Abstract
Mitochondrial and peroxisomal fission are essential processes with defects resulting in cardiomyopathy and neonatal lethality. Central to organelle fission is Fis1, a monomeric tetratricopeptide-like repeat (TPR) protein whose role in assembly of the fission machinery remains obscure. Two non-functional, Saccharomyces cerevisiae Fis1 mutants (L80P or E78D/I85T/Y88H) were previously identified in genetic screens. Here, we find that these two variants in the cytosolic domain of Fis1 (Fis1ΔTM) are unexpectedly dimeric. A truncation variant of Fis1ΔTM that lacks an N-terminal regulatory domain is also found to be dimeric. The ability to dimerize is a property innate to the native Fis1ΔTM amino acid sequence as we find this domain is dimeric after transient exposure to elevated temperature or chemical denaturants and is kinetically trapped at room temperature. This is the first demonstration of a specific self-association in solution for the Fis1 cytoplasmic domain. We propose a three-dimensional domain-swapped model for dimerization that is validated by a designed mutation, A72P, which potently disrupts dimerization of wild type Fis1. A72P also disrupts dimerization of non-functional variants indicating a common structural basis for dimerization. The obligate monomer variant A72P, like the dimer-promoting variants, is non-functional in fission consistent with a model in which Fis1 activity depends on its ability to interconvert between monomer and dimer species. These studies suggest a new functionally important manner in which TPR containing proteins may reversibly self-associate.
Organelle fission and fusion control the size and distribution of mitochondria and peroxisomes, which are fine-tuned in many adult cell types to meet their local demand for ATP or Ca2+. These membrane remodeling events may also be coordinated to ensure the fidelity of essential components that incur damage in their oxidative environments found in these organelles1-8. Supporting the importance of these processes, mutations in fission and fusion genes cause or contribute to mammalian disease9-14. For organelle fission only Fis1 and dynamin-related protein 1 (called Drp1 in mammals and Dnm1 in budding yeast) are conserved across species. Fis1 is a tail-anchored transmembrane protein that is thought to be involved in recruiting soluble fission factors, including Drp1, to sites of membrane scission from the cytoplasm2, 15-25. In budding yeast, Fis1 cooperates with Mdv1 and Caf4, proteins found only in fungi, to recruit the yeast dynamin-related protein Dnm117-19, 24-27.
Both blue-native PAGE and chemical cross-linking studies show that Fis1 can form homooligomeric complexes in mammalian cells2, 28, 29. However, seven of the eight available structures of the isolated cytoplasmic domains of Fis1 from human, mouse, and yeast are monomeric27, 30-35. In the eighth structure, human Fis1 crystallizes as a multimer, although it is a monomer in solution32. Thus, there were no previous reports that Fis1 multimerizes in solution and the exact basis of the high molecular weight species of Fis1 by blue-native PAGE analysis is unclear and may involve other proteins.
The structures of the cytoplasmic domain of Fis1 are similar; each with a core six-helix bundle, where the central four helices consist of two tandem tetratricopeptide repeat (TPR)-like motifs. The individual TPR-motifs adopt a helix-turn-helix motif and together form a right-handed superhelical structure creating a concave surface. The structures differ in the relative orientation of the Fis1 arm (residues 1-16 in budding yeast and 1-8 in human)27, 30-35. Cell biological experiments, primarily in budding yeast, have determined that both the concave surface and the Fis1 arm are functionally important. NMR and chemical modification experiments suggest that the Fis1 arm is innately dynamic and, in some structures, physically occludes the concave surface suggesting a possible role in modulating function by mediating access to the concave surface36 (Figure 1a). Consistent with this idea, removal of the Fis1 arm (ΔN) impairs yeast mitochondrial morphology producing net-like organelles from a defect in fission33, 37. An important role for the Fis1 arm is supported by two other non-functional Fis1 alleles identified in yeast genetic screens37, 38, L80P and fis1-3 (derived from E78D/I85T/Y88H) on helix 4 of the concave surface. These alleles contain substitutions to residues adjacent to the arm in the tertiary structure, and similar to ΔN-fis1, expression of either L80P or fis1-3 does not restore proper mitochondrial morphology in fis1Δ cells37, 38. Overexpression of Mdv1 can rescue the fission defect caused by L80P and ΔN37, but not fis1-3. The fis1-3 allele requires a mutant in the Mdv1 coiled coil domain (E250G) that may enhance the Fis1 interaction either directly or indirectly39. These results suggest Fis1 variants impair fission by different means. Here we isolated the cytoplasmic domains from these variants with the initial goal of determining whether they share a common mechanism for disrupting fission. Surprisingly, we found that expression of these domains revealed a previously undetected state of yeast Fis1 that is dimeric. Further, we discovered that the wild type domain also forms a dimer, but is kinetically trapped as both a monomer and a dimer. This is the first example of the cytoplasmic domain mediated dimerization in solution. We designed a point mutant (A72P) that strongly disrupts mitochondrial fission and dimerization of isolated protein. Since variants that act either as obligate monomers or dimers in vitro are impaired for fission in vivo, our results suggest a model in which the interconversion between monomeric and dimeric species may play a role in fission and reveals a new mechanism for how TPR motifs can mediate self-association.
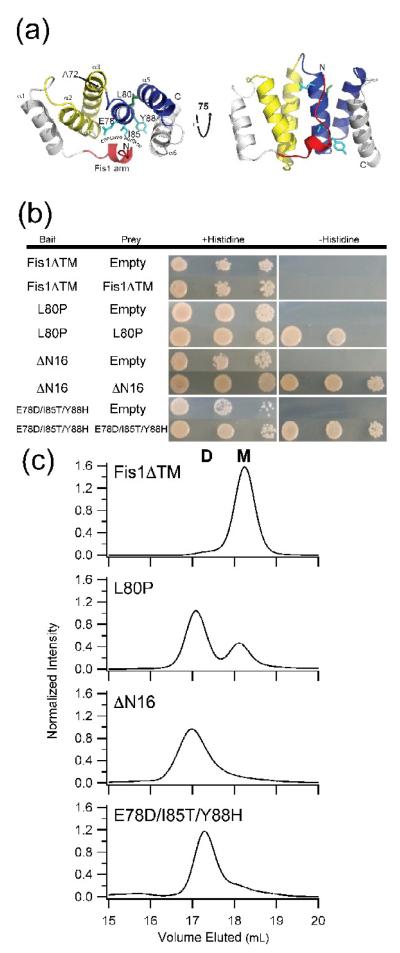
Figure 1. The cytoplasmic domains of non-functional Fis1 variants self-associate.
(a) Cartoon representation of the cytoplasmic domain of yeast Fis1 (Fis1ΔTM, 3o48.pdb). Three non-functional variants of Fis1 (L80P (green), ΔN16 (red), and Fis1ΔTM-3 (cyan, E78D/I85T/Y88H)) are highlighted by stick representation of the wild type residues. These variants have previously been reported to impair mitochondrial fission. The two TPR-like domains are colored yellow and blue. This figure was made with PyMOL86. (b) The cytoplasmic domains of non-functional Fis1 variants, but not wild type, interact by yeast two-hybrid. Yeast two-hybrid assays using the HIS3 reporter (sequential ten-fold yeast dilutions at 1, 0.1, and 0.01 OD600 are shown) with the indicated bait and prey constructs. Cells were plated onto media containing histidine (growth control) and media lacking histidine to select for yeast two-hybrid interactions and photographed after incubation for 3 days at 30 °C. (c) Fis1ΔTM variants elute by gel filtration chromatography as dimers to differing degrees. 2.5 μM of each of the indicated recombinantly expressed proteins were separated by a Superdex 200 10/300 GL at 25 °C (50 mM sodium phosphate, 184 mM NaCl, 5 mM EDTA and 2 mM DTT, pH 7.4). Each mutant is well-folded by circular dichroism spectrapolarimetry (Supplemental Figure 1).
