Abstract
A 16-amino acid oligopeptide forms a stable β-sheet structure in water. In physiological solutions it is able to self-assemble to form a macroscopic matrix that stains with Congo red. On raising the temperature of the aqueous solution above 70°C, an abrupt structural transition occurs in the CD spectra from a β-sheet to a stable α-helix without a detectable random-coil intermediate. With cooling, it retained the α-helical form and took several weeks at room temperature to partially return to the β-sheet form. Slow formation of the stable β-sheet structure thus shows kinetic irreversibility. Such a formation of very stable β-sheet structures is found in the amyloid of a number of neurological diseases. This oligopeptide could be a model system for studying the protein conformational changes that occurs in scrapie or Alzheimer disease. The abrupt and direct conversion from a β-sheet to an α-helix may also be found in other processes, such as protein folding and protein–protein interaction. Furthermore, such drastic structure changes may also be exploited in biomaterials designed as sensors to detect environmental changes.
Keywords: biomaterials, ionic residues, repeating sequences, scrapie related diseases, temperature-induced transition
Here we report an unusual phenomenon: a peptide organized in a β-sheet is able to convert directly into an α-helical conformation under the stimulus of temperature or pH changes. No evidence is found for the existence of a random-coil intermediate. The conversion is found in an oligopeptide DAR16-IV (see Table 1), which was first found to form a β-sheet. This type of structural transformation phenomenon is similar to that seen in a number of diseases, such as Alzheimer or scrapie. These diseases are associated with the accumulation of similarly stable β-sheet structures that are resistant to degradation. These substances, often called amyloid, stain with Congo red and represent a focal point in trying to understand the pathogenesis of these diseases.
Table 1.
Modulus IV self-complementary oligopeptides used in this study
| Name | Sequence | Structure |
|---|---|---|
| ++++−−−− | ||
| KAE16-IV | Ac-NH-KAKAKAKAEAEAEAEA-CONH2 | β |
| −−−−++++ | ||
| EAK16-IV | Ac-NH-AEAEAEAEAKAKAKAK-CONH2 | β |
| ++++−−−− | ||
| RAD16-IV | Ac-NH-RARARARADADADADA-CONH2 | β |
| −−−−++++ | ||
| DAR16-IV | Ac-NH-ADADADADARARARAR-CONH2 | α/β |
The oligopeptides were made by solid-phase synthesis on an Applied Biosystems model 430A. N and C termini of the oligopeptides are acetylated and amidated, respectively. The peptides were purified by HPLC. The purity was determined by analytical HPLC. The peptides were dissolved in water, and the concentrations were determined by the ninhydrin method with internal controls. Composition was confirmed by mass spectroscopy as described (16). These sequences were developed from our studies of modulus I and II molecules.
α-Helices and β-sheets are the major secondary structural motifs that organize the three-dimensional geometry of proteins (1, 2). The amino acids have been extensively studied for their ability to form either helical or sheet structures (3). Experiments have demonstrated that the sequence of amino acids plays a key role in determining the propensity for protein secondary structure (4–6). It is generally considered that secondary structures are stable once they are formed. These structures are believed to be of great importance in protein folding pathways that may be quite complex (7–10). It is well known that poly-lysine, poly-glutamate, and oligo-alanine can undergo secondary structure changes in different solvent environments, such as varying pH or salt concentration (11, 12). Furthermore, several block copolymers with identical composition but with different sequence—e.g., (Val–Lys)n, Ala20–Glu20–Phe8, Glu20–Ala20–Phe8—and others form helical structures with quite different content in the same environment (13, 14).
We have previously described a class of ionic self-complementary oligopeptides. This class of oligopeptides forms β-sheet structures that are exceedingly stable in some extreme conditions including high temperature, a wide pH range, high concentrations of denaturation reagents, and a variety of proteases (15–17). The β-sheet forming ionic complementary oligopeptides have repeating sequences and are classified by the repetitions of ionic charge groups. These β-sheets have a hydrophobic side and a charged hydrophilic side that has alternating + and − charged amino acid residues. For example, molecules of modulus I have − + − + − + − +; modulus II, − − + + − − + +; modulus IV, − − − − + + + +, etc. Several β-sheet ionic self-complementary oligopeptides including modulus I, II, and IV can spontaneously self-assemble to form macroscopic matrices in the presence of monovalent alkaline salts. These matrices are readily stained by Congo red and are visible to the naked eye (15–17). In the scanning electron microscope, it was shown that the matrices are like felt interwoven from individual fibers ≈10–20 nm in diameter with 50- to 100-nm pores. A variety of mammalian cells have been found to utilize these peptide matrices as scaffolding for cell attachment (17).
