3D-NF-GS and unseeded 3D-NF-GS were incubated with phalloidin 633 (1:50) (spectrum widow 540 to 620 nm) to visualize the scaffold architecture due to adsorption of phalloidin on the surface which was excited by HeNe 633 laser. After fixation, scaffolds were washed with 1× PBS two times for 5 minutes each and with distilled water once for 5 minutes. Then, 0.5% alizarin red (Sigma Aldrich, UK) solution (pH 7.4) was applied on the scaffolds for 30 seconds to 1 minute. The scaffolds were rinsed with distilled water 5 times for 5 minutes each. These alizarin labeled 3D-NF-GS were then analyzed with the Leica SP5 confocal microscope where phalloidin was excited by 633 wavelength (HeNe) laser (absorption 540 to 640) and alizarin was excited by 488 (Argon) wavelength laser (absorption window 514–546). The pixel intensity for 488 photon laser (alizarin) was then calculated to quantify mineralization of the 3D-NF-GS. The experiment was repeated 5 times.
Von Kossa staining was performed on 3D-NF-GS cultured with osteoblast for 14 days and unseeded 3D-NF-GS to serve as controls. Samples were fixed in 4% PFA and washed three times in double distilled water for 5 minutes each. Then they were incubated with 2% silver nitrate solution (Sigma Aldrich, UK) and exposed to ultraviolet (UV) light for 30 minutes. The samples were washed with double distilled water three times for 5 minutes each. Un-reacted silver was removed by incubating the samples with 5% sodium thiosulfate for 5 minutes. Samples were washed with double distilled water three times for 5 minutes each.
Calcium content assay
The 3D-NF-GS cultured with osteoblasts for 2 weeks were examined for calcium deposition by use of a total calcium LiquiColor- kit (Stanbio laboratory, TX). The 3D-NF-GS constructs were washed three times for 5 min each in double-distilled water and then homogenized with a tissue tearor in 1 mL of double-distilled water. The suspension was incubated in 500 µL of 0.5 N hydrochloric acid (HCL) overnight to extract calcium. Then calcium reagent working solution was added to each sample according to the manufacturer’s instruction. The absorbance was measured at 550 nm and the calcium content was calculated against standard solution provided in the kit.
Statistical analysis
Data are plotted as mean ± standard error of the mean (SEM). All analyzed data were subjected to an analysis of variance (ANOVA) followed by a Bonferroni’s t-test for multiple comparisons between pairs of means. Student’s t-test was used where ANOVA was not applicable and a value of p<0.05 was considered to be statistically significant. All tests were performed using SPSS statistical software.
RESULTS
Osteoblasts morphology on 3D-NF-GS
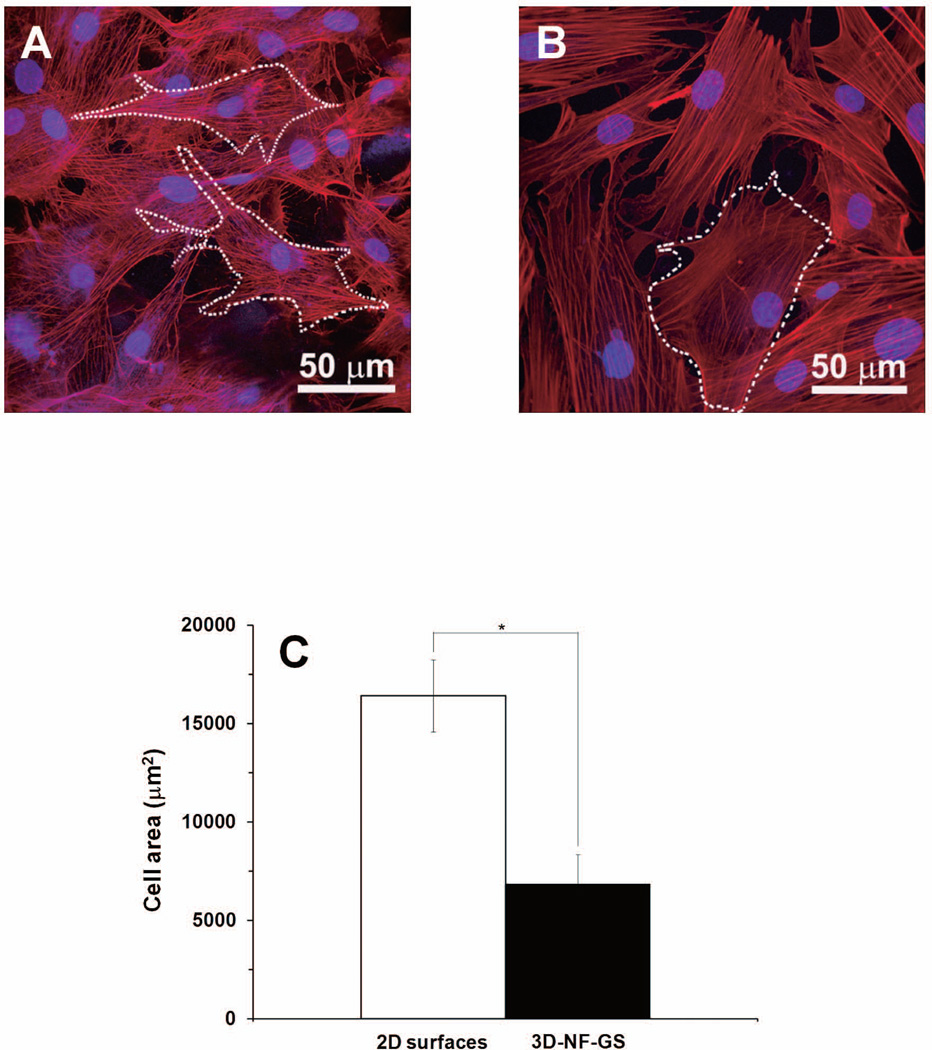
Using confocal microscopy and the LASAF software, osteoblasts attached on 3D-NF-GS were outlined and the total cell area was calculated. The osteoblasts on 3D-NF-GS appeared stellate with numerous filamentous extensions (Fig. 1A). In contrast, the osteoblasts on 2D gelatin surfaces were cuboidal and were spread broader and more flattened (Fig. 1B). Quantitative analysis indicated that osteoblasts cultured for 5 days on the 2D matrix were spread two-fold more than on 3D-NF-GS (p<0.001) (Fig. 1C).