Results
Non-functional variants of Fis1ΔTM form homotypic interactions
TPR motifs, such as those found in Fis1, have been implicated in a number of modes of protein-protein interactions40-48; therefore, we tested whether the soluble domains of wild type yeast Fis1 and the three non-functional alleles (L80P, ΔN, and fis1-3) can self-associate by yeast two-hybrid experiments. In this assay the cytoplasmic domain of Fis1 (Fis1ΔTM) in bait and prey plasmids did not interact. However, the three non-functional variants (L80P, ΔN, Fis1-3) each displayed strong self-association (Figure 1b). To determine whether association is an intrinsic property, we overexpressed and purified Fis1ΔTM and the three variants in E. coli. The purified wild type and variant proteins are well folded as judged by circular dichroism spectropolarimetry (Supplementary Figure 1). However, behavior of the variant proteins in size exclusion chromatography was markedly different from wild type (Figure 1c). Wild type Fis1ΔTM elutes primarily (>97%) as a single peak at a volume (18.2 mL) expected for a globular monomer, consistent with its structure33, 34. The molecular mass of this species was determined by in-line MALS confirming its monomeric nature (16.7 kDa theoretical vs. 15.5 kDa experimental mass). By contrast, the Fis1ΔTM-L80P variant elutes in two peaks, with ~30% eluting in a volume similar to the wild type monomer, while the majority (~70%) elutes earlier consistent with a globular dimeric species. The nearly baseline separation of the peaks implies that the two species do not interconvert on the timescale of the experiment (~45 min). The ΔN variant elutes in a broader peak corresponding to the dimer position, while the triply substituted Fis1ΔTM-3 elutes predominantly at a volume expected for the dimer species. These data correlate with the interactions detected by yeast two-hybrid and suggest that the two-hybrid results probably arise from direct bait-prey contacts.
Fis1ΔTM is isolated as monomer and a non-covalent dimer that do not detectably interconvert
Closer inspection of the SEC chromatogram for wild type Fis1ΔTM revealed the presence of a barely discernible shoulder on the leading edge of the peak that eluted at a volume predicted for dimer (calculated mass of 30.9 kDa) (Figure 1c). Analysis by Western blot, SEC-MALS, AUC, and Cys mutagenesis shows that this species is a non-covalent Fis1ΔTM dimer (Figure 2). Specifically, mutation of the two Cys residues in Fis1ΔTM (Cys79 and Cys87) to Ser did not prevent dimerization. These residues also lie on helix 4 similar to the non-functional variants. To test whether wild type monomer and dimer can interconvert, we isolated the monomer and dimer Fis1ΔTM species, and determined their oligomeric state as a function of concentration by SEC. The isolated monomer remained monomeric at concentrations >1mM, and elution of the dimer was also independent of concentration, even at the lower limits of detection (sub μM) (see Figure 3a,b for representative traces). The independence of oligomeric state from mass action shows that the wild type monomer and dimer species do not equilibrate appreciably under these conditions.
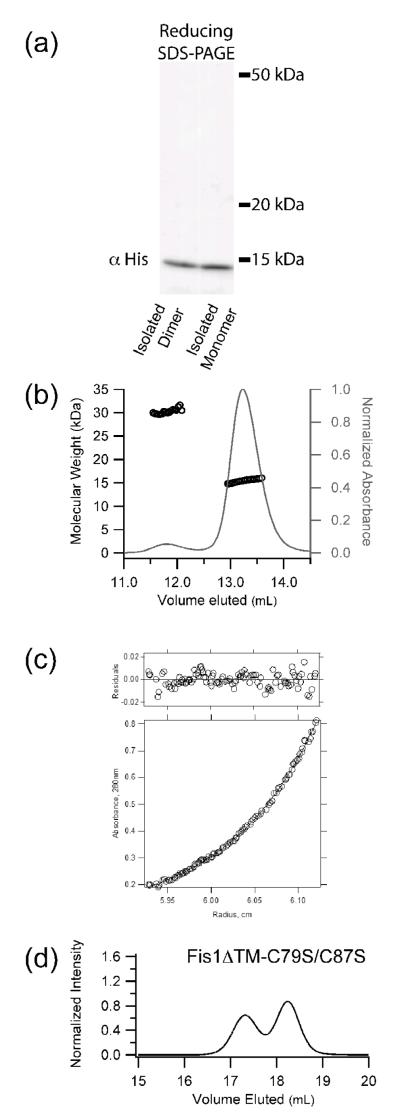
Figure 2. The wild type cytoplasmic domain of Fis1 (Fis1ΔTM) forms a noncovalent dimer.
a) Recombinant yeast Fis1ΔTM was isolated as two species by size exclusion chromatography, diluted to 2.5 μM, analyzed by non-reducing SDS-PAGE, and detected by Western blot analysis. b) Fis1ΔTM (1.3 mg/mL) eluted by size exclusion chromatography and detected by multi-angle laser light scattering has a calculated molar masses of 30,300 and 15,500 Da, which compares favorably with the theoretical molecular weights for dimer and monomer (33,400 and 16,700 Da, respectively). c) The higher order species of yeast Fis1ΔTM is a dimer. A radial distribution fit to a monomer-dimer equilibrium model is shown for a sample at 23.2 μM centrifuged at 20,000 rpm. The residuals of the fit are shown in the top panel. These data are representative of a global analysis of Fis1ΔTM samples enriched in the higher order species that were subjected to equilibrium sedimentation analysis at 16300, 20000, 24500 and 30000 rpm at 4 °C on a Beckman XLA ultracentrifuge. Samples were at 7.7 μM, 15.4 μM and 23.2 μM in 50 mM sodium phosphate and 184 mM NaCl, pH 7.4. A global fit to the data was well described by a model in which the higher order species of Fis1ΔTM forms a dimer. d) Size exclusion chromatography of a cysteine-less variant of recombinantly expressed Fis1ΔTM-C79S/C87S (2.5 μM) eluted as both monomer and dimer. Experimental conditions identical to Figure 1. Both Cys residues lie on helix 4 of the concave surface.
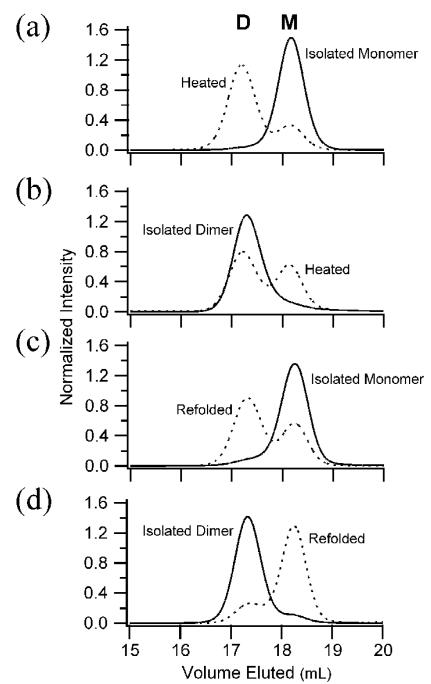
Figure 3. Fis1ΔTM monomer and dimer are kinetically trapped.
a) Heat-treated Fis1ΔTM monomer redistributes into monomer and dimer species. Size exclusion chromatography of recombinantly expressed Fis1ΔTM (2.5 μM) eluted predominantly as monomer (solid line) with a small population eluting as dimer. Upon heating to temperatures well below the thermal unfolding midpoint and subsequently cooling, Fis1ΔTM redistributed into monomer and dimer populations with a KDapp of 240 ± 80 nM. b) Heat-treated Fis1ΔTM dimer redistributes into monomer and dimer species. Upon similar heat treatment, isolated Fis1ΔTM dimer (0.4 μM) redistributed into monomer and dimer populations with a similar KDapp. c) Isolated Fis1ΔTM monomer redistributes into monomer and dimer upon refolding. Upon chemical denaturation with GdHCl and subsequent refolding at 37 μM, Fis1ΔTM distributed into monomer and dimer species (dashed line). An estimate of the apparent association constant for dimerization was determined from the relative populations upon refolding for these data and similar experiments at 2.5 μM and 25 μM (data not shown), which gave an KDapp = 15±3 μM. d) Isolated Fis1ΔTM dimer redistributes into monomer and dimer species upon refolding. Isolated Fis1ΔTM dimer remained dimeric (solid line) when diluted to 2.5 μM and reapplied. Upon chemical denaturation with GdHCl and subsequent refolding at 2.5 μM, Fis1ΔTM distributed into monomer and dimer species (dashed line) with a similar KDapp. All chromatography conditions are the same as those in Figure 1.
To determine whether the kinetic barrier between monomer and dimer can be overcome at increased temperature, we conducted concentration and temperature dependence experiments over a range of concentrations (0.2 to 25 μM) and temperatures (47-55 °C, well below the apparent thermal unfolding midpoint, Tm = 69.3 °C). In these experiments, either isolated monomeric or dimeric samples were heated at a given temperature as a function of time, followed by cooling of the sample rapidly on ice, and injecting it onto a SEC column at room temperature. As expected by mass action, the final composition was concentration dependent and a representative subset of these experiments is shown in Figure 3a in which a 2.5 μM sample of isolated monomer was incubated at 55 °C for 24 h and found to redistribute into a final composition of 23% monomer and 77% dimer A sample of isolated dimer (0.4 μM) was heated at 50 °C for 24 h and found to redistribute into 43% monomer and 57% dimer (Figure 3b). Estimating an apparent dissociation constant from these experiments yields similar values (KDapp of 240 ± 80 nM) establishing that altering sample conditions allows both the dimer and monomer states to be reversibly accessed. Similar experiments with the non-functional Fis1ΔTM variants were complicated by solubility issues, however, they were consistent with an increase in temperature allowing an increased rate of interconversion.