MATERIALS AND METHODS
Materials.
All oligopeptides used in this study (EAK16-IV, KAE16-IV, DAR16-IV, and RAD16-IV; Table 1) were synthesized by t-Boc chemistry on an Applied Biosystems automated model 430A peptide synthesizer and purified by reverse phase HPLC at the Biopolymers Laboratory at the Massachusetts Institute of Technology (16). The peptides were dissolved in deionized water and stored at room temperature. The concentrations were determined by amino acids hydrolysis as described (16).
CD Analysis.
CD spectra of serial dilution of either the α- and β-form of DAR16-IV peptide solutions showed that these structures are stable even at 0.7 μm in water. The value of mean residue ellipiticity at [θ]218nm for β and [θ]222nm for α versus the concentration remain essentially unchanged in both cases. On dilution the α-form has an isosbestic point at 198 nm and the β-form has an isosbestic point at 205 nm. Two pairs of modulus IV self-complementary oligopeptides with identical compositions and length, EAK16-IV, KAE16-IV and DAR16-IV, RAD16-IV have been systematically analyzed. To measure the effect of various pH on the peptides, they were incubated overnight at room temperature in solutions with pH range from 1–12 before measuring with the CD. For ionic strength changes, a peptide solution was divided equally into six tubes and NaCl was adjusted to a final concentration from 10 mM to 160 mM. These peptide solutions were then heated for 32 min at 75°C and cooled to 25°C before measurement.
Sedimentation Equilibrium Analysis.
For determination of molecular weight, the α-helical form of DAR16-IV oligopeptide was heated in water to 75°C for 35 min and its α-helical structure was confirmed by CD. Three different concentrations of the oligopeptide were then loaded into a hexa-cell chamber with three cells containing water as the references. The samples were then centrifuged at 50,000 rpm at 25°C for 24 h to sedimentation equilibrium. Each sample was then measured several times independently. All points were collected and analyzed. In all cases, the samples showed an average molecular mass 1870 which is close to the calculated monomeric molecular mass of 1670 Da. We therefore concluded the helical form of DAR16-IV is a monomer in water.
Data Base Search.
A search of the current protein data bases using blast through the internet did not identify a sequence identical to that of DAR16-IV. However, there are many segments containing clusters of alternating charged repeating residues in a number of proteins with charge distributions similar to DAR and EAK.
Other Analysis.
Preliminary NMR examination of DAR16-IV using either heated or unheated samples showed distinctive spectra suggesting that the secondary structures of the molecules differ from each other. A systematic analysis with NMR will be reported elsewhere.
RESULTS
Structure Properties.
We have previously described this class of ionic self-complementary oligopeptides which form remarkably stable β-sheets and can self-assemble to form macroscopic matrices that stained with Congo red (15–17). Every other residue in the peptide is hydrophobic, such as alanine, and the other residues are repeating clusters of positively and negatively charged residues. If the ionic residues alternate with two positive and two negative residues, they are described as modulus II (15–17). One of the modulus II oligopeptides, EAK16-II [AEAEAKAKAEAEAKAK], was originally found in a yeast protein, zuotin, which was characterized as a putative left-handed Z-DNA binding protein (18). Several modulus II and modulus I peptides have been systematically analyzed (S.Z., unpublished data). Recently, we have studied several modulus IV peptides containing 16 amino acids, EAK16-IV, KAE16-IV, DAR16-IV, and RAD16-IV (Table 1). All four oligopeptides displayed typical β-sheet CD spectra at ambient temperature. However, when the solutions were heated, they exhibited different behavior. For example, both EAK16-IV and KAE16-IV, similar to other members of the class, are extremely stable even at high temperature. Their β-sheet CD spectra were essentially unchanged from 20°C to 90°C (Fig. 1A). In the case of RAD16-IV, after heating at 90°C for 10 min, the room temperature CD spectrum showed that the β-sheet content remained essentially unchanged as indicated by the ellipiticity at 218 nm. However the ellipiticity at the 195-nm region was apparently reduced. This reduction probably reflects a change in the right-handed twist of the β-strands (Fig. 1B) because almost all β-strands in proteins are believed to possess some degrees of right-handed twist (19, 20).