Figure 1.
Projected confocal laser scanning microscopy (CLSM) images of osteoblasts on (A) 3D-NF-GS after cultured for 5 days, and (B) 2D gelatin surface. The actin was labeled red and nuclei were blue. (C) Quantification of cell areas on 3D-NF-GS and 2D substrate (*denotes significant difference, p<0.001).
Osteoblast-matrix adhesions on 3D-NF-GS
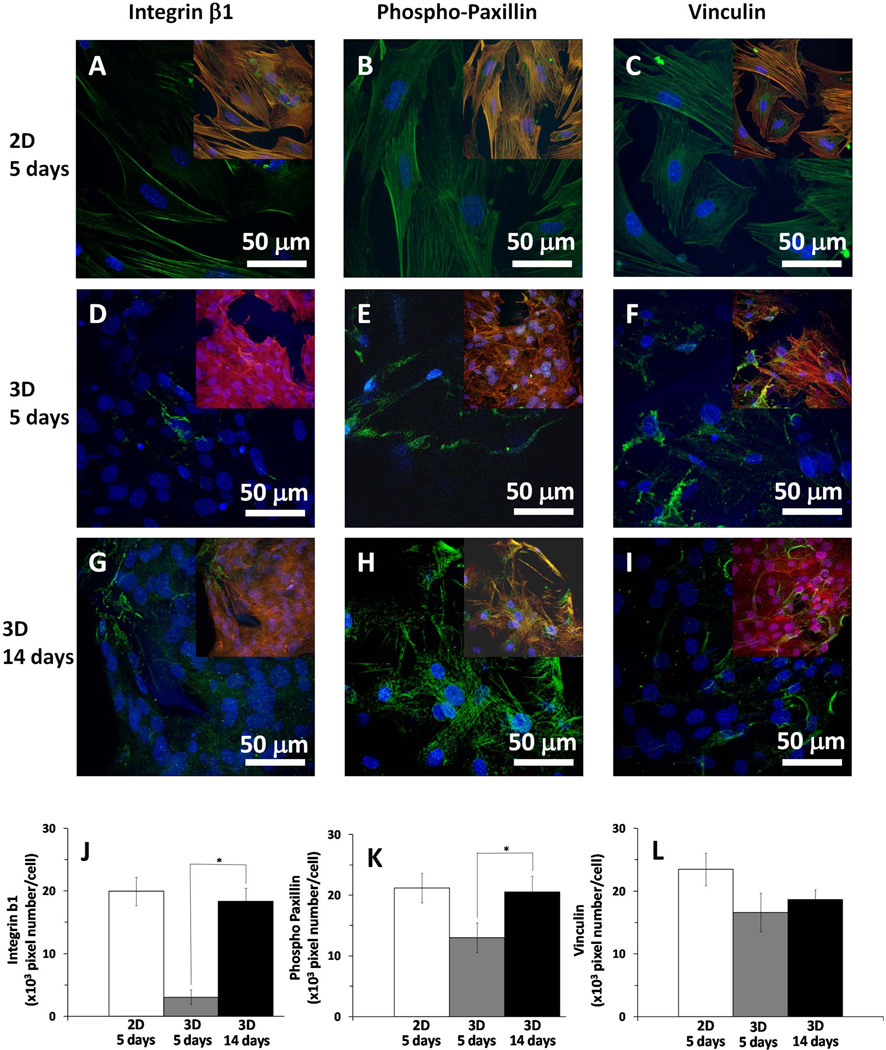
To study the cell-matrix adhesion structures exhibited by osteoblasts adhering to the 3D-NF-GS as well as on the 2D gelatin substrates, β1 integrin, phospho-paxillin and vinculin protein distributions were assessed. In the images, the nuclei are blue, F-actin is red and focal adhesions are green. To clearly show the distribution of the focal adhesion proteins, the larger images are 2 channels containing the data for nuclei (blue) and focal adhesion proteins (green). F-actin (red) was overlayed as in the third channel in the inset images. Osteoblasts on 2D surfaces had strong β1 integrin, phospho-paxillin, and vinculin expression, and contained typical focal adhesion structures (green) (Fig. 2A–2C). Osteoblasts seeded on the 3D-NF-GS expressed phospho-paxillin and vinculin after culture for 5 days (Fig. 2E–2F); however, β1 integrin was difficult to detect (Fig. 2D). The sparse β1 integrin was overlapped with F-actin and had elongated and fibrillar morphology rather than classical focal adhesion structures (inset Fig. 2D). After 14 days of culture on the 3D-NF-GS, integrin β1 was more prominent (Fig. 2G). Quantitative analysis indicated that the expression of β1 integrin on day 14 was more than five-fold higher than on day 5 (Fig. 2J). Also, phospho-paxillin expression increased more than 50% from 5 to 14 days (Fig. 2K). However, vinculin adhesions did not show an increase but were more fibrillar in phenotype (Fig. 2I&2L).
Figure 2.
Focal adhesions of osteoblasts on 3D-NF-GS and 2D gelatin substrate cultured for 5 and 14 days. (A–C) projected z stack CLSM images of osteoblasts cultured for 5 days on 2D surface; (D–F) projected z stack CLSM images of osteoblasts cultured for 5 days on 3D-NF-GS; (G–I) projected z stack CLSM images of osteoblasts cultured for 14 days on 3D-NF-GS. The cell/substrate was stained for focal adhesions using integrin β1 (A, D, G), phospho-paxillin (B, E, H) and vinculin (C, F, I) antibodies (red-actin, blue-nuclei, green-focal adhesions, yellow-overlap of focal adhesions on actin). Quantification showing significant increase in the adhesions from 5 days to 14 days of culture on the 3D-NF-GS: (J) integrin β1 (p<0.001) and (K) phospho-paxillin (p<0.001), but not significant for (L) vinculin (p=0.085).