We also tested whether chemical denaturation might lower the apparent activation energy for interconversion by refolding Fis1ΔTM from high concentrations of guanidine hydrochloride (>4 M GdHCl). Upon dialysis and refolding, either isolated monomer or isolated dimer redistributes (Figure 3c,d) suggesting that chemical denaturation enhances the rate of local structural fluctuations in wild type Fis1ΔTM that are necessary for interconversion.
To estimate an apparent dissociation constant for the wild type dimer, Fis1ΔTM monomer samples were equilibrated by chemical denaturation at several protein concentrations, refolded and resolved by SEC, and the observed monomer and dimer populations were fit to a two-state formalism. These results gave an apparent dissociation constant of KDapp = 15±3 μM. An equilibration experiment starting with Fis1ΔTM dimer (Figure 3d) gave a similar apparent dissociation constant. These results suggest that chemical denaturation permits rapid and complete interconversion of monomer and dimer and show that the wild type Fis1ΔTM sequence has a strong propensity to dimerize if the kinetic barrier to association is removed.
The Fis1ΔTM monomer and dimer have similar secondary and tertiary structure
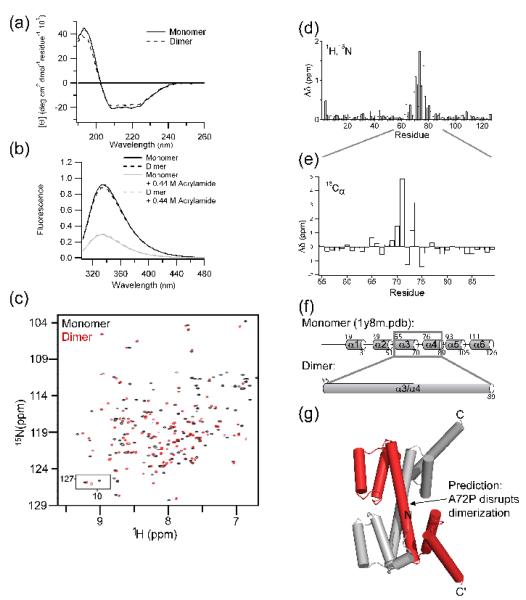
We exploited the slow interconversion between wild type Fis1ΔTM monomer and dimer to isolate these kinetically stable species and compare their structural properties. Both the monomer and dimer have nearly identical spectra by either circular dichroism or fluorescence spectroscopy, demonstrating minimal differences in the global secondary and tertiary structure (Figure 4a,b). To probe for structural differences at the individual amino acid level, the 1H-15N HSQC NMR spectra for the monomer and dimer samples was collected and compared (Figure 4c). Both have essentially the same number of crosspeaks, indicating the formation of a symmetric homodimer (Figure 4c). We assigned the monomer and dimer backbone chemical shifts from a 2H, 15N, 13C sample using standard methods. Plotting the differences in amide nitrogen and proton chemical shifts between monomer and dimer reveals that most residues have similar chemical shifts in the two states indicating similar structural environments (Figure 4d). However, significant differences occur for residues 63-81 of helices α3 and α4 with the largest differences localized to residues 69-77 of the intervening turn. Chemical shift index analysis of the backbone 13Cα chemical shifts of the turn (residues 72-75) between α3 and α4 in the monomer indicates that these residues are helical in the dimer (Figure 4e). From the structural and mutagenesis data, we hypothesize that the α3-turn-α4 in monomeric Fis1ΔTM adopts a single α-helix that allows the formation of a domain-swapped dimer (Figure 4f,g). In this model, the concave surface would be maintained by the first TPR (α2-turn-α3) of one monomer and the second TPR (α4-turn-α5) of the other (Figure 4g). This arrangement would preserve the environment of Trp7 in the Fis1 arm and Trp47 on α2 and places the two transmembrane domains on the same face of the molecule.
Figure 4. Fis1ΔTM forms a domain-swapped dimer.
a) Far-UV circular dichroism of Fis1ΔTM monomer (—) and refolded dimer (--) samples indicate little change in 2° structure between monomer and dimer. b) Intrinsic tryptophan fluorescence emission of Fis1ΔTM monomer (—) and refolded dimer (--) samples in the absence (black) and presence of 0.44 M acrylamide (grey) indicate little change in 3° structure between monomer and dimer. c) An overlay of the 1H-15N HSQC of Fis1 monomer (black) and refolded dimer (red) demonstrate that the amide backbones are largely in similar environments. d) The largest chemical shift differences, Δδ, between monomer and dimer are localized to the intervening turn between the first and second tetratricopeptide repeats of monomer Fis1 (α3-turn-α4). Δδ was calculated from the data in c) after chemical shift assignment using Δδ ={(1Hδmonomer − 1Hδdimer)2 + [1/4(15Nδmonomer − 15Nδdimer)]2}1/2. e) The 13Cα chemical shift differences, Δδ, between monomer and dimer suggest that the α3-turn-α4 adopts a single α-helix in the dimer, which is shown schematically (f) and confirmed by chemical shift index analysis (not shown). g) A model of the domain-swapped Fis1ΔTM dimer consistent with the CD, fluorescence, and NMR data. In this model, a symmetrical arrangement of helices 1-3 of one monomer associate with helices 4-6 of a second monomer to form a near identical tertiary structure to that found in the monomer structure. From this model, we predict that replacing Ala72 with a Pro will disfavor helix formation necessary for dimerization and favor the monomeric conformation.
A designed point mutant, A72P, disrupts dimerization and mitochondrial fission
The proposed model for dimerization implies that formation of the extended (α3/α4)-helix is a prerequisite for dimer formation, thus mutations that affect this conformation will oppose dimerization. To test this notion, we designed a variant where Ala72, the first residue in the turn between α3 and α4, is replaced with a proline to inhibit dimerization. Proline at this position should favor the helix-turn-helix conformation of the monomer due to the low helical propensity of proline compared to alanine49. Analysis of the Fis1ΔTM-A72 variant revealed that it adopted a native structure by circular dichroism and the presence of well-dispersed NMR chemical shifts similar to wild type Fis1ΔTM (Supplemental Figure 3,4). Consistent with our model-based prediction, this variant remained a monomer by SEC-MALS following heat treatment (Figure 5a). We also tested A72P in our refolding from GdHCl assay and found no evidence for the presence of dimer at a concentration (2.5 μM) where wild type Fis1ΔTM is partially dimeric after refolding (Figure 5b). Increasing the A72P concentration to 100 μM in refolding experiments did not change the elution profile, thus A72P can be thought of as an obligate monomer (lower limit of apparent KDapp ~ 3 mM). Further, when the three variants of Fis1ΔTM that favor dimerization were created in an A72P background, each eluted at volumes consistent with wild type monomer (Figure 5b). The abolition of dimerization of all three variants by the same A72P mutation suggests that each variant associates through the same domain-swapped dimer structure inferred for wild type.
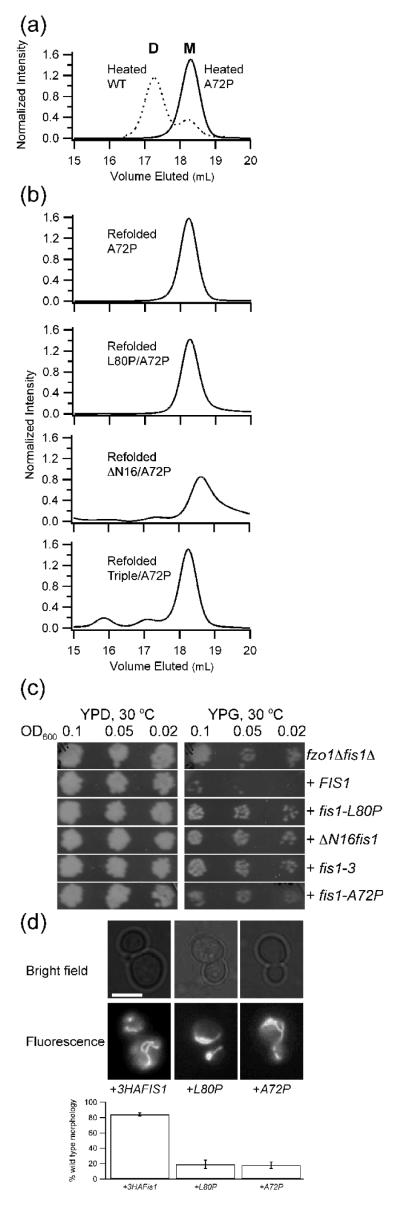
Figure 5. Designed mutant A72P impairs dimerization of Fis1ΔTM and mitochondrial fission.