Figure 1.
Thermal stability of ionic self-complementary oligopeptides (Table 1). (A) CD spectra of EAK16-IV and KAE16-IV at 20°C and 90°C after heating for 10 min. These spectra are superimposable except slight variations in the 195-nm region. ○ and •, EAK16-IV at 20°C and 90°C, respectively; ⋄ and ♦, KAE16-IV at 20°C and 90°C, respectively. (B) CD spectra of RAD16-IV at 20°C before heating and at 20°C after heating at 90°C for 10 min. It has a β-sheet spectrum at 20°C (○; however, after heating at 90°C (•), the CD spectrum of RAD16-IV has a reduced ellipiticity near 195 nm, probably due to a change in the twist on the β-sheet backbone.
An Abrupt Structure Conversion.
A more surprising result was found with the peptide DAR16-IV. On incubation at elevated temperature, it underwent a structural transition from a β-sheet directly to an α-helix without an observable random-coil intermediate. Thus the secondary structure of the oligopeptide had drastically changed as a function of temperature (Fig. 2). The β to α transition was first observed by measuring the CD spectra of the peptide solution at different temperatures in the heating chamber of the instrument. At elevated temperature, the β-sheet spectrum was replaced by an α-helix spectrum. We also examined the DAR16-IV solution by heating it at 90°C for 10 min in a water bath and took its CD spectrum at 20°C; a distinctive structure transition was observed (Fig. 2A). To find out more about the transition, several identical samples were heated at 75°C for various time periods and their spectra were measured at 20°C (Fig. 2B). No transition was observed on heating from 1 to 8 min. However, changes were seen after a longer period of heating and the transition was complete after 32 min of heating. Prolonged incubation did not further promote the transition. Once the α-helical structure is formed, it is quite stable. When the DAR16-IV oligopeptide in helical form was re-examined by reheating at different temperatures, the helical content was gradually reduced as a function of incubation temperature but it did not return to the β-sheet form nor completely denature into a random coil (Fig. 2C). The mean residue ellipiticity [θ]222nm showed that ≈60% of peptide was in the α-helical form at 0°C and 30% remained at 90°C. Furthermore, after the helical structure is formed, it is very stable at room temperature. After several weeks only a small fraction had returned to the β-sheet form. This delayed hysteresis suggests that the structural conversion is kinetically irreversible. It should be noted that slow formation of a stable β-sheet is found in the pathogenesis of a number of protein conformational diseases.
Figure 2.
Temperature-induced secondary structural transition in DAR16-IV. (A) CD spectra were measured at 20°C (○) with an identical sample heated at 90°C (•) for 10 min. The heated sample has an apparent α-helical CD spectrum as indicated. (B) Seven identical samples of DAR16-IV were heated at 75°C for different time periods in minutes as indicated and returned to 20°C. Small differences are seen in the spectra between unheated and those heated for 1, 2, and 4 min. The transition started after 8 min of heating and was completed after 32 min of heating. No further changes were observed for longer period of heating. (C) Stability of the α-helical structure. The helical form of oligopeptide DAR16-IV was heated and the spectra were measured at 10°C intervals up to 90°C. The peptide solution remained at each temperature for ≈10 min, and the temperature was held constant for each spectrum. (Inset) Decrease of the mean residue ellipiticity at [θ]222nm (103 deg·cm2/decimole) as a function of temperature. At 0°C, ≈60% of the peptide was in the helical form, while at 90°C, it was reduced to 30%. Thus the helical form was not completely denatured at the high temperature.
Structural Stability at Low Concentrations.
