Abstract
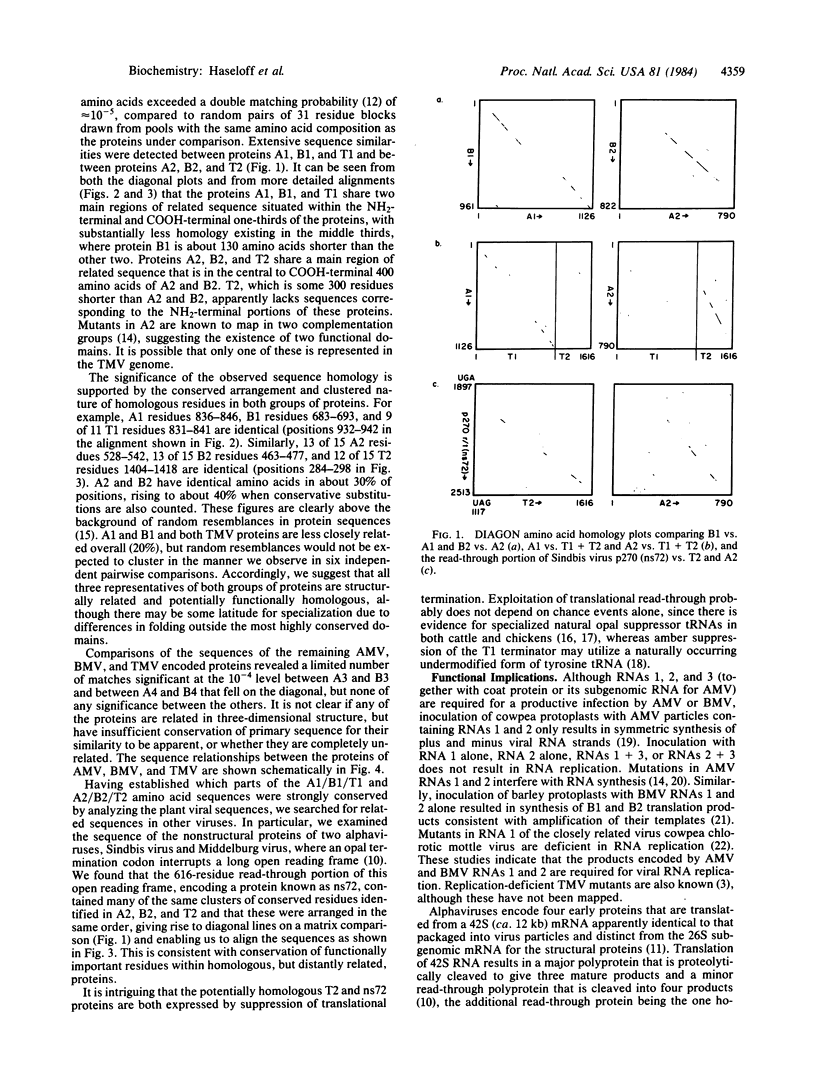
The plant viruses alfalfa mosaic virus (AMV) and brome mosaic virus (BMV) each divide their genetic information among three RNAs while tobacco mosaic virus (TMV) contains a single genomic RNA. Amino acid sequence comparisons suggest that the single proteins encoded by AMV RNA 1 and BMV RNA 1 and by AMV RNA 2 and BMV RNA 2 are related to the NH2-terminal two-thirds and the COOH-terminal one-third, respectively, of the largest protein encoded by TMV. Separating these two domains in the TMV RNA sequence is an amber termination codon, whose partial suppression allows translation of the downstream domain. Many of the residues that the TMV read-through domain and the segmented plant viruses have in common are also conserved in a read-through domain found in the nonstructural polyprotein of the animal alphaviruses Sindbis and Middelburg. We suggest that, despite substantial differences in gene organization and expression, all of these viruses use related proteins for common functions in RNA replication. Reassortment of functional modules of coding and regulatory sequence from preexisting viral or cellular sources, perhaps via RNA recombination, may be an important mechanism in RNA virus evolution.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Dasgupta R., Kaesberg P. Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J Mol Biol. 1984 Feb 5;172(4):369–383. doi: 10.1016/s0022-2836(84)80012-1. [DOI] [PubMed] [Google Scholar]
- Ahlquist P., Luckow V., Kaesberg P. Complete nucleotide sequence of brome mosaic virus RNA3. J Mol Biol. 1981 Nov 25;153(1):23–38. doi: 10.1016/0022-2836(81)90524-6. [DOI] [PubMed] [Google Scholar]
- Barker R. F., Jarvis N. P., Thompson D. V., Loesch-Fries L. S., Hall T. C. Complete nucleotide sequence of alfalfa mosaic virus RNA3. Nucleic Acids Res. 1983 May 11;11(9):2881–2891. doi: 10.1093/nar/11.9.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen B. J., Brederode F. T., Moormann R. J., Bol J. F. Complete nucleotide sequence of alfalfa mosaic virus RNA 1. Nucleic Acids Res. 1983 Mar 11;11(5):1253–1265. doi: 10.1093/nar/11.5.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen B. J., Brederode F. T., Veeneman G. H., van Boom J. H., Bol J. F. Complete nucleotide sequence of alfalfa mosaic virus RNA 2. Nucleic Acids Res. 1983 May 25;11(10):3019–3025. doi: 10.1093/nar/11.10.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R. K. Identification of a unique guanine-7-methyltransferase in Semliki Forest virus (SFV) infected cell extracts. Virology. 1983 Oct 30;130(2):452–463. doi: 10.1016/0042-6822(83)90099-5. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A., Dudock B., Hatfield D. Structure and properties of a bovine liver UGA suppressor serine tRNA with a tryptophan anticodon. Cell. 1981 Aug;25(2):497–506. doi: 10.1016/0092-8674(81)90068-4. [DOI] [PubMed] [Google Scholar]
