Abstract
Background
Clinical studies have identified several regions of the genome with copy number variations (CNVs) associated with diverse neurodevelopmental behavioral disorders.
Methods
We analyzed 1M SNP genotype arrays (Illumina BeadArrays) for evidence of previously reported recurrent CNVs and enriched genome wide CNV burden in DNA from 600 brains, including 441 individuals with various psychiatric diagnoses. We explored gene expression in the dorsolateral prefrontal cortex in selected cases with CNVs and in other subjects using Illumina BeadArrays (568 subjects in total), and additionally in 66–92 subjects using quantitative real-time PCR.
Results
CNVs in previously reported genomic regions were identified in 4/193 patients with the diagnosis of schizophrenia (1q21.1, 11q25, 15q11.2, 22q11), 4/238 patients with mood disorders (11q25, 15q11.2, 22q11), and 1/10 patients with autism (2p16.3). No evidence of increased genome wide CNV burden was observed in cases with schizophrenia or mood disorders although the study is underpowered to observe rare events. mRNA expression patterns suggested incomplete molecular penetrance of observed CNVs.
Conclusions
Our data confirm in brain DNA the presence of certain recurrent CNVs in a small percentage of patients with psychiatric diagnoses.
Keywords: copy number variations, major depressive disorder, schizophrenia, autism, gene expression, postmortem brain, prefrontal cortex
Introduction
Rare structural chromosomal anomalies are associated with neurodevelopmental disorders, including mental deficiency, autism, and schizophrenia. Recent studies have identified several recurrent CNVs in as much as 2–4% of patients with schizophrenia, and even more frequently in patients with autism and mental deficiency. Moreover, the overall genome-wide CNV burden in patients with neurodevelopmental disorders, such as autism and schizophrenia, has been reported to be increased in some, but not all, studies [1–7], suggesting the possibility of a general mass effect of structural variation in the genomes of these conditions. All prior CNV studies have been performed on peripheral cells or transformed cell lines, which undergo repeated replication throughout life. Because there is a possibility that CNVs can be generated during cell division [8], it is important to confirm that such anomalies are found in brain DNA in association with clinical illness. The replication history of brain cells is considerably more constricted than that of any readily available peripheral cell. It is also important to document that these structural anomalies impact on expression of genes in the affected regions, as their pathogenicity is presumably related to such effects. While it might seem obvious that the effects of these relatively large genetic variations would penetrate to the level of gene expression, it is conceivable that some might be compensated for by the intact chromosome. We have performed a high density SNP array analysis of CNVs from brain DNA in a relatively large sample of controls and patients with psychiatric disorders and have examined effects of selected CNVs on gene expression in brain. Our study is the first analysis of CNV burden and of the presence of CNVs previously associated with schizophrenia and autism in brain DNA and the first to explore their effect on expression of relevant genes in human brain tissue.
Material and Methods
The sample consisted of 600 brains >13 years of age collected as previously described (9), and detailed in the table. 149 of the brain samples were provided by the Stanley Medical Research Institute; the rest were from CBDB/NIMH. DNA was extracted from cerebellar tissue and RNA for the expression study was extracted from the dorsolateral prefrontal cortex according to standard protocols (Qiagen). All samples were genotyped with Illumina 1M Duo SNP chips. Log R ratios of SNP probe intensities and B allele frequencies were called using Illumina GenomeStudio V2010.1 software. CNVs were determined using two algorithms, PennCNV2010 (10) and QuantiSNP (11), which use a hidden Markov model and an objective Bayes hidden Markov model, respectively, to identify duplications and deletions based on a number of parameters, including total signal intensity and allelic intensity ratio for each SNP, total number of SNPs, distance between SNPs, and allele frequency. Standard PennCNV quality control checks were used to exclude samples for which PennCNV calling would be considered unreliable, these included LRR standard deviation > 0.28, BAF median > 0.55 or < 0.45, BAF drift > 0.002 or WF > 0.04 or <−0.04. Additionally, to ensure only high-confidence CNVs were included in the analysis, we excluded any CNV for which the difference of the log likelihood of the most likely copy number state and the less likely copy number state was less than 10 (generated using the -conf function in PennCNV). We adopted a conservative approach to CNV calling, opting to minimize type I error at the expense of Type II error. Thus, we first examined CNVs only if they were called with moderately high confidence (confidence criteria >10) by both algorithms and overlapped by at least 50% and contained at least 10 SNPs. Confidence scores are a measure that reflects how confident the CNV-detection algorithm is in calling the integer copy number. We also examined our data after increasing the threshold of confidence for CNV calls in each algorithm to 50 and also after restricting CNV calls to regions with a minimum of 20 SNPs. CNV calls were visually confirmed by inspection of the Log R plots. Expression analyses in tissue serve as a functional confirmation of the validity of the CNV call procedure. Our procedure is likely to miss some real CNVs, but is relatively robust to false positives and not likely to miss differences in genome-wide CNV burden between diagnostic groups based solely on the call algorithm. Because prior work has suggested that CNVs involving genes [1] are particularly enriched in association with schizophrenia, we only analyzed CNVs within 100 kb of annotated genes, unless the CNV had been previously associated with schizophrenia or autism. Finally, we examined separately CNVs greater than 100kb and CNVs between 10 and 100 kb. The whole-genome burden (i.e. total number of CNVs of a given size threshold identified in an individual genome) was calculated separately for groups of subjects with autism, bipolar disorder, major depression and schizophrenia and compared with controls using Students t-tests. The maps in figures S11–15 were created using the Database of Genomic Variants (DGV) http://projects.tcag.ca/cgi-bin/variation/gbrowse/hg18/, build 36, hg18.
