Abstract
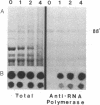
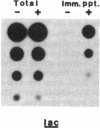
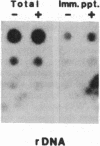
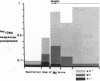
We present an approach for determining the in vivo distribution of a protein on specific segments of chromosomal DNA. First, proteins are joined covalently to DNA by irradiating intact cells with UV light. Second, these cells are disrupted in detergent, and a specific protein is immunoprecipitated from the lysate. Third, the DNA that is covalently attached to the protein in the precipitate is purified and assayed by hybridization. To test this approach, we examine the cross-linking in Escherichia coli of RNA polymerase to a constitutively expressed, lambda cI gene, and to the uninduced and isopropyl beta-D-thiogalactoside (IPTG)-induced lac operon. As expected, the recovery of the constitutively expressed gene in the immunoprecipitate is dependent on the irradiation of cells and on the addition of RNA polymerase antiserum. The recovery of the lac operon DNA also requires transcriptional activation with IPTG prior to the cross-linking step. After these initial tests, we examine the distribution of RNA polymerase on the leucine operon of Salmonella in wild-type, attenuator mutant, and promoter mutant strains. Our in vivo data are in complete agreement with the predictions of the attenuation model of regulation. From these and other experiments, we discuss the resolution, sensitivity, and generality of these methods.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Bremer H., Yuan D. RNA chain growth-rate in Escherichia coli. J Mol Biol. 1968 Dec 14;38(2):163–180. doi: 10.1016/0022-2836(68)90404-x. [DOI] [PubMed] [Google Scholar]
- Bretscher A. Purification of an 80,000-dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J Cell Biol. 1983 Aug;97(2):425–432. doi: 10.1083/jcb.97.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Cao T. M., Sung M. T. Ultraviolet light induced preferential cross-linking of histone H3 to deoxyribonucleic acid in chromatin and nuclei of chicken erythrocytes. Biochemistry. 1982 Jul 6;21(14):3419–3427. doi: 10.1021/bi00257a027. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. The selectivity of transcription. Annu Rev Biochem. 1974;43(0):721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Friedberg D., Mikulka T. W., Jones J., Calvo J. M. flrB, a regulatory locus controlling branched-chain amino acid biosynthesis in Salmonella typhimurium. J Bacteriol. 1974 Jun;118(3):942–951. doi: 10.1128/jb.118.3.942-951.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill R. M., Jones J. W., Haughn G. W., Calvo J. M. Transcription initiation sites of the leucine operons of Salmonella typhimurium and Escherichia coli. J Mol Biol. 1983 Oct 15;170(1):39–59. doi: 10.1016/s0022-2836(83)80226-5. [DOI] [PubMed] [Google Scholar]
- Gemmill R. M., Wessler S. R., Keller E. B., Calvo J. M. leu operon of Salmonella typhimurium is controlled by an attenuation mechanism. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4941–4945. doi: 10.1073/pnas.76.10.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M., Oppenheim A., Court D. Retroregulation: control of gene expression from sites distal to the gene. Cell. 1982 Jul;29(3):727–728. doi: 10.1016/0092-8674(82)90434-2. [DOI] [PubMed] [Google Scholar]
- Harrison C. A., Turner D. H., Hinkle D. C. Laser crosslinking of E. coli RNA polymerase and T7 DNA. Nucleic Acids Res. 1982 Apr 10;10(7):2399–2414. doi: 10.1093/nar/10.7.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havron A., Sperling J. Specificity of photochemical cross-linking in protein-nucleic acid complexes: identification of the interacting residues in RNase- pyrimidine nucleotide complex. Biochemistry. 1977 Dec 13;16(25):5631–5635. doi: 10.1021/bi00644a038. [DOI] [PubMed] [Google Scholar]
- KEPES A. KINETICS OF INDUCED ENZYME SYNTHESIS. DETERMINATION OF THE MEAN LIFE OF GALACTOSIDASE-SPECIFIC MESSENGER RNA. Biochim Biophys Acta. 1963 Oct 15;76:293–309. [PubMed] [Google Scholar]
- Keller E. B., Calvo J. M. Alternative secondary structures of leader RNAs and the regulation of the trp, phe, his, thr, and leu operons. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6186–6190. doi: 10.1073/pnas.76.12.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Szybalski W. Orientation of transcription of the lac operon and its repressor gene in Escherichia coli. J Mol Biol. 1969 Feb 28;40(1):145–151. doi: 10.1016/0022-2836(69)90303-9. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pace N. R. Structure and synthesis of the ribosomal ribonucleic acid of prokaryotes. Bacteriol Rev. 1973 Dec;37(4):562–603. doi: 10.1128/br.37.4.562-603.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. S., Hillel Z., Wu C. W. Molecular mechanism of promoter selection in gene transcription. I. Development of a rapid mixing-photocrosslinking technique to study the kinetics of Escherichia coli RNA polymerase binding to T7 DNA. J Biol Chem. 1982 Jun 25;257(12):6944–6949. [PubMed] [Google Scholar]
- Payvar F., Firestone G. L., Ross S. R., Chandler V. L., Wrange O., Carlstedt-Duke J., Gustafsson J. A., Yamamoto K. R. Multiple specific binding sites for purified glucocorticoid receptors on mammary tumor virus DNA. J Cell Biochem. 1982;19(3):241–247. doi: 10.1002/jcb.240190305. [DOI] [PubMed] [Google Scholar]
- Polisky B., Bishop R. J., Gelfand D. H. A plasmid cloning vehicle allowing regulated expression of eukaryotic DNA in bacteria. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3900–3904. doi: 10.1073/pnas.73.11.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabkin S. D., Moore P. D., Strauss B. S. In vitro bypass of UV-induced lesions by Escherichia coli DNA polymerase I: specificity of nucleotide incorporation. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1541–1545. doi: 10.1073/pnas.80.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Searles L. L., Wessler S. R., Calvo J. M. Transcription attenuation is the major mechanism by which the leu operon of Salmonella typhimurium is controlled. J Mol Biol. 1983 Jan 25;163(3):377–394. doi: 10.1016/0022-2836(83)90064-5. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Simpson R. B. The molecular topography of RNA polymerase-promoter interaction. Cell. 1979 Oct;18(2):277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- Smith K. C., Meun D. H. Kinetics of the photochemical addition of [35S] cysteine to polynucleotides and nucleic acids. Biochemistry. 1968 Mar;7(3):1033–1037. doi: 10.1021/bi00843a023. [DOI] [PubMed] [Google Scholar]
- Smith K. C. Photochemical addition of amino acids to 14C-uracil. Biochem Biophys Res Commun. 1969 Feb 7;34(3):354–357. doi: 10.1016/0006-291x(69)90840-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]