Osteoblasts migration and proliferation in 3D-NF-GS
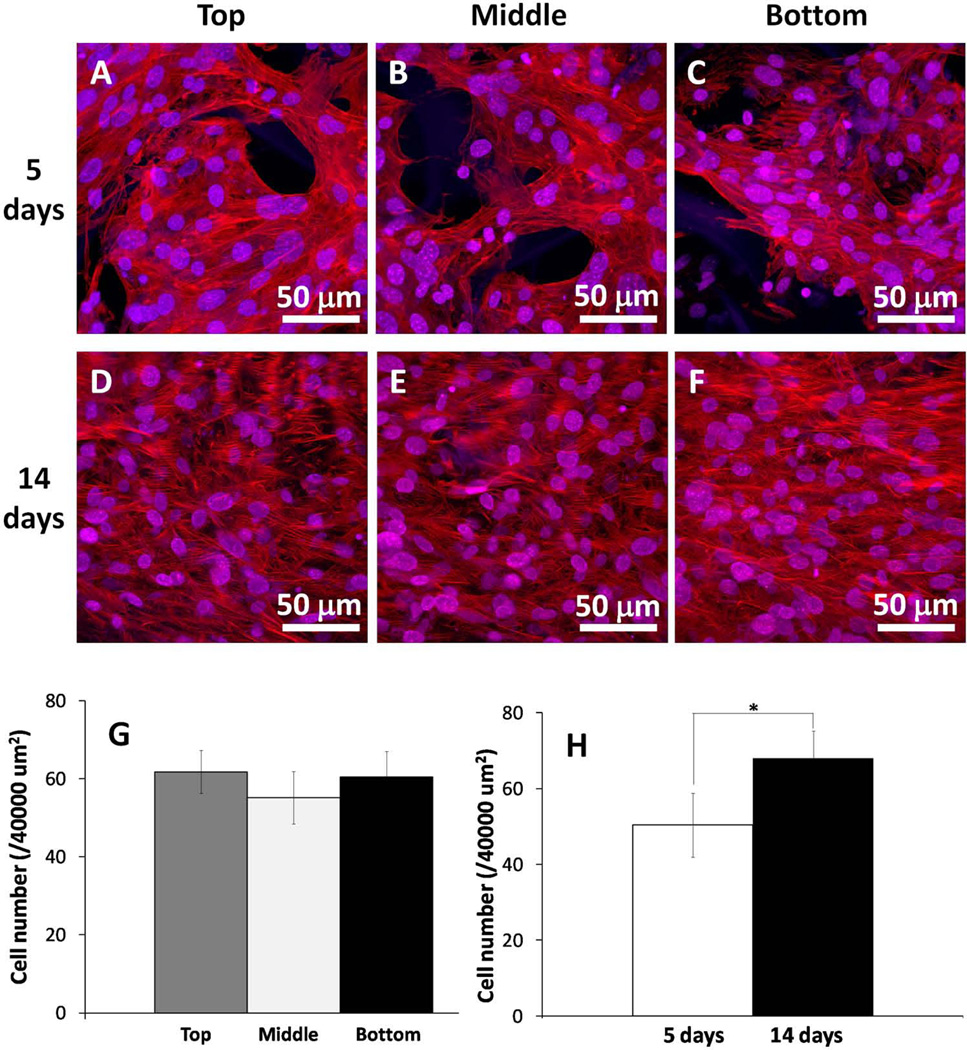
Using confocal microscopy, we analyzed the migration and distribution of osteoblasts (F-actin and nuclear staining) on 3D-NF-GS (thickness = 1 mm) at the top, middle and bottom levels of the 3D-NF-GS after 5 and 14 days of culture (Fig. 3A–3F). At day 5, the osteoblasts were present throughout the 3D-NF-GS, demonstrating they had migrated through the whole scaffold (Fig. 3A–3C). Further quantitative analyses showed that the cell numbers among the three areas (top, middle and bottom) were not significantly different, indicating that the osteoblasts were evenly distributed throughout the entire 3D-NF-GS (Fig. 3G). The cell density increased from 5 to 14 days, suggesting that the osteoblasts were healthy and proliferating (p<0.001) (Fig. 3H). Meanwhile, the uniform distribution of the osteoblasts was maintained throughout the 14 days (Fig. 3D–3F).
Figure 3.
Osteoblast migration and proliferation on 3D-NF-GS. (A–C) Projected z stack CLSM images of osteoblasts cultured on 3D-NF-GS for 5 days. (D–F) Projected z stack CLSM images of osteoblasts cultured on 3D-NF-GS for 14 days. (G) Quantification of cell numbers in different areas (top, middle, and bottom) of the scaffold at day 5. (H) Quantification of average cell numbers in the samples at 5 and 14 days.
Gene expression and protein synthesis
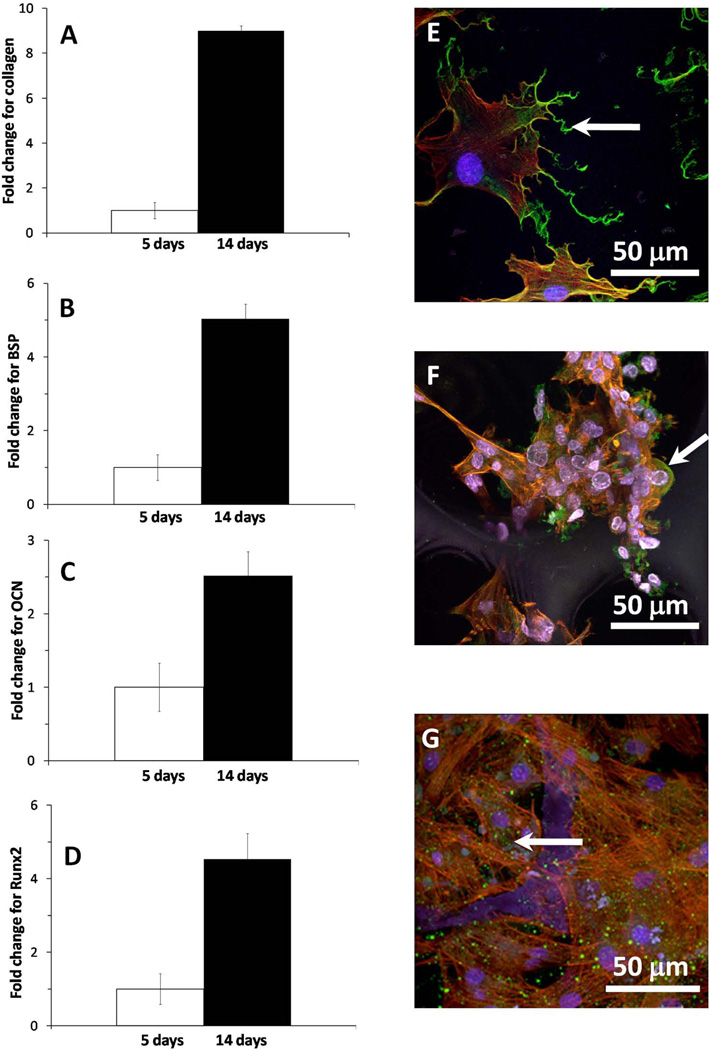
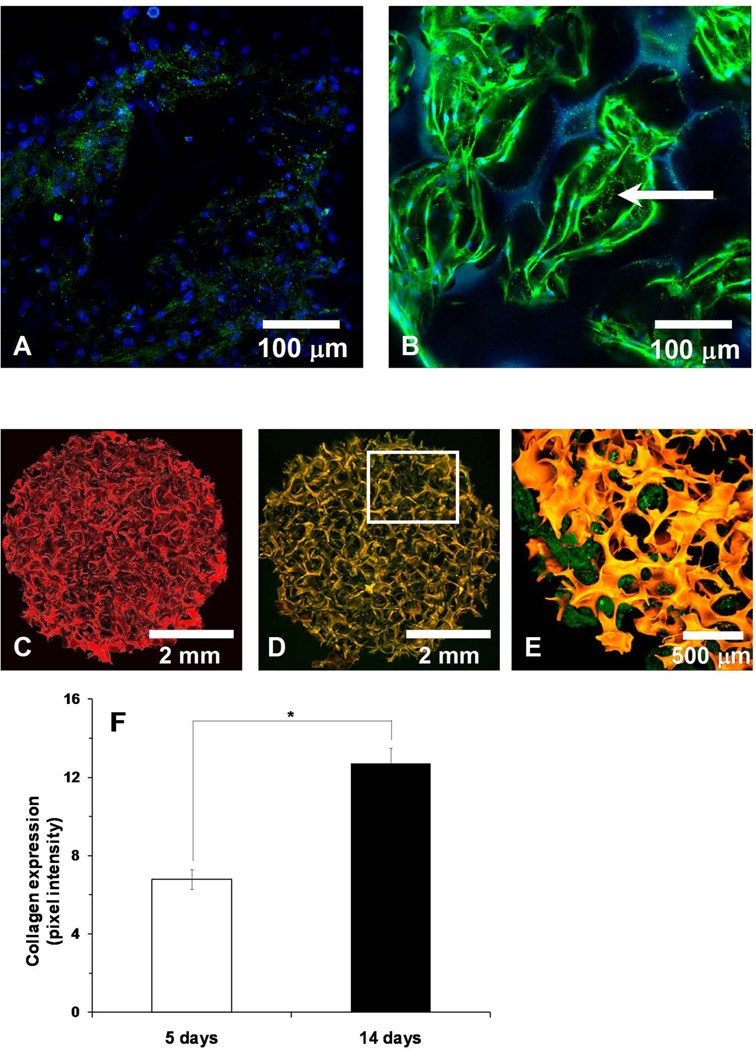
Expression of genes associated with the osteoblastic differentiation on the 3D-NF-GS was examined using real time PCR at 5 and 14 days. All osteoblastic differentiation markers were elevated to significantly higher levels with the culture time (Fig. 4A–4D). For example, the expression of Col I and BSP on the 3D-NF-GS at 14 days was more than 8-fold and 4-fold higher than at 5 days, respectively (Fig. 4A–4B). The synthesis and distribution of BSP on the 3D-NF-GS was also detected via immunofluorescence. Interestingly, BSP on the 3D-NF-GS was present mainly in the cell cytoplasm at 5 days (Fig. 4F-arrow) and inside secretory vesciles at 2 weeks (Fig. 4G-arrow); whereas most of the BSP on the 2D substrates was concentrated either in cell interface towards the periphery or at local adhesion sites (Fig. 4E-arrow).
