Abstract
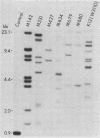
DNA from Escherichia coli strains in a reference collection of 72 recent natural isolates (ECOR strains) and 25 natural isolates from the "pre-antibiotic" period 1930-1940 (Murray strains) were studied to determine the genomic abundance of insertion element IS5 and the size of genomic restriction fragments carrying sequences homologous to IS5. Among the ECOR strains, nearly two-thirds lack DNA sequences that hybridize with IS5, and one-half of the remainder have only one copy. Among strains in which IS5 is present, extensive variation in the size of IS5-bearing restriction fragments occurs, in many cases allowing distinction among strains that are judged to be nearly identical in genotype because of the identical electrophoretic mobility of the enzyme coded by each of 11 chromosomal loci. Among the Murray strains in which IS5 is present, the average number of elements per strain is larger, but not markedly so, than among recent isolates. Comparison between duplicate strains in the Murray collection suggests that the rate of accumulation of IS5 elements in prolonged storage in stab tubes corresponds to an apparent probability of transposition of approximately 0.008 +/- 0.002 per IS5 element per year. Because of the extensive genetic variation among strains, insertion elements such as IS5 would seem to be convenient genetic markers with which to detect recent common ancestry among strains.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anilionis A., Riley M. Conservation and variation of nucleotide sequences within related bacterial genomes: Escherichia coli strains. J Bacteriol. 1980 Jul;143(1):355–365. doi: 10.1128/jb.143.1.355-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Campbell A. Evolutionary significance of accessory DNA elements in bacteria. Annu Rev Microbiol. 1981;35:55–83. doi: 10.1146/annurev.mi.35.100181.000415. [DOI] [PubMed] [Google Scholar]
- Ciampi M. S., Schmid M. B., Roth J. R. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler J. A., van Bree M. P. The nucleotide sequence and protein-coding capability of the transposable element IS5. Gene. 1981 Aug;14(3):155–163. doi: 10.1016/0378-1119(81)90111-6. [DOI] [PubMed] [Google Scholar]
- Harshman L., Riley M. Conservation and variation of nucleotide sequences in Escherichia coli strains isolated from nature. J Bacteriol. 1980 Nov;144(2):560–568. doi: 10.1128/jb.144.2.560-568.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Dykhuizen D. E., Miller R. D., Green L., de Framond J. Transposable element IS50 improves growth rate of E. coli cells without transposition. Cell. 1983 Dec;35(2 Pt 1):503–510. doi: 10.1016/0092-8674(83)90184-8. [DOI] [PubMed] [Google Scholar]
- Hu M., Deonier R. C. Comparison of IS1, IS2 and IS3 copy number in Escherichia coli strains K-12, B and C. Gene. 1981 Dec;16(1-3):161–170. doi: 10.1016/0378-1119(81)90072-x. [DOI] [PubMed] [Google Scholar]
- Hughes V. M., Datta N. Conjugative plasmids in bacteria of the 'pre-antibiotic' era. Nature. 1983 Apr 21;302(5910):725–726. doi: 10.1038/302725a0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kröger M., Hobom G. Structural analysis of insertion sequence IS5. Nature. 1982 May 13;297(5862):159–162. doi: 10.1038/297159a0. [DOI] [PubMed] [Google Scholar]
- Nasoff M. S., Wolf R. E., Jr Molecular cloning, correlation of genetic and restriction maps, and determination of the direction of transcription of gnd of Escherichia coli. J Bacteriol. 1980 Aug;143(2):731–741. doi: 10.1128/jb.143.2.731-741.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman K., Ohtsubo H., Davison D., Ohtsubo E. Distribution of insertion element IS1 in natural isolates of Escherichia coli. Mol Gen Genet. 1983;189(3):516–518. doi: 10.1007/BF00325920. [DOI] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984 Feb;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak B., Lusky M., Hable M. Expression of two proteins from overlapping and oppositely oriented genes on transposable DNA insertion element IS5. Nature. 1982 May 13;297(5862):124–128. doi: 10.1038/297124a0. [DOI] [PubMed] [Google Scholar]
- Reynolds A. E., Felton J., Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981 Oct 22;293(5834):625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- Saedler H., Cornelis G., Cullum J., Schumacher B., Sommer H. IS1-mediated DNA rearrangements. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):93–98. doi: 10.1101/sqb.1981.045.01.017. [DOI] [PubMed] [Google Scholar]
- Schoner B. Escherichia coli K-12 F' plasmids carrying insertion sequences IS1 and IS5. Gene. 1983 Mar;21(3):203–210. doi: 10.1016/0378-1119(83)90003-3. [DOI] [PubMed] [Google Scholar]
- Schoner B., Kahn M. The nucleotide sequence of IS5 from Escherichia coli. Gene. 1981 Aug;14(3):165–174. doi: 10.1016/0378-1119(81)90112-8. [DOI] [PubMed] [Google Scholar]
- Schoner B., Schoner R. G. Distribution of IS5 in bacteria. Gene. 1981 Dec;16(1-3):347–352. doi: 10.1016/0378-1119(81)90093-7. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Hoopes B. C., McClure W. R., Kleckner N. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983 Sep;34(2):673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Whittam T. S., Ochman H., Selander R. K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]