a) A 2.5 μM sample of Fis1ΔTM-A72P was heated, cooled, and analyzed by size exclusion chromatography as described in Figure 3. For comparison purposes, a similar experiment with wild type Fis1ΔTM is overlaid as a dashed line (from Fig 3a). b) A 2.5 μM sample of Fis1ΔTM-A72P was chemically unfolded in GdHCl, refolded, and analyzed by size exclusion chromatography as described in Figure 3. Introducing A72P into each of the non-functional Fis1ΔTM variants and refolding from chemical denaturant also strongly favored the monomeric state. These variants were well folded (Supplemental Figure 3) indicating a common structural basis for dimerization of each variant. c) Yeast expression of either obligate dimers (ΔN, fis1-3) or obligate monomer (A72P) do not rescue mitochondrial fission. In this standard growth assay, functional fission is signified by no growth on YP+Glycerol (YPG) plates. Note that these are full-length Fis1 constructs that include the C-terminal transmembrane domain. d) Expression of full-length Fis1-A72P in fis1Δ yeast cells impairs mitochondrial morphology to a similar extent as full-length Fis1-L80P expression. Representative epifluorescence microscopy images from fis1Δ yeast cells expressing the indicated plasmids and stained with MitoTracker Red CMXRos. Percentage of cells with wild type morphology is shown based on scoring >100 cells from three independent experiments. Expression of full-length 3xHA-FIS1 rescued 84 ± 2 % of cells compared to 19 ± 5 % for 3xHA-FIS1-L80P and 18 ± 4 % for 3xHA-FIS1-A72P. Scale bar is 5 μm.
We next tested the functionality of the A72P variant introduced into full-length Fis1 in a fission-dependent growth assay. Cells lacking the fusion fzo1 and fission fis1 genes are viable on glycerol. However, cells expressing an additional functional copy of FIS1 will not survive on glycerol because excessive fission causes loss of mtDNA and an inability to respire50, 51. This assay is similar to the screen employed to identify the FIS1 and MDV1 genes and was previously used to test the temperature sensitive allele fis1-3 for altered activity17, 37, 38. Plasmids encoding the galactose-inducible wild type and mutant fis1 genes were individually transformed into fzo1Δfis1Δ cells and tested for growth on glucose and glycerol plates. As a negative control, we expressed wild type, full-length Fis1 in yeast cells, which grew on glucose but not on glycerol demonstrating that mitochondrial fission is restored (Figure 5c). As positive controls, we expressed full-length constructs of L80P, ΔN, and Fis1-3 in fzo1Δfis1Δ cells33, 37, 38, which grew on both glucose and glycerol showing that these variants disrupt Fis1 activity consistent with previous reports33, 37, 38. Cells expressing full-length A72P also grew on glycerol demonstrating that this mutation disrupts mitochondrial fission. Introducing the A72P mutation into fis1-L80P, ΔNfis1, and fis1-3 also impaired mitochondrial fission indicating that this mutant did not rescue the phenotype (data not shown).
Fis1 is also necessary for proper mitochondrial morphology, which can be monitored by fluorescence microscopy using mitochondrial-specific fluorescent dyes. Deleting the fis1 gene causes altered, net-like morphology that can be rescued upon plasmid expression of wild type Fis1 (Figure 5d). By contrast, expression of full-length Fis1-A72P was unable to rescue morphology in over 80% of cells similar to the non-functional full-length control L80P, which was not due to poor expression or mislocalization (data not shown). We conclude that mutations that enhance (L80P) or inhibit (A72P) in vitro dimerization of Fis1ΔTM when introduced into full-length Fis1 in yeast, disrupt mitochondrial fission.
Discussion
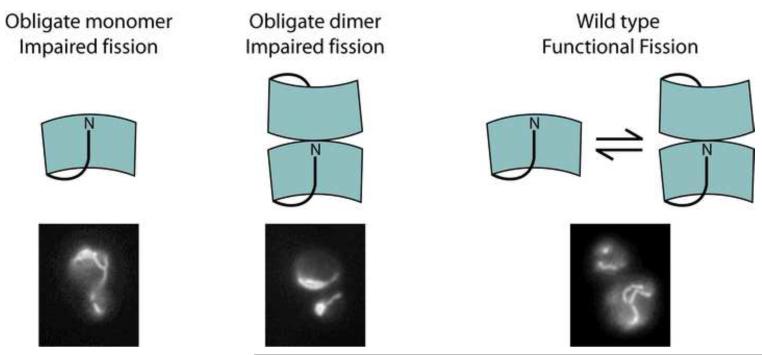
Our analysis of non-functional variants of Fis1 (L80P, ΔN, and Fis1-3), two of which were identified by genetic methods, revealed that the cytoplasmic domain can adopt a dimeric state and that E. coli expressed protein can access this state. These results indicate that the propensity for dimer formation resides within the native polypeptide sequence, and thus may contribute to the function of Fis1. At room temperature, the interconversion between wild type monomer and dimer is sufficiently slow to allow isolation of both species and subsequent analyses led us to hypothesize a structural model for the Fis1 dimer. This model involves three-dimensional domain-swapping, which is distinct from that observed in the human Fis1ΔTM crystal structure. Using a rational design approach, we identified a point mutant, A72P, which potently disrupts homodimerization of wild type and the Fis1ΔTM variants, suggesting a common mode of dimerization. Notably, we observed that A72P alone, which is an obligate monomer, impairs mitochondrial fission. Thus, Fis1 mutants that are obligate monomers (A72P) or constitutive dimers (ΔN and Fis1-3) in vitro are non-functional in vivo.
While the exact oligomeric state of yeast Fis1 on the mitochondrial outer membrane is unknown, several lines of evidence support the hypothesis that the oligomeric state of Fis1 may be important for fission. In mammals, higher order species of Fis1 were proposed to be responsible for a dominant negative phenotype observed upon expression of human Fis1 with substitutions in the TPR motif 52. Also, chemical cross-linking and blue-native PAGE analyses in either mammalian cells or on isolated rat mitochondria demonstrated the presence of higher order species of Fis12, 28, 29. The basis of this higher-order species is unclear, but may involve helix 1 that mediates higher-order assemblies in the crystalline lattice of a high-resolution x-ray structure and has also been shown to be essential for function28, 29, 32. Human Fis1 crosslinks as trimers and dimers in one study and substitutions to residues in the loop between α3 and α4 reduce dimerization and were found to impair fission29. These results in light of the current study suggest that Fis1 dimerization may be evolutionarily conserved between species. While the exact role of each oligomeric species of Fis1 in fission remains obscure, our data that both obligate monomers and obligate dimers are non-functional support a model where the interconversion of at least the monomeric and dimeric states are important, although we cannot yet exclude the possibility that the observed conformational change between α3 and α4 is itself required for fission.
The possibility that A72P is non-functional from a different mechanism, such as direct disruption of Mdv1 or Dnm1 binding, cannot be excluded by our data. However, the ability of the wild type cytoplasmic domain to adopt both monomer and dimer species raises the possibility that they may differ in their relative affinities for Mdv1 and Dnm1. Previously, the mutant alleles L80P, ΔN, and fis1-3 were reported to disrupt fission through mislocalization of Mdv1 that presumably occurred via weakened Fis1-Mdv1 interactions,38,33,37 which our data here indicate may involve Fis1 dimerization. These data lead to the possibility that the Fis1 monomer, when compared to the dimer, binds Mdv1 more tightly. This possibility is supported by biochemical pull-down experiments in which Mdv1 affinity is higher for Fis1ΔTM than ΔN-Fis1ΔTM25. In contrast, this decreased affinity might be explained by disruption of a direct interaction between the Fis1 arm and Mdv1, which is observed in a co-crystal structure with monomer Fis1 and a fragment derived from Mdv127. In a different co-crystal structure of Fis1ΔTM with a longer fragment of Mdv1, the Mdv1 coiled coil domain additionally appears to span two Fis1 monomers like a bridge that would appear to sterically occlude Fis1 homodimerization35. Thus the decreased affinity of ΔN-Fis1 for Mdv1 observed earlier is perhaps a consequence of dimerization. In contrast, Dnm1 shows a hundred-fold higher affinity to ΔN than to wild type Fis1ΔTM25, which we interpret in light of the current study to support the hypothesis that a Fis1 dimer increases binding to Dnm1 while decreases binding to Mdv1.