Figure 4.
Gene expression and BSP synthesis after osteoblasts were cultured for 5 and 14 days. (A) Col I; (B) BSP; (C) OCN, (D) Runx2; (E) BSP expression and distribution on 2D substrate at day 5; (F) BSP expression and distribution on 3D-NF-GS at day 5; (F) BSP expression and distribution on 3D-NF-GS at day 14.
Matrix deposition on 3D-NF-GS
To detect type I collagen (Col I) production on the 3D-NF-GS, it was labeled with an antibody specific for mouse Col I (green). After culture for 5 days, osteoblasts inside the 3D-NF-GS were arranged in layers (Fig 5A). Meanwhile, scattered Col I secreted from osteoblasts was detected around the pore wall of the 3D-NF-GS (Fig. 5A). By 14 days, osteoblasts were organized as nodular aggregations inside the scaffold pores and the amount of the deposited Col I was significantly increased (Fig 5B). To enhance the contrast and visualize the deposition and distribution of Col I in the scaffold, 3D-NF-GS were labeled with TRITC (Fig 5C). In scaffolds cultured for 14 days, the whole construct was orange (overlap of the red from TRITC of the 3D-NF-GS and the green from Col I), indicating that Col I was evenly deposited and distributed on the surfaces of the 3D-NF-GS (Fig 5D). Higher magnification images clearly showed that Col I was uniformly deposited inside the pore wall of the scaffold (green) (Fig 5E). The Col I production on the 3D-NF-GS was quantified using green pixel intensity values and was significantly increased from 5 to 14 days (p<0.001) (Fig. 5F).
Figure 5.
Collagen type I (Col I) production by the osteoblasts on 3D-NF-GS. (A) Projected z stack CLSM image of osteoblasts cultured on 3D-NF-GS for 5 days, showing scattered collagen deposition. (B) Projected z stack CLSM image of osteoblasts cultured on 3D-NF-GS for 14 days, showing nodule aggregation and greater collagen deposition (blue-nuclues, green-Col I, dotted line-pore of scaffold). (C) Projected z stack CLSM image of an unseeded TRITC labeled 3D-NF-GS. (D) Osteoblasts seeded on TRITC labeled 3D-NF-GS and cultured for 14 days (red-scaffold, green-Col I). (E) Higher magnification picture of area highlighted in (D). (F) Quantification of Col I expression on the 3D-NF-GS.