Table 1.
Results of CNV analyses and sample characteristics
| Diagnosis | N | Males | Race/AA/C/AS/HISP | Mean age | Mean/dels/subj | Mean/dups/subj | Mean del size | Mean dup size |
|---|---|---|---|---|---|---|---|---|
| A. Summary of CNVs >100kb called by both algorithms. Confidence parameter is 10 | ||||||||
| Autism | 10 | 8 | 2/8/0/0 | 29.8 | 1.8 | 3.9 | 314506.9 | 1544605 |
| Bipolar | 84 | 48 | 7/71/3/3 | 46.1 | 1.5 | 3.2 | 281145.4 | 902227.3 |
| MDD | 154 | 90 | 17/132/2/3 | 44.3 | 1.3 | 3.3 | 253754.3 | 1071212 |
| Schizo | 193 | 121 | 82/103/3/5 | 49.9 | 1.2 | 3 | 292690.8 | 946572.3 |
| Control | 159 | 114 | 77/71/6/5 | 44.5 | 1.4 | 2.8 | 272851.2 | 839455.9 |
| B. Summary of CNVs 10kb–100kb. Confidence parameter is 10 | ||||||||
| Autism | 10 | 8 | 2/8/0/0 | 29.8 | 2.2 | 1.4 | 108379.1 | 100344 |
| Bipolar | 81 | 46 | 7/68/3/3 | 45.4 | 2.1 | 2.3 | 90982.5 | 116176.4 |
| MDD | 154 | 90 | 18/131/2/3 | 44.3 | 1.8 | 2.1 | 81915.1 | 113914.9 |
| Schizo | 189 | 119 | 80/101/3/5 | 50 | 1.9 | 2.4 | 90494.9 | 136542.8 |
| Control | 153 | 111 | 76/67/6/4 | 44.7 | 1.8 | 2.6 | 83934.7 | 124391.5 |
| C. Summary of CNVs >10kb. Confidence parameter is 50 | ||||||||
| Autism | 10 | 8 | 2/8/0/0 | 29.8 | 2.3 | 3.4 | 283170.1 | 837499.5 |
| Bipolar | 80 | 47 | 7/67/3/3 | 46.1 | 2 | 2.7 | 232715 | 686375.7 |
| MDD | 155 | 90 | 18/132/2/3 | 44.5 | 1.7 | 2.7 | 188946.1 | 830051 |
| Schizo | 192 | 119 | 82/102/3/5 | 50 | 1.8 | 2.3 | 252047 | 623722.1 |
| Control | 155 | 112 | 77/67/6/5 | 44.6 | 1.9 | 2.5 | 223198.7 | 575786.7 |
CNV = copy number variations, dels = deletions, dups=duplications, subj=subject
AA =African American, C=Caucasian, AS=Asian, HISP=Hispanic
Expression data obtained from Human HT-12_V3 Illumina BeadArrays were generated by Illumina Beadstudio. Only probes expressed above the background (p<0.05) in at least 80% subjects were further analyzed. We normalized the data using the lumi R package (12). The ComBat R package was used for batch effect correction (13). We applied surrogate variable analysis using SVA R package (14) with age, sex, race and diagnosis as primary variables. A step-wise model selection was used for each gene and a multiple linear regression analysis performed using the best fit model. After removing the effects of sex, race and surrogate variables, the residuals were used for comparing the expression levels between subjects. We also performed quantitative PCR (qPCR) to measure mRNA expression of selected genes in the CNV regions in the dorsolateral prefrontal cortex of three patients with schizophrenia and one patient with major depressive disorder (MDD), each found to have a recurrent CNV, and in a subsample of 66–92 subjects without CNVs in these regions (PRKA2B in 1q21.1, Taqman assay Hs00271294_m1; COMT in 22q11, assay Hs00241349_m1; NIPA1 in 15q11, assay Hs00331974_m1), according to previously described protocols (9). mRNA levels were normalized to the geometric mean of three “housekeeping” genes (ACTB, B2M and GUSB) for analysis of the qPCR expression levels.