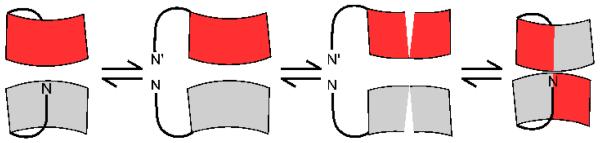
Our data support a plausible physical basis for the kinetic stabilization of monomer and dimer. In order for dimerization to occur, the Fis1 arm that sterically occludes the concave surface in the monomeric structure would need to be physically displaced on two different Fis1 molecules, and these two “open” Fis1 molecules would then have to productively collide during the time frame of their coincident opening (Figure 6). This model predicts that the N-terminal arm would be docked on its own concave surface in a “closed” conformation in the equilibrium resting states of monomer and dimer. Indeed, we have recently addressed the question of whether the Fis1 arm exists in a dynamic equilibrium between closed and open states36. We found that the Fis1 arm lies in a predominantly closed state at room temperature, which is consistent with the idea that energy needs to be added to overcome the barrier between the two states and displace the Fis1 arm producing the open conformation. In this model, dimer formation and/or equilibration between monomer and dimer requires conditions in which the Fis1 arm would have a high probability of being open, as would be expected from elevated temperature, addition of chemical denaturant or mutations that delete (ΔN) or may displace (E78D/I85T/Y88H) the arm from its closed state. Mutations that disrupt the interface between the TPR1 and TPR2 (L80P and C79S/C87S) also would be expected to favor dimer formation, as we observe. A consequence of the model is that non-functional variants should lower the barrier to interconversion. Our initial heat-treatment experiments with the non-functional variants were indeed qualitatively consistent with this model; however, detailed analysis was not possible due to their limited solubility at elevated temperatures. In refolding experiments with the variants, they appear to still have a barrier to interconversion (not shown). We conclude that our data on the wild type and variants support a model in which the Fis1 arm acts as a gatekeeper to lock in the ground states by helping stabilize the α3/α4 interface. The conformational change involving α3/α4 of Fis1 resembles the opening of a “jackknife”, which was previously identified in the Cyp40 and Pex5 TPR containing proteins under crystallization conditions53-55. In these proteins the opening of an extended helix occurred within the helix-turn-helix motif and not between TPR motifs as we find for Fis1. Whether these other TPR containing proteins are kinetically trapped as monomers in solution is not known.
Figure 6. Model for Fis1 dimerization.
In this cartoon model of dimerization, Fis1 is depicted as red and gray crescents and the Fis1 arm as a solid, curved line. For dimerization to occur, the interface between the Fis1 arm and concave surface must be disrupted, and the interface between TPR1 and TPR2 must be disrupted. Mutations that disrupt interactions between either the arm and concave surface (ΔN, fis1-3) or TPR1 and TPR2 (L80P, C79S/C87S) allow dimerization of Fis1.
An important question raised by the current study is how does the interconversion between the Fis1 monomer and dimer occur in vivo? From our data these species do not readily interconvert at room temperature and thus are kinetically trapped. However, many factors missing in our experiments could control this interconversion in vivo. First, the Fis1 transmembrane domain, which is absent in our in vitro studies, may influence the distribution of monomer and dimer by acting to concentrate and limit the orientation of the protein at the membrane. The transmembrane domain may influence the collision rate between two monomers or directly mediate self-association as proposed for the tail-anchored TPR domain protein TM163456. In addition, other physiological factors important for fission that are not present in the current experiments could facilitate equilibration between monomer and dimer, including the membrane itself57, phosphorylation, or ligand binding.
Our identification of the self-association of yeast Fis1 joins a group of emerging examples of TPR domains that mediate self-association41-47. Three examples demonstrate how TPR motif proteins can homooligomerize, a requirement for function. The TPR domain of rapsyn self-associates in clustering nicotinic acetylcholine receptors that is required for proper signaling at the neuromuscular junction. The exact mechanism and oligomeric state of this self-association is unknown but appears to involve reversible interconversion of different oligomeric states40. The conserved and essential eukaryotic protein Sgt1 has also been shown to reversibly dimerize43, which is required for yeast kinetochore assembly58. While the topology of the Sgt1 dimer is unknown and may also involve domain-swapping, other means of TPR-mediating self-association are possible. The bacterial TPR protein YbgF forms a stable trimer whose topology is unknown but likely is mediated by both head-to-head and head-to-tail interactions involving the capping helices that flank the TPR domains48. In this system, trimerization was found to be mediated by conserved Tyr residues that lie between TPR motifs and were found to be both necessary and sufficient for trimerization since a designed, monomeric TPR domain CTPR3 could be “switched” from a monomer to a trimer by introducing two Tyr residues48. Taken together with our findings, these data suggest that the residues that link TPR domains can be modulated to control self-association in different manners that allow exposure of distinct binding surfaces. It is plausible that many TPR-mediated signaling events might have another level of regulation involving TPR self-association as recently proposed48, 59 and supported by our data here.
Domain-swapped dimers play important functional roles in many biological processes60, and many of these dimers require reversible interconversion of oligomeric states via domain-swapping61-65. For example, the cadherins reversibly self-associate through a domain-swap of their IC1 domains to mediate cell adhesion66, 67 and reversible swapping is also found in the interleukin family. Another example is the anti-apoptotic protein Bcl-xL that is predominantly found as a dimer in the cytoplasm68. This protein is predominantly a monomer upon isolation from recombinant expression, but appears kinetically trapped for dimer formation via a similar conformational change to Fis1 involving a helix-turn-helix in the monomer that forms a single, extended helix in the dimer69, 70. The ability of a single polypeptide sequence to adopt more than one conformation is becoming increasingly appreciated with several examples identified through molecular evolution and structural biological approaches71-75. Here we describe an example in which this second conformation is achieved through domain-swapped dimerization, which we propose provides a means to regulate Fis1 activity as is evidenced by nonfunctional variants that favor the monomer or dimer.
Material and methods
Cloning
Recombinant Fis1ΔTM and variants were primarily produced using a pET-29b vector (EMDBiosciences) containing the gene encoding Fis1(1–127) from Saccharomyces cerevisiae, which was kindly provided by Drs. Emily Coonrod and Dr. Janet M. Shaw (University of Utah) and contains at the N-terminus a single, non-native Met and a C-terminal hexahistidine tag that was cleavable by tobacco etch virus (TEV) protease. For purposes of NMR spectroscopy, a Fis1ΔTM construct encoding residues 1-126 without a linker or affinity tag was used. Protein derived from these two constructs showed little difference in their structural properties by NMR spectroscopy (data not shown). Finally, for the variant lacking the N-terminal arm (ΔN16Fis1ΔTM) we used a plasmid encoding residues 17-128 which was subcloned into a pET-29b plasmid. This construct has Leu17 replaced with a Met and contains a linker and thrombin cleavage site preceding a C-terminal 6x His affinity tag.
Protein Expression and Purification
For protein expression, we transformed DNA plasmids into the Rosetta strain of Escherichia coli (EMD Biosciences). An overnight starter culture was used to inoculate 0.5 L of Luria Broth medium with 30 μg/mL of kanamycin and grown at 37 °C. Protein expression was induced at an OD600 of ~0.7 by the addition of 0.4 mM isopropyl β-D-1-thiogalactopyranoside, and the culture temperature was lowered to 20 °C. After 16 to 24 hours, the cells were collected by centrifugation at 6100 x g for 10 minutes. The cells were resuspended in 20 mM sodium phosphate, 250 mM NaCl and 20 mM imidazole, pH 7.4. Cells were homogenized using an EmulsiFlex-C3 cell homogenizer (Avestin). The lysed cells supernatant was applied to a column with Ni Sepharose™ fast flow beads (GE Healthcare) at 4 °C, and protein was eluted with a gradient of 20 to 500 mM imidazole. Wild type Fis1ΔTM protein was further purified by size exclusion chromatography (SEC) using a HiLoad 16/60 Superdex 75 prep grade (S-75) column (GE Healthcare) in Buffer A (50 mM sodium phosphate, 184 mM NaCl, 5 mM ethylenediaminetetraacetic acid (EDTA) and 2 mM dithiothreitol (DTT), pH 7.4). 2H, 15N, 13C labeled Fis1ΔTM (1-126) was expressed similarly in M9 minimal media with 1 g/L 15N ammonium chloride (Sigma-Aldrich), 4 g/L of 13C-glucose (Sigma-Aldrich) in 99.8% deuterium oxide (Sigma-Aldrich). Lysate of 2H, 15N, 13C labeled Fis1ΔTM (1-126) was dialyzed into 50 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.0 (Buffer B). Protein was purified using an SP ion exchange column (GE Healthcare) and eluted by a salt gradient from 0 to 1 M NaCl in Buffer B. Protein concentrations were determined by absorbance at 280 nm using molar extinction coefficients estimated from the primary sequence. For western blot analysis, the C-terminal His tag was detected with a primary mouse monoclonal anti-His tag antibody 1:2000 (R & D Systems) and amplified using a horseradish peroxidase conjugated secondary sheep anti-mouse antibody 1:5000 (GE Healthcare). Wild type samples were at least 99% pure by SEC-HPLC. The mass of each purified construct was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF).