Mineralization on 3D-NF-GS
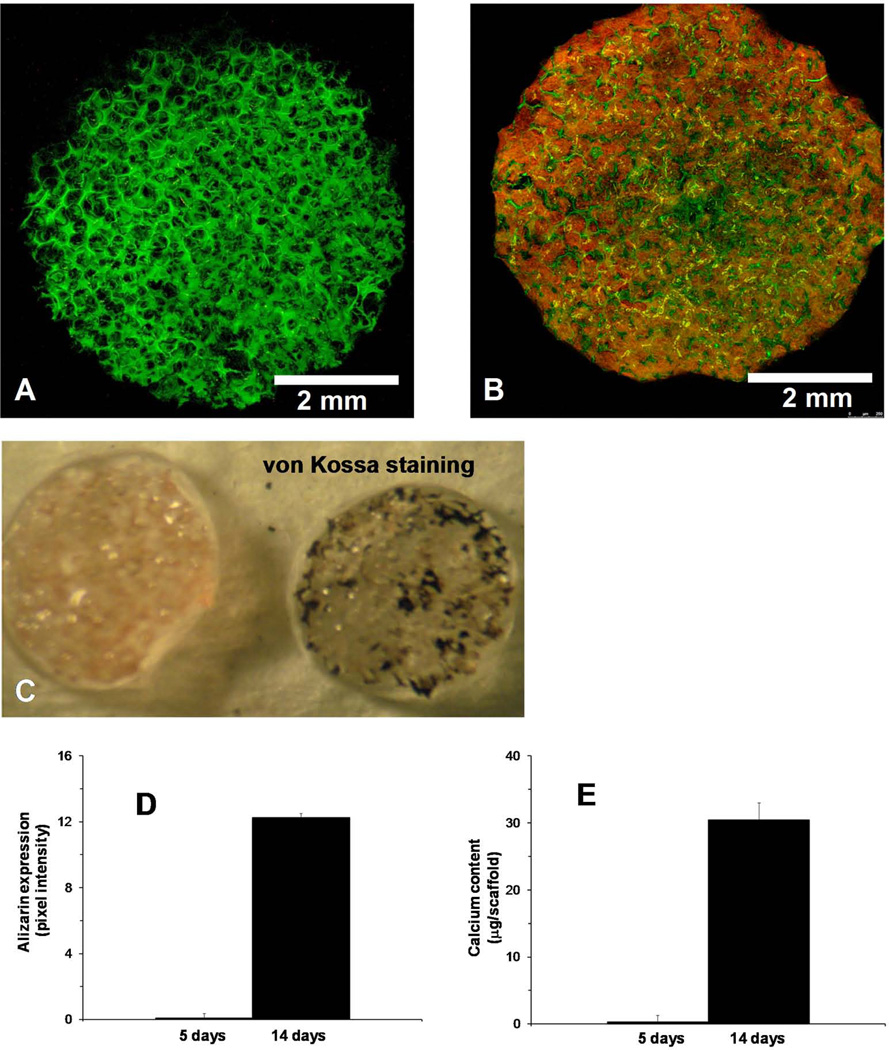
Mineralization of osteoblasts on the 3D-NF-GS was detected via alizarin red and von Kossa staining. In order to identify the scaffold architecture and to prevent the unspecified adsorption of dyes on the 3D-NF-GS, the scaffold was pre-adsorbed with phalloidin 633 (green) before alizarin staining (Fig. 6A). Confocal analysis revealed that the mineral deposited on 3D-NF-GS for 14 days were abundantly stained with alizarin red throughout the entire scaffold (Fig. 6B). The alizarin amount was quantified using pixel intensity for Argon laser excitation (absorption from 510–546 nm) and significantly higher amount of alizarin was detected for osteoblasts cultured on 3D-NF-GS for 14 days than for 5 days (Fig. 6D). Van Kossa staining also demonstrated that numerous mineralized nodules of osteoblasts uniformly distributed inside the entire 3D-NF-GS (Fig. 6C). Calcium content analysis quantitatively demonstrated that each osteoblasts/3D-NF-GS construct produced significantly more calcium (30 µg) after 14 days of culture, while only trace amount of calcium was detected at 5 days (Fig. 6E).
Figure 6.
Mineral deposition by the osteoblasts cultured on 3D-NF-GS for 14 days. (A) Projected z stack CLSM image of a 3D-NF-GS pre-adsorbed with phalloidin 633 (green); (B) Projected z stack CLSM images of alizarin expression on a 3D-NF-GS pre-adsorbed with phalloidin 633. (C) von Kossa staining of osteoblasts cultured on 3D-NF-GS for 14 days. (D) Quantification of alizarin expression of osteoblasts cultured on 3D-NF-GS for 14 days. (E) Calcium content deposition on the 3D-NF-GS (6 mm × 1 mm) after 14 days of osteoblasts culture.
Osteoblasts remodel the 3D-NF-GS
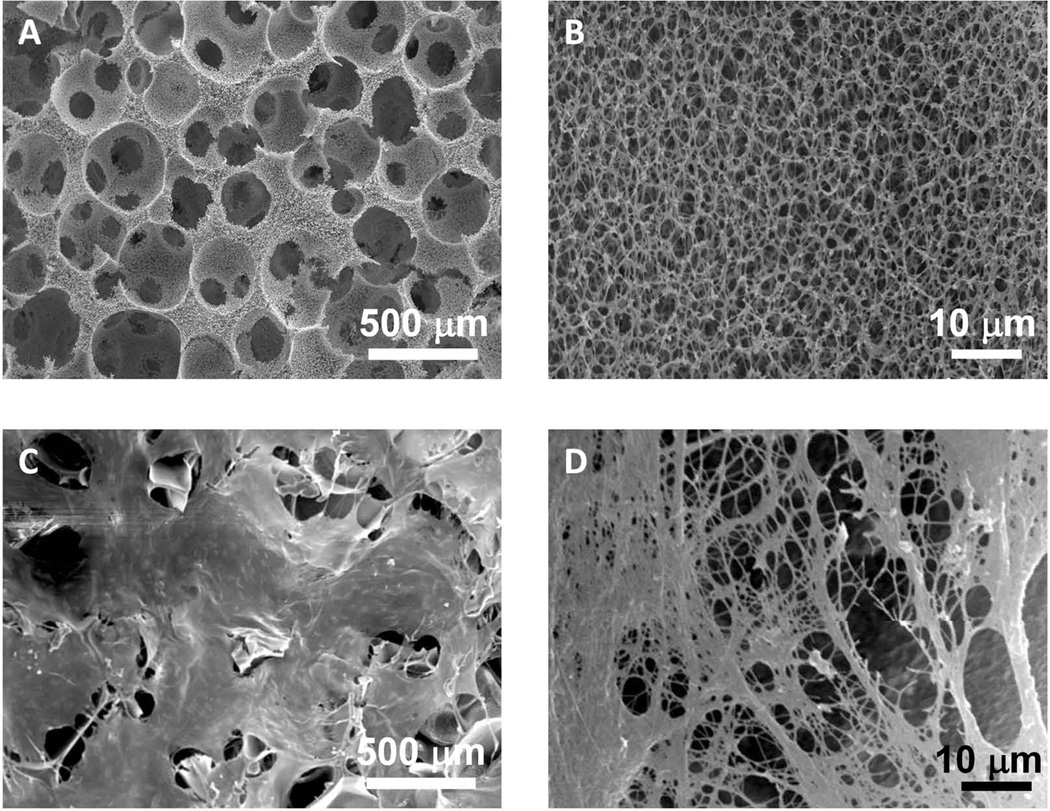
SEM imaging demonstrated that the matrix configuration of the 3D-NF-GS had significantly changed after the osteoblast culture on the scaffold for 14 days (Fig 7). At a low SEM magnification, the collagen fibers and other cell secretions spread and covered the entire surfaces of the 3D-NF-GS (Fig. 7C). A higher SEM magnification image clearly demonstrated dense collagen nanofibers deposited on the scaffold (Fig. 7D). In fact, most of the secreted collagen fibers were bound together to form large collagen bundles.