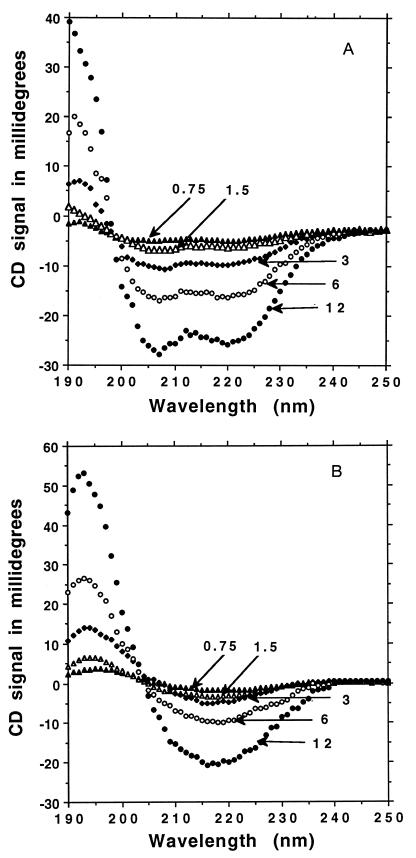
Both the α-helical and the β-sheet forms of DAR16-IV are stable at submicromolar concentrations. The oligopeptide solutions were serially diluted with water and their CD spectra were measured. The spectra showed a typical α-helical profile with an isosbestic point at 198 nm for the helical form (Fig. 3A). This result is not surprising because many short α-helical structures are monomeric and are not sensitive to concentration changes. This finding is in agreement with other helical oligopeptides of similar length (21, 22). The existence of DAR16-IV as a monomer in the α-helical form is consistent with the analytical equilibrium sedimentation centrifugation as described in Materials and Methods. Interestingly, the β-sheet structure was likewise unaffected by dilution of the oligopeptide and CD spectra had an isosbestic point at 205 nm (Fig. 3B). This is unusual because β-sheet structures of oligopeptides are in general affected by changes in concentration. β-Sheet structures consist of short sequences largely stabilized by intermolecular interactions that usually disassociated upon dilution. However, this result is in a good agreement with our previous analysis of this class of β-sheet ionic self-complementary oligopeptides in which very stable β-sheet structures are largely independent of peptide concentration (16). This may be related to their architecture in which repetitive + and − charged residues can form ionic bonds on one side of the sheet while the alanines on the opposite hydrophobic side form staggered van de Waals interactions; part of the structure resembles interactions found in silk (23, 24).
Figure 3.
Structural stability at various concentrations from 12 to 0.75 μm. (A) DAR16-IV after heating forms a stable α-helical structure at serial dilution with an isosbestic point at 198 nm. (B) DAR16-IV without heating forms a stable β-sheet structure at serial dilution with an isosbestic point at 204 nm.
Ionic Effect.
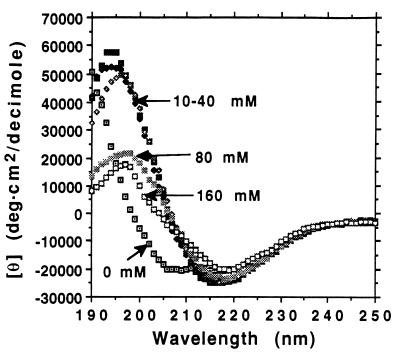
Ionic strength can inhibit the structural transition. Addition of NaCl to 10 mM or greater effectively inhibited the β- to α-structure transition (Fig. 4). NaCl at 10, 20, and 40 mM inhibited the structural conversion. However, increasing the NaCl concentration to 80 and 160 mM resulted in reducing the β-sheet structure CD spectra in the 195-nm region. This change may reflect the slight structural alteration of the backbone twists of the β-sheet peptide. Salts such as NaCl and KF have been shown to stabilize the β-sheet structures, and for some peptides they even induce formation of a β-sheet from a random coil or an α-helix (13, 14). In this case, NaCl not only inhibits the structure conversion, but also induces the peptide self-assembly into a peptide matrix that can be stained Congo red (data not shown) (15–17).
Figure 4.
Effect of ionic strength. Six identical tubes containing DAR16-IV peptide were incubated with various concentration of NaCl and heated at 75°C for 45 min. The NaCl effectively inhibited the conversion of the structure from β-sheet to α-helix. Without NaCl, it has a helical spectrum. Ten, 20, and 40 mM NaCl stabilized the β-sheet structure. Higher concentration of NaCl (80 and 160 mM) not only stabilized the β-sheet, but also altered the twist of the peptide backbone. We also observed that when DAR16-IV is in β-sheet structure, it forms a macroscopic matrix, whereas it does not when it is in the helical form.
pH Effect.