Dimer Formation and Quantification of Oligomeric Species
For thermal perturbation experiments, samples of yeast Fis1ΔTM were dialyzed into Buffer A, incubated at the indicated temperature overnight, and then cooled to 4 °C before returning to room temperature for analysis. For chemical refolding experiments, samples were diluted to the indicated concentration in unfolding buffer (6 M guanidine hydrochloride, 100 mM sodium phosphate, 50 mM NaCl, 10 mM DTT, pH 7.4) and incubated at room temperature before dialysis in Buffer A to remove the chemical denaturant. In these experiments, exact protein concentrations are difficult to control as some dilution is unavoidable during dialysis. We report the initial concentration of the sample before dialysis and estimate the amount of dilution to be <10% from control experiments. Samples were analyzed at room temperature using size exclusion chromatography with detection by intrinsic fluorescence emission at 350 nm (excitation at 280 nm). The resulting chromatograms were fit to determine relative monomer and dimer populations using a modified Gaussian equation: in Igor Pro version 4 (WaveMetrics, Inc.). These populations were used to determine the concentrations of each species for estimation of the apparent dissociation constant for dimerization, KDapp, by: . For ease of comparison the chromatograms are normalized.
Sedimentation Equilibrium Analytical Ultracentrifugation
Sedimentation equilibrium analytical ultracentrifugation data were collected on isolated dimer samples at 7.7 μM, 15.4 μM and 23.2 μM in 50 mM sodium phosphate and 184 mM NaCl, pH 7.4.at rotor speeds of 16.3, 20.0, 24.5, and 30.0 × 103 rpm at 4 °C using a Beckman XLA analytical ultracentrifuge according to published protocols76, 77. Edited sedimentation profiles were analyzed using the Windows version of NONLIN78 or using a software package written by Dr. James Lear for global analysis79. The solvent density, ρ, monomer molecular weight, M, and partial specific volume, , of the protein was calculated from the amino acid sequence using SEDNTERP80, which was also used to estimate the solvent density.
Multi-angle Laser Light Scattering detection of size exclusion chromatography
Fis1ΔTM (1-127) at 75 μM (1.3 mg/mL) was applied onto a Superdex 75 size exclusion column (GE Healthcare) in Buffer A at 0.4 mL/min at room temperature with eluent detection by DAWN HELEOS for 18 light scattering angles and Optilab for refractive index (Wyatt Technologies). Molar masses were determined by fitting the resulting data to the Zimm formalism using the ASTRA software.
Circular Dichroism Spectropolarimetry
Spectra were collected at room temperature using 5 μM samples in 25 mM sodium phosphate and 150 mM sodium fluoride, pH 7.4 in a 0.1 cm path length cuvette. Spectra represent the average of 5 scans from 260 to 190 nm at 50 nm/min with a bandwidth of 0.5 nm, pitch of 0.5 nm and a response of 2 seconds on a J710 spectropolarimeter (JASCO, Inc).
Fluorescence Spectroscopy
The intrinsic tryptophan fluorescence emission spectra was collected of 5 μM yeast Fis1ΔTM in Buffer A using the SLM-48000 spectrofluorometer (SLM Instruments) with an excitation wavelength of 295 nm. The excitation monochromator bandwidth was 2 nm, while that of the emission monochromator was 8 nm to enhance light detection. The excitation polarizer was set at 55° from the vertical, while the emission polarizer was set at 0° from the vertical to minimize the Wood’s anomaly. Each spectrum is the average of two wavelength scans sampled every 1 nm from 306 to 480 nm at 25 °C with each emission wavelength sampled four times. For acrylamide quenching experiments, isolated monomer and dimer samples were incubated in 0.44 M acrylamide in Buffer A for 1.5 hours at room temperature before data collection.
Nuclear Magnetic Resonance Spectroscopy
For monomer samples, HSQC spectra were recorded at 14.1 T on a Bruker Avance600 spectrometer at 25 °C with 16 scans per increment, 640 (t2) x 110 (t1) complex points with acquisition times of 64 ms (1H) and 64 ms (15N). For dimer samples, TROSY-HSQC spectra were recorded at 18.8 T on a Varian INOVA800 spectrometer at 40 °C with 4 scans per increment, 768 (t2) x 283 (t1) complex points with acquisition times of 64 ms (1H) and 100 ms ( 15N). Analyses of TROSY HNN and TROSY HNCACB experiments on dimer samples in conjunction with known chemical shift assignments of monomer were used to determine backbone 1H,13C, and 15N chemical shift assignments of dimer. The TROSY HNN was collected at 8 scans per increment, 720 (t3) x 42 (t2) x 34 (t1) complex points with acquisition times of 60 ms (1H), 18.5 (15N) and 15 ms (15N). The TROSY HNCACB was collected at 16 scans per increment, 720 (t3) x 26 (t2) x 64 (t1) complex points with acquisition times of 60 ms (1H), 17 ms ( 15N) and 5 ms (13C). NMR data processing was carried out using NMRPipe81 and subsequently analyzed using NMRView82, 83.
Yeast two-hybrid
Yeast two-hybrid was performed as previously described using the L40 yeast strain with selection by HIS3 induction84, 85. Two-hybrid constructs were subcloned into the plasmids pGADT7 (prey) and pBHA (bait). L40 transformed with bait (pBHA) and prey (pGADT7) plasmids were grown at 30 °C overnight in synthetic complete media supplemented with 2% glucose lacking leucine and tryptophan. Cells at 1, 0.1, and 0.01 OD600/mL were then replica plated onto the same medium supplemented with 10 mM 3-amino-1,2,4-triazole and lacking histidine. Plates were analyzed for growth after 3 days at 30 °C and are representative of at least 3 independent experiments.
Yeast Growth Assay
Yeast growth assays were performed as described17. Constructs were subcloned into a galactose-inducible pGAL plasmid with a URA3 selection marker and transformed into fzo1Δfis1Δ cells (strain ADM574 gift of Dr. Janet Shaw)17. The cells were grown were grown at 30 °C overnight in synthetic complete media supplemented with 2% glucose lacking uracil. The cells were diluted to an OD600 of 0.1, 0.05 and 0.02 and replica plated onto YP medium containing either 2% glucose or 3% glycerol. Plates were analyzed for growth after 5 days at 30 °C and are representative of at least 3 independent experiments.
Mitochondrial morphology
For mitochondrial morphology 3xHAFIS1 and mutant constructs were subcloned into pRS416MET25 and transformed into fis1Δ yeast (ADM552 gift of Dr. Janet Shaw)17 . The yeast were grown at 30 °C in synthetic complete media supplemented with 2% glucose lacking uracil and leucine to log-phase (0.55 to 0.8 OD600). Cells were stained with 2 μM MitoTracker Red CMXRos (Invitrogen) for 20 minutes in the dark followed by two washes and resuspension in synthetic complete media with 2% glucose lacking uracil and leucine. At least 100 cells per strain were counted from three independent growths using a BX51TRF microscope (Olympus) with the bright field or fluorescence settings. Images were taken with MagnaFire (Karl Storz Imaging) and processed using ImageJ 1.45S (NIH).
Supplementary Material
Fis1 is a tetratricopeptide repeat protein involved in organelle homeostasis.
Nonfunctional variants of yeast Fis1 are unexpectedly dimeric
Dimerization is an intrinsic property of Fis1 but is kinetically trapped
Both a designed obligate monomer and obligate dimer are each nonfunctional
TPR proteins may reversibly self-associate in a new and functionally important manner
Acknowledgements
We thank Kevin R. MacKenzie for insightful discussions and critical readings of the manuscript. This work was supported by the National Institutes of Health Grant RO1GM067180 and American Cancer Society Award IRG-58-005-41. SC was a recipient of a post-doctoral fellowship from the Spanish Government. JPBL, LKP and AET were supported by a training grant from NIH (2T32GM007231). CMM was supported by an NSF pre-doctoral award. KGF acknowledges funding from the NSF (MCB0423807). We thank Dr. Arne Schon for commenting on an earlier draft of this manuscript. We would like to thank Dr. Ananya Majumdar of the JHU Biomolecular NMR Center for invaluable advice and assistance, Dr. Ludwig Brand and Dmitri Toptygin for use of their fluorometer. We thank Drs. Janet Shaw and Emily Coonrod for graciously providing a plasmid that encodes Fis1ΔTM with a 6x His affinity tag at the C-terminus and yeast strain JSY5663 for the growth assay. We also thank Dr. Craig Blackstone for the pGADT7, pBHA yeast two-hybrid vectors and L40 yeast strain. We also thank Dr. Marie Hardwick for the pGAL plasmid that was used for cloning for the growth assay. We would like to thank Drs. Edward Hedgecock and Rui Proenca for use of their microscope and general lab equipment.