Figure 7.
SEM images of osteoblasts/3D-NF-GS constructs before and after cultured for 14 days. (A) SEM image of a representative 3D-NF-GS, showing well-defined pore structure and high pore interconnectivity. (B) High magnification of (A), showing nanofibrous architecture of the pore wall of the 3D-NF-GS. (C) SEM image of osteoblasts/3D-NF-GS construct after cultured for 14 days, showing the entire surface of the 3D-NF-GS has been covered with ECM secreted by osteoblasts. (D) A high magnification of (C), showing ECM nanofibers deposited on the scaffold.
DISCUSSION
Although it has been well recognized that cells cultured in a 3D environment simulate a more physiologic behavior than those cultured on 2D planar substrates or films,4,12,28,35 our understanding of osteoblast-matrix interactions on a 3D biomimetic scaffold is limited. Most of the 3D scaffolds currently used for bone regeneration are not good candidates to study osteoblast-matrix interactions: a) they cannot mimic the nano-structured architecture of ECM; and b) they do not have well-defined inter-connected open pore structure inside the 3D scaffold, which limits the use of light microscopy to characterize cell-material interactions. The 3D-NF-GS recently developed in our lab is an ECM-like scaffold which mimics both the nanofibrous architecture and the chemical composition of natural collagen.30 Furthermore, the 3D-NF-GS has well-defined macropore structure with high interconnectivity, which allows for the use light microscopy (e.g. confocal microscopy) to visualize cell behaviors on the scaffold. Therefore, our 3D-NF-GS serve as an ideal substrate to mimic in vivo osteoblast-matrix interactions.
Cell-matrix adhesions comprise a major part of cell-matrix interactions that control cell behavior on 3D matrices. Cell adhesions are attachment and signaling organelles generally mediated through specific class of transmembrane adhesion receptors known as integrins. Numerous structural proteins such as vinculin, talin, and paxillin act as scaffolding proteins that strengthen cell adhesion by anchoring them to the actin cytoskeleton.36 These adhesion molecules also serve as signal transduction for communication between cells and the ECM in a bi-directional manner.4,37–39 Using single photon confocal microscopy, we observed that the osteoblast adhesion on 3D-NF-GS was very different compared to 2D gelatin substrates. The osteoblasts on the 3D-NF-GS assumed a stellate appearance with numerous filamentous extensions; whereas, cells on the 2D substrates were more flattened and spread (Fig. 1). This result was consistent with our previous report that osteoblasts respond to surface morphology of biomaterials, and are more spread on smooth surface than on nanofibrous ones.40 During the first 5 days, the phospho-paxillin focal adhesion structures on the 3D-NF-GS were less than on 2D substrates (Fig. 2B&2E), and β1 integrin expression was barely detectable (Fig. 2D). The few observed β1 integrin adhesions were overlapped with actin and had a fibrillar morphology rather than classical focal adhesion structures. Both scaffolding pore structure and nanofibrous architecture has been reported to affect cell adhesion.15,17,40 and the cell adhesion structures on our 3D-NF-GS are more likely due to the combination of these two factors. Previous studies have shown that the typical focal adhesion structure formation observed in cells on 2D culture substrates are rare in vivo.4,17 In this regard, the 3D-NF-GS may provide osteoblasts with an environment closer to natural ECM than the 2D counterparts.
Cell migration and distribution inside 3D scaffolds is the key to form uniform tissues. The traditional method of assessing cell distribution into 3D matrices uses histological sectioning, which is time consuming and may cause a loss of integrity of the sample. Moreover, the section thickness is often less than the width of a single cell which may compromise the results as well. The use of confocal imaging allows the analysis of 3D matrices to greater depths (~400 µm in this study), providing an accurate representation of cell distribution within the scaffold without going through the laborious process of embedding and sectioning. The confocal images clearly showed that osteoblasts were present throughout the 3D-NF-GS, demonstrating they had migrated through the whole scaffold (Fig. 3A–3F). The quantitative analyses further confirmed that the cells were evenly distributed throughout the entire scaffold after 5 days culture (Fig. 3G). Meanwhile, we found that cell number increased with culture time (Fig. 3). These results indicate that the architecture of the 3D-NF-GS supports cell migration, proliferation, and even distribution inside these scaffolds.