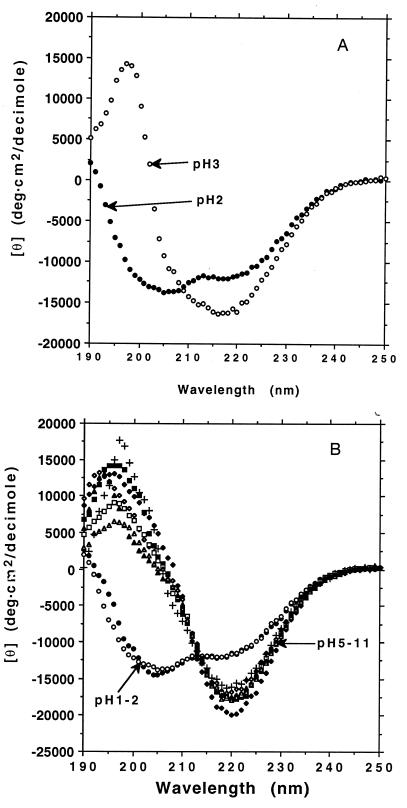
We also investigated secondary structural conformational changes as a function of pH. Our previous study of EAK16-II and several other ionic self-complementary oligopeptides at various pH values showed that their β-sheet conformation was quite stable over a wide range (16). When oligopeptide DAR16-IV was incubated at a pH of 1 and 2, it showed a helical structure. When DAR16-IV was incubated at pH 3 and 4, the CD spectra displayed a β-sheet form but without a clear isosbestic point, suggesting that there may be some structural intermediates near pH 3 (Fig. 5A). However, when the peptide was incubated in solutions from pH 5–11, the CD spectra exhibited a typical β-sheet form with an isosbestic point at 212 nm and a gradual decrease of ellipiticity as a function of increasing pH (Fig. 5B). These observations are consistent with previous reports that polymeric peptides, such as polylysine can readily undergo secondary structural transitions as a function of pH (11, 12).
Figure 5.
The effect of pH on secondary structure. DAR16-IV can adopt two distinctive secondary structures at different pH conditions. (A) DAR16-IV was incubated at pH 2 and at pH 3 overnight at 23°C. It shows an α-helical CD spectrum at pH 2 but remained in the β-form at pH 3. (B) CD spectra of DAR16-IV at pH values from 1, 2, to 5–11. The α-helical spectra of DAR16-IV at pH 1 and pH 2 are similar. The β-form spectra in pH 5–11 are quite similar as indicated by the isosbestic point at 212 nm. The differences suggest a change in the backbone twist of the β-sheet from 5 to 11. Symbols and pH values are as follows: •, pH 1; ○, pH 2; ♦, pH 5; ⋄, pH 6; ▪, pH7; □, pH 8; ▵, pH 9; ▴, pH 10; +, pH 11.
DISCUSSION
Almost all β-sheet ionic self-complementary oligopeptides studied to date spontaneously assemble to form a macroscopic matrix in the presence of monovalent salts (15–17). We also tested the property of matrix formation of DAR16-IV in the β-sheet form. When DAR16-IV was tested in phosphate-based saline at room temperature, it readily formed a visible matrix that stained with Congo red; however, it failed to form such a matrix after the same sample was heated at 75°C or higher temperature for 30 min (data not shown). These observations are consistent with our previous observations that the β-sheet structure is an important structural component in facilitating its self-assembly to a macroscopic structure. The unique structural feature of this class of ionic β-sheet oligopeptides is characterized by hydrophobic residues entirely on one side of the β-sheet (alanine, in this case) and both positively and negatively charged residues on the other side which vary in an alternating repeating pattern. In the case of DAR16-IV there are four negatively charged aspartic acid residues at the N terminus and four positively charged arginines at the C terminus. It is interesting that this β to α conversion does not occur when the polarity of the polypeptide is reversed, as in the case of RAD16-IV. This suggests that the orientation of the α-helix dipole moment may contribute to stabilization of the α-helix in one case but not in the other. Previous reports have shown that the α-helix has a dipole moment with the positive pole at the N terminus and the negative pole at the C terminus (25). It is interestingly that negatively charged residues, Asp and Glu, are more frequently found located at the N terminus of the helices in proteins, whereas the positively charged residues, Arg, Lys, and His, tend to be found at the C terminus of the helices (25, 26).
Structure Conversion.
There appears to be a threshold effect involved in the conversion as shown in Fig. 2B. Heating DAR16-IV at 75°C produces very little change in the CD spectra for the first 8 min, but the change to an α-helix is essentially completed after 32 min of heating. Thus it takes some time to thermally disrupt the β-sheet lattice to free peptides to form the α-helix. Likewise, when the β-sheet is destabilized by lowering the pH, the destabilization only begins to become evident in the CD spectra when the pH is lowered below 4, and is essentially complete when the pH is dropped to 2. It is likely in this case that the aspartate residues are gradually becoming protonated, and that they destroy the stability of the ionic, self-complementary β-sheet interactions.