Footnotes
Author Contributions J.P.L participated in research design, data collection of all experiments, excluding yeast two-hybrid and analytical ultracentrifugation experiments, and analyzed the data. C.M.M. collected and analyzed yeast two-hybrid data. S.C.A participated in research design, collection and analysis of the NMR chemical shift data. A.E.T. and K.G.F. participated in research design, collection and analysis of the analytical ultracentrifugation data. J.M.F. participated in research design, data collection and analysis of the SEC-MALS data. L.K.P. participated in research design, data collection and analysis of NMR chemical shifts and growth assay data. R.B.H. participated in research design and data analysis, as well as NMR and SEC-MALS data collection. The manuscript was written primarily by J.P.L. and R.B.H. with input from all co-authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Camoes F, Bonekamp NA, Delille HK, Schrader M. Organelle dynamics and dysfunction: A closer link between peroxisomes and mitochondria. J. Inherit. Metab. Dis. 2009;32:163–180. doi: 10.1007/s10545-008-1018-3. 10.1007/s10545-008-1018-3. [DOI] [PubMed] [Google Scholar]
- 2.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell. Sci. 2008;121:1633–1640. doi: 10.1242/jcs.026344. 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi S, Tanaka A, Fujiki Y. Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 2007;313:1675–1686. doi: 10.1016/j.yexcr.2007.02.028. 10.1016/j.yexcr.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen T, Bjorkman J, Paton BC, Crane DI. Failure of microtubule-mediated peroxisome division and trafficking in disorders with reduced peroxisome abundance. J. Cell. Sci. 2006;119:636–645. doi: 10.1242/jcs.02776. jcs.02776 [pii]; 10.1242/jcs.02776 [doi] [DOI] [PubMed] [Google Scholar]
- 6.Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M. Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 2003;278:8597–8605. doi: 10.1074/jbc.M211761200. 10.1074/jbc.M211761200 [doi]; M211761200 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 2003;278:17012–17020. doi: 10.1074/jbc.M212031200. 10.1074/jbc.M212031200 [doi]; M212031200 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. E05-02-0159 [pii]; 10.1091/mbc.E05-02-0159 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank S. Dysregulation of mitochondrial fusion and fission: an emerging concept in neurodegeneration. Acta Neuropathol. (Berl) 2006;111:93–100. doi: 10.1007/s00401-005-0002-3. 10.1007/s00401-005-0002-3 [doi] [DOI] [PubMed] [Google Scholar]
- 10.Santel A. Get the balance right: mitofusins roles in health and disease. Biochim. Biophys. Acta. 2006;1763:490–499. doi: 10.1016/j.bbamcr.2006.02.004. S0167-4889(06)00032-2 [pii]; 10.1016/j.bbamcr.2006.02.004 [doi] [DOI] [PubMed] [Google Scholar]
- 11.Amiott EA, Lott P, Soto J, Kang PB, McCaffery JM, Dimauro S, Abel ED, Flanigan KM, Lawson VH, Shaw JM. Mitochondrial fusion and function in Charcot-Marie-Tooth type 2A patient fibroblasts with mitofusin 2 mutations. Exp. Neurol. 2008 doi: 10.1016/j.expneurol.2008.01.010. 10.1016/j.expneurol.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. 356/17/1736 [pii]; 10.1056/NEJMoa064436 [doi] [DOI] [PubMed] [Google Scholar]
- 13.Chan DC. Mitochondrial dynamics in disease. N Engl J Med. 2007;356:1707. doi: 10.1056/NEJMp078040. [DOI] [PubMed] [Google Scholar]
- 14.Ashrafian H, Docherty L, Leo V, Towlson C, Neilan M, Steeples V, Lygate CA, Hough T, Townsend S, Williams D, Wells S, Norris D, Glyn-Jones S, Land J, Barbaric I, Lalanne Z, Denny P, Szumska D, Bhattacharya S, Griffin JL, Hargreaves I, Fernandez-Fuentes N, Cheeseman M, Watkins H, Dear TN. A mutation in the mitochondrial fission gene Dnm1l leads to cardiomyopathy. PLoS Genet. 2010;6:e1001000. doi: 10.1371/journal.pgen.1001000. 10.1371/journal.pgen.1001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fekkes P, Shepard KA, Yaffe MP. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakobs S, Martini N, Schauss AC, Egner A, Westermann B, Hell SW. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J. Cell. Sci. 2003;116:2005–2014. doi: 10.1242/jcs.00423. 10.1242/jcs.00423 [doi]; jcs.00423 [pii] [DOI] [PubMed] [Google Scholar]
- 22.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. 10.1074/jbc.M303758200 [doi]; M303758200 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell. Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. 10.1242/jcs.01058 [doi]; 117/7/1201 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Schauss AC, Bewersdorf J, Jakobs S. Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface. J. Cell. Sci. 2006;119:3098–3106. doi: 10.1242/jcs.03026. jcs.03026 [pii]; 10.1242/jcs.03026 [doi] [DOI] [PubMed] [Google Scholar]
- 25.Wells RC, Picton LK, Williams SC, Tan FJ, Hill RB. Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J. Biol. Chem. 2007;282:33769–33775. doi: 10.1074/jbc.M700807200. 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. jcb.200503148 [pii]; 10.1083/jcb.200503148 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jofuku A, Ishihara N, Mihara K. Analysis of functional domains of rat mitochondrial Fis1, the mitochondrial fission-stimulating protein. Biochem. Biophys. Res. Commun. 2005;333:650–659. doi: 10.1016/j.bbrc.2005.05.154. S0006-291X(05)01103-4 [pii]; 10.1016/j.bbrc.2005.05.154 [doi] [DOI] [PubMed] [Google Scholar]
- 29.Serasinghe MN, Yoon Y. The mitochondrial outer membrane protein hFis1 regulates mitochondrial morphology and fission through self-interaction. Exp. Cell Res. 2008 doi: 10.1016/j.yexcr.2008.09.009. 10.1016/j.yexcr.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohashi W, Hirota H, Yamazaki T, Koshiba S, Hamada T, Yoshida M, Yokoyama S. Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA. 2002.