- Domingo E., Sabo D., Taniguchi T., Weissmann C. Nucleotide sequence heterogeneity of an RNA phage population. Cell. 1978 Apr;13(4):735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. Similar amino acid sequences: chance or common ancestry? Science. 1981 Oct 9;214(4517):149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- Franssen H., Leunissen J., Goldbach R., Lomonossoff G., Zimmern D. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984 Apr;3(4):855–861. doi: 10.1002/j.1460-2075.1984.tb01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goelet P., Lomonossoff G. P., Butler P. J., Akam M. E., Gait M. J., Karn J. Nucleotide sequence of tobacco mosaic virus RNA. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5818–5822. doi: 10.1073/pnas.79.19.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D. L., Dudock B. S., Eden F. C. Characterization and nucleotide sequence of a chicken gene encoding an opal suppressor tRNA and its flanking DNA segments. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4940–4944. doi: 10.1073/pnas.80.16.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth L., Richards K. E. Tobacco mosaic virus: model for structure and function of a simple virus. Adv Virus Res. 1981;26:145–199. doi: 10.1016/s0065-3527(08)60423-6. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Hollis G. F., Hieter P. A., McBride O. W., Swan D., Leder P. Processed genes: a dispersed human immunoglobulin gene bearing evidence of RNA-type processing. Nature. 1982 Mar 25;296(5855):321–325. doi: 10.1038/296321a0. [DOI] [PubMed] [Google Scholar]
- King A. M., McCahon D., Slade W. R., Newman J. W. Recombination in RNA. Cell. 1982 Jul;29(3):921–928. doi: 10.1016/0092-8674(82)90454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovaara P., Söderlund H., Keränen S., Pettersson R. F., Käriäinen L. 18S defective interfering RNA of Semliki Forest virus contains a triplicated linear repeat. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5353–5357. doi: 10.1073/pnas.78.9.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe S. S., Schlesinger S. RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5' ends. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3279–3283. doi: 10.1073/pnas.80.11.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. G., Strauss J. H. Replication strategies of the single stranded RNA viruses of eukaryotes. Curr Top Microbiol Immunol. 1983;105:1–98. doi: 10.1007/978-3-642-69159-1_1. [DOI] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Tolskaya E. A., Romanova L. A., Kolesnikova M. S., Agol V. I. Intertypic recombination in poliovirus: genetic and biochemical studies. Virology. 1983 Jan 15;124(1):121–132. doi: 10.1016/0042-6822(83)90295-7. [DOI] [PubMed] [Google Scholar]