We examined the expressions of Runx2, Col I, BSP, and OCN on the 3D-NF-GS constructs. Runx2 is an osteoblast-specific transcription factor that works intracellularly to upregulate a host of bone-specific genes (e.g. Col I and OCN). Runx2 protein is first detected in pre-osteoblasts, and the expression is upregulated in immature osteoblasts, but downregulated in mature osteoblasts.41 Col I gene produces type I collagen, a major ECM component of bone tissue.42,43 Also, bone sialoprotein (BSP) and osteocalcin (OCN) are considered the most sensitive markers for bone specific tissue formation.44,45 Col I and BSP are early markers of osteoblast differentiation, while OCN appears late, concomitantly with mineralization.46 On the 3D-NF-GS constructs, the expression of osteogenic genes (Runx2, Col I, BSP, and OCN) significantly increased when osteoblasts on the scaffold were cultured from 5 to 14 days (Fig. 4). Surprisingly, the distribution of BSP at protein level was distinct on the 3D-NF-GS as compared to 2D substrates. BSP on the scaffold was primarily present in the cell cytoplasm at 5 days and inside secretory vesicles at 2 weeks; whereas, most of the BSP on the 2D substrates was concentrated at the cell interface or on the focal adhesion sites (Fig. 4E–4G). This finding shows that the morphology of substrates (2D vs. 3D) affects not only the expression of cell adhesion proteins (e.g. β1 integrin, vinculin, and paxillin) on the materials, but also the distribution of other proteins (e.g. BSP) on the substrates. Further studies are under way to fully delineate the underlying mechanism and will be reported elsewhere.
In conventional 2D culture, primary osteoblasts start making nodules around 21 days.47 In our 3D-NF-GS, the cells were confluent by 14 days and started organizing as nodular aggregations inside the scaffold pores (Fig. 5B). The earlier nodule formation on the 3D-NF-GS is attributed to the nanofibrous architecture of the scaffold as our previous study has indicated that nanofibrous structure can significantly enhance osteoblast differentiation compared to smooth surfaces.40 This result confirmed that the 3D-NF-GS is a favorable substrate to enhance osteoblasts differentiation. Using TRITC-labeled 3D-NF-GS and confocal microscopy, we visualized the deposition and uniform distribution of Col I on the 3D scaffold. Based on the confocal microscopy, we further developed a technique to quantify the amount of deposited cellular matrix product on the 3D-NF-GS (Fig. 5F). For the mineralization of osteoblasts on the 3D-NF-GS, we incorporated a similar approach to both visualize and quantify the amount of alizarin on the 3D matrix (Fig. 6). Significantly higher amount of alizarin was detected for osteoblasts cultured on 3D-NF-GS for 14 days than for 5 days, which was confirmed by the calcium content analysis. Both alizarin and von Kossa staining showed that the mineralized nodules of osteoblasts uniformly distributed inside the entire 3D-NF-GS (Fig. 6).
In summary, our results demonstrate that the 3D-NF-GS are attractive scaffolds to support osteoblasts growth and uniform bone tissue formation. Furthermore, the uses of confocal microscopy to visualize, analyze, and quantify the cell adhesion, migration, proliferation, differentiation, and tissue formation on biomimetic 3D scaffolds provides a new approach to study cell-matrix interactions. This approach will help better understand osteoblast-material interactions in 3D nano-structured scaffolds and ultimately guide to develop optimal scaffolds for bone tissue engineering. Meanwhile, our image processing method to quantify osteoblast-matrix interactions can be also applied to other cell types.
CONCLUSIONS
Nanofibrous gelatin scaffolds (3D-NF-GS), which mimic the architecture of natural ECM, were developed and used as a biomimetic substrate to study osteoblast-matrix interactions in vitro. Using confocal microscopy, we visualized osteoblasts adhesion, migration, proliferation, differentiation, and mineralization on the 3D-NF-GS. Unlike the quick and numerous focal adhesion formation on the 2D substrates, osteoblasts seeded on the 3D-NF-GS showed less focal adhesions for the first 5 days. On the other hand, the migration and proliferation of osteoblasts on the 3D-NF-GS was considerably fast and cells distributed evenly throughout the 3D-NF-GS within 5 days. By 14 days, osteoblasts were organized as nodular aggregations that resided in the scaffold pores. Meanwhile, a large amount of type I collagen and other cell secretions covered and remodeled the surfaces of the 3D-NF-GS. The osteoblasts also mineralized the nodules inside the 3D-NF-GS after culture for 2 weeks. Our findings will aid a better understanding of osteoblast-material interactions in 3D nano-structured scaffolds and provide a guide to develop optimal scaffolds for bone tissue engineering.
ACKNOWLEDGEMENTS
This study was supported by financial support from the National Institute of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) - P30 DE020742. The authors thank Dr. Chunlin Qin for contribution of BSP mouse monoclonal antibody.
REFERENCES
- 1.Hynes RO. The Extracellular Matrix: Not Just Pretty Fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hutmacher DW. Scaffolds in tissue engineering bone and cartilage. Biomaterials. 2000;21:2529–2543. doi: 10.1016/s0142-9612(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 3.Owen SC, Shoichet MS. Design of three-dimensional biomimetic scaffolds. J Biomed Mater Res Part A. 2010;94A:1321–1331. doi: 10.1002/jbm.a.32834. [DOI] [PubMed] [Google Scholar]
- 4.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 5.Yamada KM. Cell- surface interactions with extracellular materials. Annu Rev Biochem. 1983;52:761–799. doi: 10.1146/annurev.bi.52.070183.003553. [DOI] [PubMed] [Google Scholar]
- 6.Fraley SI, Feng YF, Wirtz D, Longmore GD. Reply: reducing background fluorescence reveals adhesions in 3D matrices. Nat Cell Biol. 2011;13:3–5. doi: 10.1038/ncb0111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruoslahti E. Fibronectin and its receptors. Annu Rev Biochem. 1988;57:375–413. doi: 10.1146/annurev.bi.57.070188.002111. [DOI] [PubMed] [Google Scholar]
- 8.Adams JC, Watt FM. Regulation of development and differentiation by the extracellular-matrix. Development. 1993;117:1183–1198. doi: 10.1242/dev.117.4.1183. [DOI] [PubMed] [Google Scholar]
