Abstract
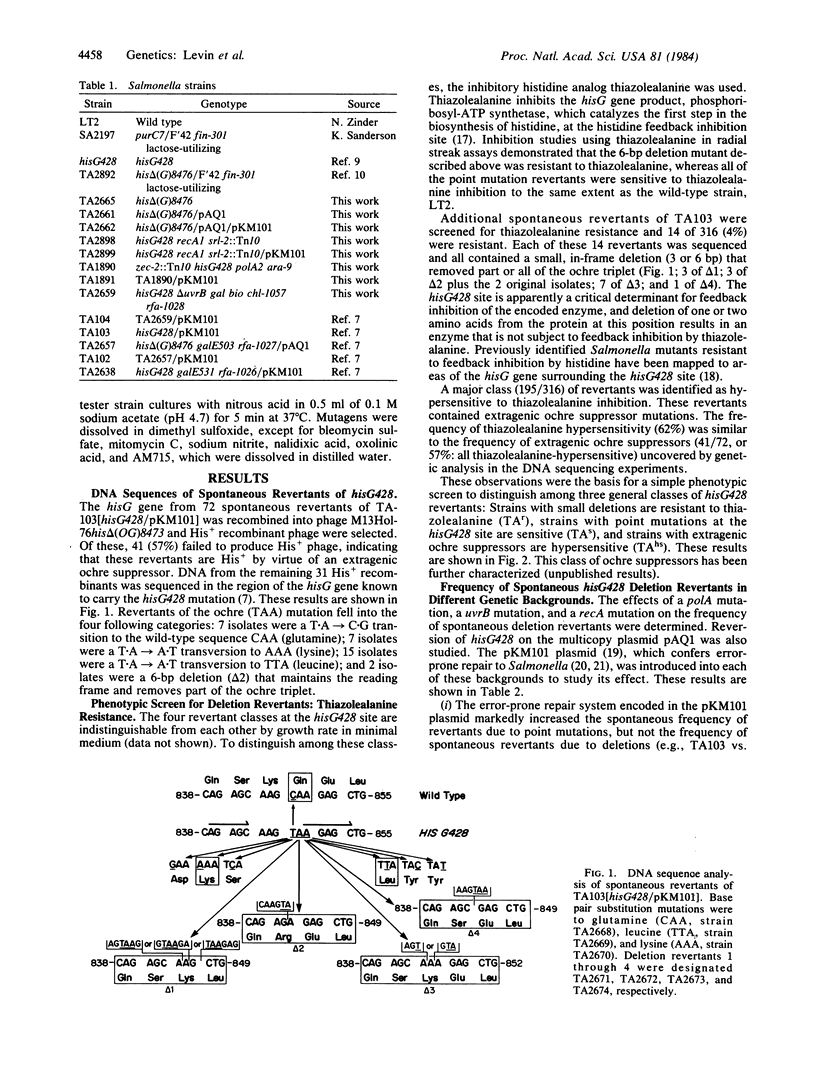
Salmonella tester strain TA102 carries the hisG428 ochre mutation on the multicopy plasmid pAQ1. DNA sequence analysis of 45 spontaneous revertants of hisG428 on the chromosome in the presence of pKM101 (strain TA103) indicates that hisG428 revertants fall into three major categories: (i) small, in-frame deletions (3 or 6 base pairs) that remove part or all of the ochre triplet; (ii) base substitution mutations at the ochre site; (iii) extragenic ochre suppressors. Deletion revertants are identified in a simple phenotypic screen by their resistance to the inhibitory histidine analog thiazolealanine, which feedback inhibits the wild-type hisG enzyme but not the enzyme resulting from the deletions. The effect of various genetic backgrounds on the generation of spontaneous deletion revertants was examined. The error-prone repair system encoded in the pKM101 plasmid markedly increased the frequency of total spontaneous reversion events in all genetic backgrounds except recA but did not affect the frequency of spontaneous deletion revertants in any background except polA. The presence of a polA mutation increased the frequency of spontaneous deletion revertants by 2-fold in the absence of pKM101 and by 20-fold with pKM101. The presence of a uvrB mutation or a recA mutation suppressed the generation of spontaneous deletion revertants to approximately 1/2.5. When hisG428 was in multiple copies on pAQ1, the frequency of spontaneous deletion revertants increased by 40-fold, which is the approximate copy number of pAQ1. Mutagenic agents that induce single-strand breaks in DNA (e.g., x-rays, bleomycin, and nalidixic acid) induced deletion revertants in TA102. These agents induced deletion revertants only in hisG428 on pAQ1 and only in the presence of pKM101. Deletion revertants were not induced by frameshift mutagens (i.e., ICR-191 and 9-aminoacridine). These results indicate that different pathways exist for the generation of spontaneous and mutagen-induced deletion revertants of hisG428.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AMES B. N., MARTIN R. G., GARRY B. J. The first step of histidine biosynthesis. J Biol Chem. 1961 Jul;236:2019–2026. [PubMed] [Google Scholar]
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Alper M. D., Ames B. N. Positive selection of mutants with deletions of the gal-chl region of the Salmonella chromosome as a screening procedure for mutagens that cause deletions. J Bacteriol. 1975 Jan;121(1):259–266. doi: 10.1128/jb.121.1.259-266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B. N., Mccann J., Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res. 1975 Dec;31(6):347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- Barnes W. M. Cloning and restriction map of the first part of the histidine operon of Salmonella typhimurium. J Bacteriol. 1981 Jul;147(1):124–134. doi: 10.1128/jb.147.1.124-134.