Implications in Amyloid Formation.
Once the α-helix is formed, it takes many weeks at room temperature to slowly reform the β-sheet structure. The cycle of β-sheet formation disrupted by heat to form an α-helix, and then upon cooling the slow reformation of the β-sheet shows considerable hysteresis; the process is not kinetically reversible. The rate of reforming the sheet at room temperature is much slower than the rate at which the helix forms once the β-sheet is disrupted. This slow process of going from an α-helix to β-sheet is one that is studied widely at present because it is believed to form the molecular basis of a number of disorders, including amyloid formation in neurological tissues in diseases such as scrapie, bovine spongiform encephalitis, or Alzheimer disease (27–34). There it is postulated that secondary structural conformation changes occur in which a cleaved segment of a protein that is normally found in an α-helical form, is converted to a stable β-sheet. Many investigators believe that these protein conformation changes are central to the disease process. Thus the progression of the disease may be related to changes in protein secondary structure leading to the formation of insoluble β-sheet plaques. In the infectious diseases such as scrapie, a segment of the protein in the β-sheet form (a prion) is believed to catalyze the further conversion of segments from α-helix to the β-sheet form (31, 32). By further studying DAR16-IV and its derivatives we may perhaps gain some insight into factors that influence these conformational changes and this might have relevance to the pathogenesis of these diseases. It is interesting that these diseases seem to be characterized by a slow conversion of protein into an insoluble β-sheet form, and DAR16-IV shows similarly slow kinetics of conversion.
In contrast, the β to α conversion is fast, acting like a molecular switch with a dramatic conformational change. This type of conformational switch may also occur in proteins. It could be an element in protein folding, or it might even occur in some proteins during the mechanism of their action. An example of a large conformational change is found in the hemagglutinin molecule where membrane fusion is brought about through a pH drop in endosomes accompanied by conversion of an unstructured peptide segment into an α-helix (35, 36). The β to α conversion we are describing here results in a considerable geometrical change; DAR16-IV has an extended β-sheet length of ≈5 nm compared with the helical length of ≈2.5 nm. Thus, changes of this type might occur in protein mechanisms with significant shape changes.
Structural transitions also have been found in other systems. Perutz and coworkers (37, 38) have reported that glutamine repeats found in Huntington and several other neurological diseases form β-sheets with side chains forming complementary hydrogen bonds. One of these repeats changes its conformation from a β-sheet to a type I β-turn at high temperature (37, 38). These are some of the examples in which protein conformation can be altered as a consequence of perturbations in the environment of a particular sequence within a protein. This phenomenon may be more widespread than previously realized, as nature is likely to exploit these properties.
Applications in Biomedical Engineering.
Such a drastic structural change from a β-sheet to an α-helix found in DAR16-IV, with a 50% change in distance, from 5 nm to 2.5 nm in a 16-residue peptide, may also be exploited for biomedical engineering applications. For example, one can design peptide biosensors so that they can rapidly response to in vivo or in vitro environmental pH or temperature changes. This type of biosensor may be further developed for a variety of diagnostic devices.
Acknowledgments
We thank Drs. Peter Kim, Jonathan King, Martin Egli, Michael Hecht, Joël Janin, Joel Sussman, and Jan Hermans for helpful discussions. We also thank Richard Cook and Heather LeBanc for helping the synthesis and purification of the oligopeptides, and Massachusetts Institute of Technology undergraduate student Anya Hawrylchak for some technical assistance. This work is supported by grants from Hercules, Inc., and the U.S. Army Research Office.