- 31.Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N. The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J. Mol. Biol. 2003;334:445–458. doi: 10.1016/j.jmb.2003.09.064. S002228360301221X [pii] [DOI] [PubMed] [Google Scholar]
- 32.Dohm JA, Lee SJ, Hardwick JM, Hill RB, Gittis AG. Cytosolic domain of the human mitochondrial fission protein Fis1 adopts a TPR fold. Proteins. 2004;54:153–156. doi: 10.1002/prot.10524. 10.1002/prot.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki M, Neutzner A, Tjandra N, Youle RJ. Novel structure of the N terminus in yeast Fis1 correlates with a specialized function in mitochondrial fission. J. Biol. Chem. 2005;280:21444–21452. doi: 10.1074/jbc.M414092200. M414092200 [pii]; 10.1074/jbc.M414092200 [doi] [DOI] [PubMed] [Google Scholar]
- 34.Tooley JE, Khangulov V, Lees JP, Schlessman JL, Bewley MC, Héroux A, Bosch J, Hill RB. 1.75 Å crystal structure of fission protein Fis1 from Saccharomyces cerevisiae reveals elusive interactions of the autoinhibitory domain. Acta Cryst. 2011;F67:1310–1315. doi: 10.1107/S1744309111029368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Chan NC, Ngo HB, Gristick H, Chan DC. Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (mdv1) coiled coil. J. Biol. Chem. 2012;287:9855–9861. doi: 10.1074/jbc.M111.329359. 10.1074/jbc.M111.329359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picton LK, Casares S, Monahan AC, Majumdar A, Hill RB. Evidence for conformational heterogeneity of fission protein Fis1 from Saccharomyces cerevisiae. Biochemistry. 2009;48:6598–6609. doi: 10.1021/bi802221h. 10.1021/bi802221h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karren MA, Coonrod EM, Anderson TK, Shaw JM. The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J. Cell Biol. 2005;171:291–301. doi: 10.1083/jcb.200506158. jcb.200506158 [pii]; 10.1083/jcb.200506158 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. 10.1083/jcb.200205031 [doi]; jcb.200205031 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koirala S, Bui HT, Schubert HL, Eckert DM, Hill CP, Kay MS, Shaw JM. Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes. J. Cell Biol. 2010;191:1127–1139. doi: 10.1083/jcb.201005046. 10.1083/jcb.201005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramarao MK, Bianchetta MJ, Lanken J, Cohen JB. Role of rapsyn tetratricopeptide repeat and coiled-coil domains in self-association and nicotinic acetylcholine receptor clustering. J. Biol. Chem. 2001;276:7475–7483. doi: 10.1074/jbc.M009888200. 10.1074/jbc.M009888200. [DOI] [PubMed] [Google Scholar]
- 41.Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- 42.Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 43.Nyarko A, Mosbahi K, Rowe AJ, Leech A, Boter M, Shirasu K, Kleanthous C. TPR-Mediated Self-Association of Plant SGT1. Biochemistry. 2007;46:11331–11341. doi: 10.1021/bi700735t. [DOI] [PubMed] [Google Scholar]
- 44.Han D, Kim K, Kim Y, Kang Y, Lee JY, Kim Y. Crystal structure of the N-terminal domain of anaphase-promoting complex subunit 7. J. Biol. Chem. 2009;284:15137–15146. doi: 10.1074/jbc.M804887200. 10.1074/jbc.M804887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graf C, Stankiewicz M, Nikolay R, Mayer MP. Insights into the conformational dynamics of the E3 ubiquitin ligase CHIP in complex with chaperones and E2 enzymes. Biochemistry. 2010;49:2121–2129. doi: 10.1021/bi901829f. 10.1021/bi901829f. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z, Roe SM, Diogon M, Kong E, El Alaoui H, Barford D. Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture. J. Mol. Biol. 2010;397:1316–1328. doi: 10.1016/j.jmb.2010.02.045. 10.1016/j.jmb.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 48.Krachler AM, Sharma A, Kleanthous C. Self-association of TPR domains: Lessons learned from a designed, consensus-based TPR oligomer. Proteins. 2010;78:2131–2143. doi: 10.1002/prot.22726. 10.1002/prot.22726. [DOI] [PubMed] [Google Scholar]
- 49.O’Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 50.Hermann GJ, Shaw JM. Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- 51.Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- 52.Yu T, Fox RJ, Burwell LS, Yoon Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J. Cell. Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. jcs.02537 [pii]; 10.1242/jcs.02537 [doi] [DOI] [PubMed] [Google Scholar]
- 53.Kumar A, Roach C, Hirsh IS, Turley S, deWalque S, Michels PA, Hol WG. An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei. J. Mol. Biol. 2001;307:271–282. doi: 10.1006/jmbi.2000.4465. 10.1006/jmbi.2000.4465. [DOI] [PubMed] [Google Scholar]
- 54.Sampathkumar P, Roach C, Michels PA, Hol WG. Structural insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5. J. Mol. Biol. 2008;381:867–880. doi: 10.1016/j.jmb.2008.05.089. 10.1016/j.jmb.2008.05.089. [DOI] [PubMed] [Google Scholar]
- 55.Taylor P, Dornan J, Carrello A, Minchin RF, Ratajczak T, Walkinshaw MD. Two structures of cyclophilin 40: folding and fidelity in the TPR domains. Structure. 2001;9:431–438. doi: 10.1016/s0969-2126(01)00603-7. [DOI] [PubMed] [Google Scholar]
- 56.McCleverty CJ, Columbus L, Kreusch A, Lesley SA. Structure and ligand binding of the soluble domain of a Thermotoga maritima membrane protein of unknown function TM1634. Protein Sci. 2008;17:869–877. doi: 10.1110/ps.083432208. 10.1110/ps.083432208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wells RC, Hill RB. The cytosolic domain of Fis1 binds and reversibly clusters lipid vesicles. PLoS One. 2011;6:e21384. doi: 10.1371/journal.pone.0021384. 10.1371/journal.pone.0021384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bansal PK, Nourse A, Abdulle R, Kitagawa K. Sgt1 Dimerization Is Required for Yeast Kinetochore Assembly. J. Biol. Chem. 2009;284:3586–3592. doi: 10.1074/jbc.M806281200. 10.1074/jbc.M806281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allan RK, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16:353–367. doi: 10.1007/s12192-010-0248-0. 10.1007/s12192-010-0248-0; 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bennett M, Schlunegger M, Eisenberg D. 3d Domain Swapping - a Mechanism for Oligomer Assembly. Protein Science. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fletterick R, Bazan J. When One and One are Not 2 Rid B-4562-2010. Nat. Struct. Biol. 1995;2:721–723. doi: 10.1038/nsb0995-721. 10.1038/nsb0995-721. [DOI] [PubMed] [Google Scholar]
- 62.Saint-Jean A, Phillips K, Creighton D, Stone M. Active monomeric and dimeric forms of Pseudomonas putida glyoxalase I: Evidence for 3D domain swapping. Biochemistry (N. Y. ) 1998;37:10345–10353. doi: 10.1021/bi980868q. 10.1021/bi980868q. [DOI] [PubMed] [Google Scholar]
- 63.Schymkowitz J, Rousseau F, Wilkinson H, Friedler A, Itzhaki L. Observation of signal transduction in three-dimensional domain swapping. Nat. Struct. Biol. 2001;8:888–892. doi: 10.1038/nsb1001-888. 10.1038/nsb1001-888. [DOI] [PubMed] [Google Scholar]
- 64.Rousseau F, Schymkowitz J, Itzhaki L. The unfolding story of three-dimensional domain swapping. Structure. 2003;11:243–251. doi: 10.1016/s0969-2126(03)00029-7. 10.1016/S0969-2126(03)00029-7. [DOI] [PubMed] [Google Scholar]
- 65.Czjzek M, Letoffe S, Wandersman C, Delepierre M, Lecroisey A, Izadi-Pruneyre N. The crystal structure of the secreted dimeric form of the hemophore HasA reveals a domain swapping with an exchanged heme ligand. J. Mol. Biol. 2007;365:1176–1186. doi: 10.1016/j.jmb.2006.10.063. 10.1016/j.jmb.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 66.Nagar B, Overduin M, Ikura M, Rini J. Structural basis of calcium-induced E-cadherin rigidification and dimerization RID C-1297-2010. Nature. 1996;380:360–364. doi: 10.1038/380360a0. 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 67.Vendome J, Posy S, Jin X, Bahna F, Ahlsen G, Shapiro L, Honig B. Molecular design principles underlying beta-strand swapping in the adhesive dimerization of cadherins. Nat. Struct. Mol. Biol. 2011;18:693–700. doi: 10.1038/nsmb.2051. 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeong SY, Gaume B, Lee YJ, Hsu YT, Ryu SW, Yoon SH, Youle RJ. Bcl-x(L) sequesters its C-terminal membrane anchor in soluble, cytosolic homodimers. Embo J. 2004;23:2146–55. doi: 10.1038/sj.emboj.7600225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Neill JW, Manion MK, Maguire B, Hockenbery DM. BCL-XL dimerization by three-dimensional domain swapping. J. Mol. Biol. 2006;356:367–381. doi: 10.1016/j.jmb.2005.11.032. 10.1016/j.jmb.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 70.Denisov AY, Sprules T, Fraser J, Kozlov G, Gehring K. Heat-induced dimerization of BCL-xL through alpha-helix swapping. Biochemistry. 2007;46:734–740. doi: 10.1021/bi062080a. 10.1021/bi062080a. [DOI] [PubMed] [Google Scholar]
- 71.DeAntoni A, Sala V, Musacchio A. Explaining the oligomerization properties of the spindle assembly checkpoint protein Mad2. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2005;360:637–47. doi: 10.1098/rstb.2004.1618. discussion 447-8. 10.1098/rstb.2004.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 75.Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS. Metamorphic proteins mediate evolutionary transitions of structure. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7287–7292. doi: 10.1073/pnas.0912616107. 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fleming KG, Ackerman AL, Engelman DM. The effect of point mutations on the free energy of transmembrane alpha-helix dimerization. J. Mol. Biol. 1997;272:266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- 77.Stanley AM, Chuawong P, Hendrickson TL, Fleming KG. Energetics of outer membrane phospholipase A (OMPLA) dimerization. J. Mol. Biol. 2006;358:120–131. doi: 10.1016/j.jmb.2006.01.033. 10.1016/j.jmb.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 78.Yphantis DA. Equilibrium Ultracentrifugation of Dilute Solutions. Biochemistry. 1964;3:297–317. doi: 10.1021/bi00891a003. [DOI] [PubMed] [Google Scholar]
- 79.Salom D, Hill BR, Lear JD, DeGrado WF. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laue T, Shah B, Ridgeway R, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe AJ, Horton JC, editors. Analytical ultra-centrifugation in biochemistry and polymer. Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- 81.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 82.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 83.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4 doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 84.Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, Blackstone C. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J. Biol. Chem. 2010;285:32494–32503. doi: 10.1074/jbc.M110.142430. 10.1074/jbc.M110.142430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J. Biol. Chem. 2004;279:35967–35974. doi: 10.1074/jbc.M404105200. 10.1074/jbc.M404105200 [doi]; M404105200 [pii] [DOI] [PubMed] [Google Scholar]
- 86.DeLano WL. PyMOL. 2002 http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.