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W. M. Construction of an M13 histidine-transducing phage: a single-stranded cloning vehicle with one EcoRI site. Gene. 1979 Feb;5(2):127–139. doi: 10.1016/0378-1119(79)90098-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cairns J. The origin of human cancers. Nature. 1981 Jan 29;289(5796):353–357. doi: 10.1038/289353a0. [DOI] [PubMed] [Google Scholar]
- Crooke S. T., Bradner W. T. Bleomycin, a review. J Med. 1976;7(5):333–428. [PubMed] [Google Scholar]
- Dizdaroglu M., von Sonntag C., Schulte-Frohlinde D. Letter: Strand breaks and sugar release by gamma-irradiation of DNA in aqueous solution. J Am Chem Soc. 1975 Apr 16;97(8):2277–2278. doi: 10.1021/ja00841a051. [DOI] [PubMed] [Google Scholar]
- Echols H. SOS functions, cancer and inducible evolution. Cell. 1981 Jul;25(1):1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Walker G. C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983 Feb 25;164(2):175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Fogliano M., Schendel P. F. Evidence for the inducibility of the uvrB operon. Nature. 1981 Jan 15;289(5794):196–198. doi: 10.1038/289196a0. [DOI] [PubMed] [Google Scholar]
- Fowler R. G., McGinty L., Mortelmans K. E. Spontaneous mutational specificity of drug resistance plasmid pKM101 in Escherichia coli. J Bacteriol. 1979 Dec;140(3):929–937. doi: 10.1128/jb.140.3.929-937.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W. Replication of the DNA of the colicinogenic factor E 1 (Col E 1 ) at the restrictive temperature in a DNA replication mutant thermosensitive for DNA polymerase. 3. Nat New Biol. 1972 May 17;237(72):67–70. doi: 10.1038/newbio237067a0. [DOI] [PubMed] [Google Scholar]
- Hartman P. E., Hartman Z., Stahl R. C. Classification and mapping of spontaneous and induced mutations in the histidine operon of Salmonella. Adv Genet. 1971;16:1–34. doi: 10.1016/s0065-2660(08)60352-1. [DOI] [PubMed] [Google Scholar]
- Heidecker G., Messing J., Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980 Jun;10(1):69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- Ishii Y., Kondo S. Spontaneous and radiation-induced deletion mutations in Escherichia coli strains with different DNA repair capacities. Mutat Res. 1972 Sep;16(7):13–25. doi: 10.1016/0027-5107(72)90059-0. [DOI] [PubMed] [Google Scholar]
- Levin D. E., Hollstein M., Christman M. F., Ames B. N. Detection of oxidative mutagens with a new Salmonella tester strain (TA102). Methods Enzymol. 1984;105:249–254. doi: 10.1016/s0076-6879(84)05032-1. [DOI] [PubMed] [Google Scholar]