References
- 1.Pauling L C. The Nature of Chemical Bond. 3rd Ed. New York: Cornell Univ. Press; 1961. [Google Scholar]
- 2.Creighton T E. Proteins: Structures and Molecular Principles. 2nd Ed. New York: Freeman; 1993. [Google Scholar]
- 3.Chou P Y, Fasman G D. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 4.Williams R W, Chang A, Juretic D, Loughran S. Biochim Biophys Acta. 1987;916:200–204. doi: 10.1016/0167-4838(87)90109-9. [DOI] [PubMed] [Google Scholar]
- 5.Minor D, Kim P S. Nature (London) 1994;371:264–267. doi: 10.1038/371264a0. [DOI] [PubMed] [Google Scholar]
- 6.Xiong H, Buckwalter B L, Shieh H M, Hecht M H. Proc Natl Acad Sci USA. 1995;92:6349–6353. doi: 10.1073/pnas.92.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim P S, Baldwin R L. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- 8.Fersht A R. Proc Natl Acad Sci USA. 1995;92:10869–10873. doi: 10.1073/pnas.92.24.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dill K A, Fiebig K M, Chan H S. Proc Natl Sci USA. 1993;90:1924–1928. doi: 10.1073/pnas.90.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dill K A. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 11.Greenfield N, Fasman G. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 12.Hartman R, Schwaner R C, Hermans J. J Mol Biol. 1974;90:415–429. doi: 10.1016/0022-2836(74)90225-3. [DOI] [PubMed] [Google Scholar]
- 13.Brack A, Orgel L. Nature (London) 1975;256:383–387. doi: 10.1038/256383a0. [DOI] [PubMed] [Google Scholar]
- 14.Ihara S, Ooi T, Takahashi S. Biopolymers. 1982;21:131–145. [Google Scholar]
- 15.Zhang S, Holmes T, Lockshin C, Rich A. Proc Natl Acad Sci USA. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Lockshin C, Cook R, Rich A. Biopolymers. 1994;34:663–672. doi: 10.1002/bip.360340508. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Holmes T C, DiPersio C M, Hynes R O, Su X, Rich A. Biomaterials. 1995;16:1386–1393. doi: 10.1016/0142-9612(95)96874-y. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Lockshin C, Herbert A, Winter E, Rich A. EMBO J. 1992;11:3787–3796. doi: 10.1002/j.1460-2075.1992.tb05464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chothia C. J Mol Biol. 1973;75:295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- 20.Manning M C, Illangasekare M, Woody R W. Biophys Chem. 1974;31:77–86. doi: 10.1016/0301-4622(88)80011-5. [DOI] [PubMed] [Google Scholar]
- 21.Marqusee S, Baldwin R L. Proc Natl Acad Sci USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marqusee S, Robbins V H, Baldwin R L. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astbury W T. Nature (London) 1936;137:803–805. [Google Scholar]
- 24.Matthews C K, van Holde K E. Biochemistry. Redwood City, CA: Benjamin & Cummings; 1990. p. 179. [Google Scholar]
- 25.Hol W G J. Prog Biophys Mol Biol. 1985;145:149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 26.Chou P Y, Fasman G D. Biochemistry. 1974;13:211–221. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 27.Zagorski M G, Barrow C J. Biochemistry. 1992;31:5621–5631. doi: 10.1021/bi00139a028. [DOI] [PubMed] [Google Scholar]
- 28.Hilbich C, Kisters-Woike B, Reed J, Masters C L, Beyreuther K. J Mol Biol. 1991;50:149–165. doi: 10.1016/0022-2836(91)90881-6. [DOI] [PubMed] [Google Scholar]
- 29.Kirschner D A, Inouye H, Duffy L K, Sinclair A, Lind M, Selkoe D J. Proc Natl Acad Sci USA. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prusiner S B. Science. 1991;252:1515–1517. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 31.Gasset M, Baldwin M A, Lloyd D H, Gabriel J-M, Holtzman D M, Cohen F, Fletterick R, Prusiner S B. Proc Natl Acad Sci USA. 1992;89:10940–10944. doi: 10.1073/pnas.89.22.10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Z, Prusiner S B, Cohen F E. Folding Design. 1996;1:13–20. doi: 10.1016/S1359-0278(96)00007-7. [DOI] [PubMed] [Google Scholar]
- 33.Han H, Weinreb P H, Lansbury P T. Chem Biol. 1995;2:163–169. doi: 10.1016/1074-5521(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 34.Safar J, Roller P P, Gajdusek D C, Gibbs C J., Jr Protein Sci. 1993;2:2206–2216. doi: 10.1002/pro.5560021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carr C M, Kim P S. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 36.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 37.Perutz M. Proc Natl Acad Sci USA. 1994;91:5355–5359. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott K, Perutz M. Proc Natl Acad Sci USA. 1995;92:6509–6513. doi: 10.1073/pnas.92.14.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]