- 9.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. BBA-Mol Cell Res. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 11.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 12.Berrier AL, Yamada KM. Cell-matrix adhesion. J Cell Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 13.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 14.Guillame-Gentil O, Semenov O, Roca AS, Groth T, Zahn R, Voros J, Zenobi-Wong M. Engineering the Extracellular Environment: Strategies for Building 2D and 3D Cellular Structures. Adv Mater. 2010;22:5443–5462. doi: 10.1002/adma.201001747. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Castillejos R. Replication of the 3D architecture of tissues. Mater Today. 2010;13:32–41. [Google Scholar]
- 16.Kraning-Rush CM, Carey SP, Califano JP, Smith BN, Reinhart-King CA. The role of the cytoskeleton in cellular force generation in 2D and 3D environments. Phys Biol. 2011;8:1–9. doi: 10.1088/1478-3975/8/1/015009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 18.Zaman MH, Trapani LM, Siemeski A, MacKellar D, Gong HY, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn MS, Miller JS, West JL. Three-dimensional biochemical and biomechanical patterning of hydrogels for guiding cell behavior. Adv Mater. 2006;18:2679–2684. [Google Scholar]
- 21.Benoit DSW, Schwartz MP, Durney AR, Anseth KS. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat Mater. 2008;7:816–823. doi: 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KY, Mooney DJ. Hydrogels for tissue engineering. Chem Rev. 2001;101:1869–1879. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 23.Sikavitsas VI, Temenoff JS, Mikos AG. Biomaterials and bone mechanotransduction. Biomaterials. 2001;22:2581–2593. doi: 10.1016/s0142-9612(01)00002-3. [DOI] [PubMed] [Google Scholar]
- 24.Mooney DJ, Baldwin DF, Suh NP, Vacanti LP, Langer R. Novel approach to fabricate porous sponges of poly(D,L-lactic-co-glycolic acid) without the use of organic solvents. Biomaterials. 1996;17:1417–1422. doi: 10.1016/0142-9612(96)87284-x. [DOI] [PubMed] [Google Scholar]
- 25.Schuckert KH, Jopp S, Teoh SH. Mandibular Defect Reconstruction Using Three-Dimensional Polycaprolactone Scaffold in Combination with Platelet-Rich Plasma and Recombinant Human Bone Morphogenetic Protein-2: De Novo Synthesis of Bone in a Single Case. Tissue Eng Part A. 2009;15:493–499. doi: 10.1089/ten.tea.2008.0033. [DOI] [PubMed] [Google Scholar]
- 26.Ruhe PQ, Hedberg-Dirk EL, Padron NT, Spauwen PHM, Jansen JA, Mikos AG. Porous poly(DL-lactic-co-glycolic acid)/calcium phosphate cement composite for reconstruction of bone defects. Tissue Eng. 2006;12:789–800. doi: 10.1089/ten.2006.12.789. [DOI] [PubMed] [Google Scholar]
- 27.Kasper FK, Tanahashi K, Fisher JP, Mikos AG. Synthesis of poly(propylene fumarate) Nat Protoc. 2009;4:518–525. doi: 10.1038/nprot.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anselme K. Osteoblast adhesion on biomaterials. Biomaterials. 2000;21:667–681. doi: 10.1016/s0142-9612(99)00242-2. [DOI] [PubMed] [Google Scholar]
- 29.Liu XH, Ma PX. Polymeric scaffolds for bone tissue engineering. Ann Biomed Eng. 2004;32:477–486. doi: 10.1023/b:abme.0000017544.36001.8e. [DOI] [PubMed] [Google Scholar]
- 30.Liu XH, Ma PX. Phase separation, pore structure, and properties of nanofibrous gelatin scaffolds. Biomaterials. 2009;30:4094–4103. doi: 10.1016/j.biomaterials.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Smith LA, Hu J, Ma PX. Biomimetic nanofibrous gelatin/apatite composite scaffolds for bone tissue engineering. Biomaterials. 2009;30:2252–2258. doi: 10.1016/j.biomaterials.2008.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ecarotcharrier B, Glorieux FH, Vanderrest M, Pereira G. Osteoblasts isolated from mouse calvaria initiate matrix mineralization in culture. J Cell Biol. 1983;96:639–643. doi: 10.1083/jcb.96.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu XH, Smith L, Wei GB, Won YJ, Ma PX. Surface engineering of nano-fibrous poly(L-Lactic Acid) scaffolds via self-assembly technique for bone tissue engineering. J Biomed Nanotech. 2005;1:54–60. [Google Scholar]
- 34.Liu XH, Won YJ, Ma PX. Surface modification of interconnected porous scaffolds. J Biomed Mater Res Part A. 2005;74A:84–91. doi: 10.1002/jbm.a.30367. [DOI] [PubMed] [Google Scholar]
- 35.Feng B, Weng J, Yang BC, Qu SX, Zhang XD. Characterization of surface oxide films on titanium and adhesion of osteoblast. Biomaterials. 2003;24:4663–4670. doi: 10.1016/s0142-9612(03)00366-1. [DOI] [PubMed] [Google Scholar]
- 36.Miyamoto S, Akiyama SK, Yamada KM. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science. 1995;267:883–885. doi: 10.1126/science.7846531. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz MA. Integrin signaling revisited. Trends Cell Biol. 2001;11:466–470. doi: 10.1016/s0962-8924(01)02152-3. [DOI] [PubMed] [Google Scholar]
- 38.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 39.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 40.Hu J, Liu XH, Ma PX. Induction of osteoblast differentiation phenotype on poly(L-lactic acid) nanofibrous matrix. Biomaterials. 2008;29:3815–3821. doi: 10.1016/j.biomaterials.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komori T. Regulation of Osteoblast Differentiation by Runx2. In: Choi Y, editor. Osteoimmunology: Interactions of the Immune and Skeletal Systems Ii. Berlin: Springer-Verlag Berlin; 2010. pp. 43–49. [Google Scholar]
- 42.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 43.Olsen BR, Reginato AM, Wang WF. Bone development. Annu Rev Cell Dev Bio. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 44.Bianco P, Riminucci M, Silvestrini G, Bonucci E, Termine JD, Fisher LW, Robey PG. Localization of bone sialoprotein (BSP) to Golgi and post-Golgi secretory structures in osteoblasts and to discrete sites in early bone matrix. J Histochem Cytochem. 1993;41:193–203. doi: 10.1177/41.2.8419459. [DOI] [PubMed] [Google Scholar]
- 45.Machwate M, Jullienne A, Moukhtar M, Marie PJ. Temporal variation of c-Fos protooncogene expression during osteoblast diffferentiation and osteogenesis in developing rat bone. J Cell Biochem. 1995;57:62–70. doi: 10.1002/jcb.240570108. [DOI] [PubMed] [Google Scholar]
- 46.Lian JB, Stein GS. Development of the osteoblast phenotype: molecular mechanisms mediating osteoblast growth and differentiation. Iowa Orthop J. 1995;15:118–140. [PMC free article] [PubMed] [Google Scholar]
- 47.Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, Kennedy MB, Pockwinse S, Lian JB, Stein GS. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol. 1990;143:420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]