- Levin D. E., Hollstein M., Christman M. F., Schwiers E. A., Ames B. N. A new Salmonella tester strain (TA102) with A X T base pairs at the site of mutation detects oxidative mutagens. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7445–7449. doi: 10.1073/pnas.79.23.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Yamasaki E., Ames B. N. A new Salmonella tester strain, TA97, for the detection of frameshift mutagens. A run of cytosines as a mutational hot-spot. Mutat Res. 1982 Jun;94(2):315–330. doi: 10.1016/0027-5107(82)90294-9. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Lloyd R. S., Haidle C. W., Robberson D. L. Site specificity of bleomycin-mediated single-strand scissions and alkali-labile damage in duplex DNA. Gene. 1979 Nov;7(3-4):289–302. doi: 10.1016/0378-1119(79)90049-0. [DOI] [PubMed] [Google Scholar]
- Maron D. M., Ames B. N. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983 May;113(3-4):173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- McCann J., Spingarn N. E., Kobori J., Ames B. N. Detection of carcinogens as mutagens: bacterial tester strains with R factor plasmids. Proc Natl Acad Sci U S A. 1975 Mar;72(3):979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein V., Otto B., Bonhoeffer F., Schaller H. Function of DNA polymerase 3 in DNA replication. Nat New Biol. 1971 Dec 29;234(52):285–286. doi: 10.1038/newbio234285a0. [DOI] [PubMed] [Google Scholar]
- Perry K. L., Walker G. C. Identification of plasmid (pKM101)-coded proteins involved in mutagenesis and UV resistance. Nature. 1982 Nov 18;300(5889):278–281. doi: 10.1038/300278a0. [DOI] [PubMed] [Google Scholar]
- Sancar G. B., Sancar A., Little J. W., Rupp W. D. The uvrB gene of Escherichia coli has both lexA-repressed and lexA-independent promoters. Cell. 1982 Mar;28(3):523–530. doi: 10.1016/0092-8674(82)90207-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausville E. A., Peisach J., Horwitz S. B. A role for ferrous ion and oxygen in the degradation of DNA by bleomycin. Biochem Biophys Res Commun. 1976 Dec 6;73(3):814–822. doi: 10.1016/0006-291x(76)90882-2. [DOI] [PubMed] [Google Scholar]
- Schwartz D. O., Beckwith J. R. Mutagens which cause deletions in Escherichia coli. Genetics. 1969 Feb;61(2):371–376. doi: 10.1093/genetics/61.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanabruch W. G., Walker G. C. Localization of the plasmid (pKM101) gene(s) involved in recA+lexA+-dependent mutagenesis. Mol Gen Genet. 1980;179(2):289–297. doi: 10.1007/BF00425456. [DOI] [PubMed] [Google Scholar]
- Snyder M., Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979 Jun 25;131(2):287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. REPLICAtion of small plasmids in extracts of Escherichia coli: requirement for both DNA polymerases I and II. Mol Gen Genet. 1976 Dec 8;149(2):151–158. doi: 10.1007/BF00332883. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Wainscott V. J., Ferretti J. J. Biochemical-genetic study of the first enzyme of histidine biosynthesis in Salmonella typhimurium: substrate and feedback binding regions. J Bacteriol. 1978 Jan;133(1):114–121. doi: 10.1128/jb.133.1.114-121.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]