Abstract
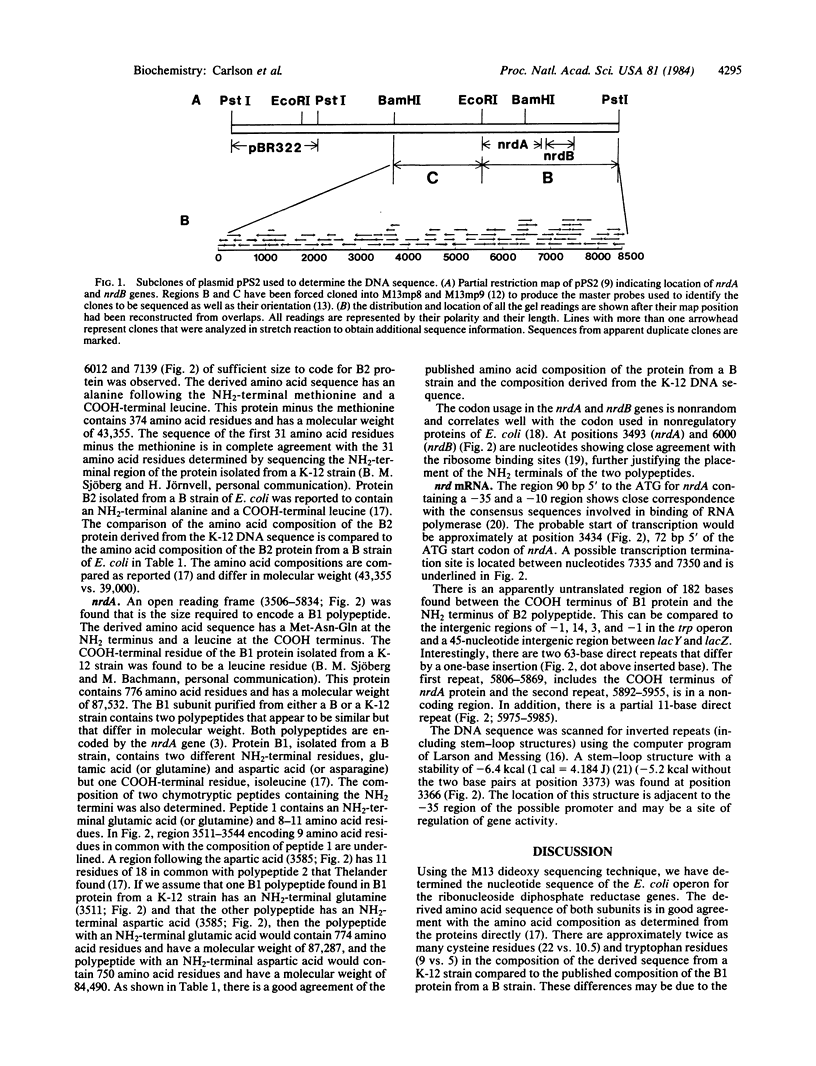
The nucleotide sequence of the Escherichia coli K-12 DNA comprising the operon for the structural genes of the subunits of ribonucleotide diphosphate reductase has been determined. The DNA sequenced maps at 48.5 minutes on the E. coli chromosome and includes a total length of 8557 nucleotides. An open reading frame between nucleotides 3506 and 5834, encoding a 776-amino acid polypeptide chain with a molecular weight of 87,532, has been identified as the nrdA gene. An open reading frame between nucleotides 6012 and 7139, encoding a 375-amino acid polypeptide with a molecular weight of 43,466, has been identified as the nrdB gene. The sequences reveal not only the primary structures for both subunits, but also some interesting aspects of potential regulatory sites.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Eriksson S. Direct photoaffinity labeling of ribonucleotide reductase from Escherichia coli. Evidence for enhanced binding of the allosteric effector dTTP by the presence of substrates. J Biol Chem. 1983 May 10;258(9):5674–5678. [PubMed] [Google Scholar]
- Eriksson S., Sjöberg B. M., Hahne S. Ribonucleoside diphosphate reductase from Escherichia coli. An immunological assay and a novel purification from an overproducing strain lysogenic for phage lambdadnrd. J Biol Chem. 1977 Sep 10;252(17):6132–6138. [PubMed] [Google Scholar]
- Fuchs J. A., Karlström H. O. A mutant of Escherichia coli defective in ribonucleosidediphosphate reductase. 2. Characterization of the enzymatic defect. Eur J Biochem. 1973 Feb 1;32(3):457–462. [PubMed] [Google Scholar]
- Fuchs J. A., Karlström H. O. Mapping of nrdA and nrdB in Escherichia coli K-12. J Bacteriol. 1976 Dec;128(3):810–814. doi: 10.1128/jb.128.3.810-814.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J. A., Karlström H. O., Warner H. R., Reichard P. Defective gene product in dnaF mutant of Escherichia coli. Nat New Biol. 1972 Jul 19;238(81):69–71. doi: 10.1038/newbio238069a0. [DOI] [PubMed] [Google Scholar]
- Hanke P. D., Fuchs J. A. Characterization of the mRNA coding for ribonucleoside diphosphate reductase in Escherichia coli. J Bacteriol. 1983 Dec;156(3):1192–1197. doi: 10.1128/jb.156.3.1192-1197.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke P. D., Fuchs J. A. Requirement of protein synthesis for the induction of ribonucleoside diphosphate reductase mRNA in Escherichia coli. Mol Gen Genet. 1984;193(2):327–331. doi: 10.1007/BF00330689. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Phillips S., Edgell M. H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978 Sep 25;253(18):6551–6560. [PubMed] [Google Scholar]
- Hög J. O., Jörnvall H., Holmgren A., Carlquist M., Persson M. The primary structure of Escherichia coli glutaredoxin. Distant homology with thioredoxins in a superfamily of small proteins with a redox-active cystine disulfide/cysteine dithiol. Eur J Biochem. 1983 Oct 17;136(1):223–232. doi: 10.1111/j.1432-1033.1983.tb07730.x. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R., Messing J. Apple II computer software for DNA and protein sequence data. DNA. 1983;2(1):31–35. doi: 10.1089/dna.1.1983.2.31. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Platz A., Sjöberg B. M. Construction and characterization of hybrid plasmids containing the Escherichia coli nrd region. J Bacteriol. 1980 Aug;143(2):561–568. doi: 10.1128/jb.143.2.561-568.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Thelander L., Larsson B. Active site of ribonucleoside diphosphate reductase from Escherichia coli. Inactivation of the enzyme by 2'-substituted ribonucleoside diphosphates. J Biol Chem. 1976 Mar 10;251(5):1398–1405. [PubMed] [Google Scholar]
- Thelander L. Physicochemical characterization of ribonucleoside diphosphate reductase from Escherichia coli. J Biol Chem. 1973 Jul 10;248(13):4591–4601. [PubMed] [Google Scholar]
- Thelander L., Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]



