Abstract
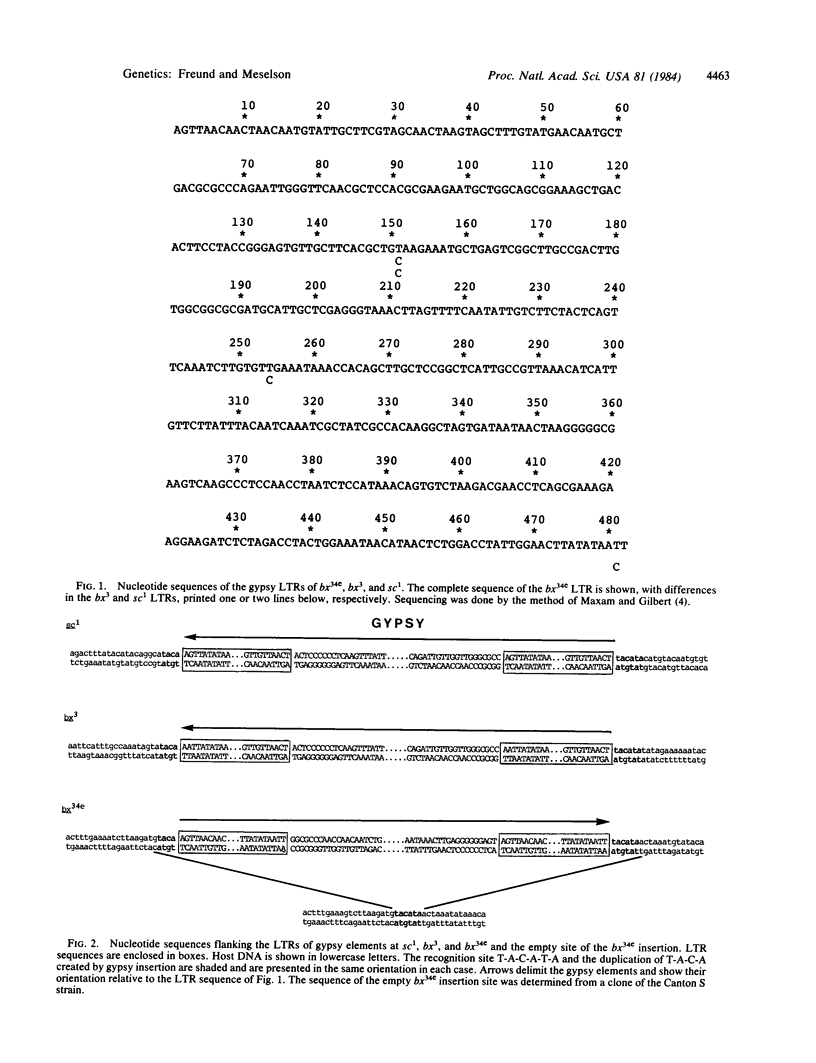
We have determined the nucleotide sequences of the long terminal repeats of the transposable element gypsy from the cloned mutant alleles sc1, bx3, and bx34e. These mutations are suppressible by the suppressor of Hairy-wing, su(Hw). The long terminal repeats are 482 base pairs long and are highly conserved. In each case, gypsy is inserted into the sequence T-A-C-A-T-A and generates a duplication of the sequence T-A-C-A. This was verified by sequencing an empty site in the wild-type bx gene. Consideration of the sequence of the long terminal repeats and their surroundings limits the possible explanations for the mechanism of mutation by these gypsy insertions and for their suppression by su(Hw).
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayev A. A., Jr, Krayev A. S., Lyubomirskaya N. V., Ilyin Y. V., Skryabin K. G., Georgiev G. P. The transposable element Mdg3 in Drosophila melanogaster is flanked with the perfect direct and mismatched inverted repeats. Nucleic Acids Res. 1980 Aug 11;8(15):3263–3273. doi: 10.1093/nar/8.15.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Dunsmuir P., Brorein W. J., Jr, Simon M. A., Rubin G. M. Insertion of the Drosophila transposable element copia generates a 5 base pair duplication. Cell. 1980 Sep;21(2):575–579. doi: 10.1016/0092-8674(80)90495-x. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Ikenaga H., Saigo K. Insertion of a movable genetic element, 297, into the T-A-T-A box for the H3 histone gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4143–4147. doi: 10.1073/pnas.79.13.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulguskin V. V., Ilyin Y. V., Georgiev G. P. Mobile dispersed genetic element MDG1 of Drosophila melanogaster: nucleotide sequence of long terminal repeats. Nucleic Acids Res. 1981 Jul 24;9(14):3451–3464. doi: 10.1093/nar/9.14.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980 Sep;21(2):581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Farabaugh P. J., Chaleff D. T., Fink G. R. The origins of gene instability in yeast. Science. 1980 Sep 19;209(4463):1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
- Scherer G., Tschudi C., Perera J., Delius H., Pirrotta V. B104, a new dispersed repeated gene family in Drosophila melanogaster and its analogies with retroviruses. J Mol Biol. 1982 May 25;157(3):435–451. doi: 10.1016/0022-2836(82)90470-3. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981 Nov;27(1 Pt 2):1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Goddard J. G., Berns A., Verma I. M. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5' long terminal repeat and adjacent cellular sequences. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3307–3311. doi: 10.1073/pnas.77.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Will B. M., Bayev A. A., Finnegan D. J. Nucleotide sequence of terminal repeats of 412 transposable elements of Drosophila melanogaster. A similarity to proviral long terminal repeats and its implications for the mechanism of transposition. J Mol Biol. 1981 Dec 25;153(4):897–915. doi: 10.1016/0022-2836(81)90458-7. [DOI] [PubMed] [Google Scholar]