Results
Despite the lack of power because of the small number of cases with the diagnosis of autism (n=10), this sample had a numerically increased burden of CNVs >100kb calculated at the threshold of confidence 10 (t=2.0, p=0.045 with the number of SNPs >10 in the calling algorithm, and t=1.8, p=0.07 with the number of SNPs >20, total sample n=600). After increasing the confidence to 50, there was still a nominally significant increase in the burden of CNVs >100 kb for the cases with autism (t=2.3, p= 0.02 with 10 SNPs in the CNV call, but t=1.6, p=0.09 when the number of SNPs was increased to >20). When the CNV size was restricted to 10–100 kb, there was no significant increase in the CNV burden in autism regardless of all the other criteria (all p values >0.2). No evidence of an increased genome-wide CNV burden was found in cases with schizophrenia (all t values <1.0, all p values >0.2), major depression (all t values <1.5, all p values >0.08) or bipolar disorder (all t values <1.5, all p values >0.14), regardless of the CNV size criteria, the number of SNPS in the CNV call (>10 or >20) or the threshold of confidence for CNV calls (10 or 50). Finally, we counted the genome wide CNV burden using calls from only PennCNV or only QuantiSNP and again, no differences were found in genome-wide CNVs between patients with schizophrenia, major depression or bipolar disorder and controls. These negative results must be viewed with caution as the sample is decidedly underpowered to observe genome wide CNV burden increases given the low frequency of CNVs found in much larger clinical populations.
Ten subjects (Samples 1–10) had a CNV called in regions previously identified as associated with schizophrenia or autism. There were four patients with schizophrenia with such recurrent CNVs: Sample 1 had a 2.7 Mb deletion in the 22q11 region associated with the velo-cardio-facial (VCFS) syndrome (Fig S1), Sample 2 had a 1.6 Mb deletion in 1q21.1, Fig S2; Sample 3 had a 53 kb deletion in 11q25, Fig S3, and Sample 4 had a 1.9 Mb duplication in 15q11.2 Fig S4 (15). A 53 kb deletion in 11q25 was also found in Sample 5, a subject with major depression (Fig S5), and a 52 kb duplication in 11q25 was found in Sample 6, a normal subject (Fig S6). Two patients with bipolar disorder had duplications in 15q11.2, Sample 7 (476 kb) and Sample 8 (325 kb), Figs S7 and S8. Sample 9, a patient with major depression, had a 2.5 Mb duplication in the 22q11 VCFS region (Fig S9). Sample 10, a patient with autism, had a 69 kb deletion in 2p16.3 affecting NRXN1 (16), Fig S10. The detailed maps of the CNVs in all ten samples, including coordinates, comparisons of sizes between the cases, indications of the annotated genes affected, as well as comparisons with previously reported CNVs in the same region are provided in figures S11–S15. No other CNVs were identified in other chromosomal regions previously associated with schizophrenia or autism, including in 2q33.3, 3q29, 5p13, 7q34, 7q36, 8q21, 15q13.3, 16p11.2, 16p13.1, 16p13.2, or 18p21 (1,3,6).
We assessed the effects of the CNVs on the expression profiles of genes within the CNVs and compared the cases with control subjects using a whole-genome approach for seven of the cases described above (samples 1–4, 6–8, on whom we had expression array data), and additionally examined four cases by qPCR (samples 1, 2, 4 and 9). In Sample 1 with a 2.7 Mb deletion in the 22q11 region, a comparison of the distribution of the expression levels of the genes in the CNV region with genes genome-wide (or outside the CNV on the same chromosome) clearly shows enrichment of relatively low expressed genes in the 22q11 region of this subject as compared to all other subjects (Fig S16). In addition to a dramatic down regulation of COMT expression, 19 other hemi-deleted genes showed extremely low levels of expression in comparison to all other subjects. Data obtained by qPCR confirmed down regulation of COMT in Sample 1: relative COMT mRNA level was 0.62 compared with an average of 1.24 in the other subjects (N=90, Figure 1). Similarly, in Sample 2 with the 1q21 deletion, there was an enrichment of relatively low expressed genes among the genes in the deleted region as compared with the whole genome (and with the genes outside the CNV on the same chromosome, data not shown) (Fig S17). As measured by qPCR, PRKAB2 mRNA expression in Sample 2 was 0.45 compared with an average 1.40 in other assayed subjects (N=67, FigS23). We also measured COMT mRNA expression by qPCR in the case with major depression (Sample 9 with the 22q11 duplication), which showed a slight increase in COMT expression (1.45 compared with an average of 1.20 for 92 other subjects, Figure 1). Although less clear, there was also enrichment of genes in the duplicated regions, which showed higher expression as compared to expression levels across the whole genome (see Figs S18–22). A few genes in the duplicated regions of 15q11.2 stand out as showing relatively high expression, including NIPA2 that shows upregulation in all three subjects with this CNV (Figs S18, S19, S20 and S22). However, the 15q duplication case (Sample S4) showed only a slight difference of NIPA1 expression compared with all other subjects (1.32 vs 1.83, N=66), Fig S22. In sum, gene expression in the deletion cases tended to be at the bottom of the entire distribution of samples, whereas the duplication cases were less consistently separated from the rest of the samples.
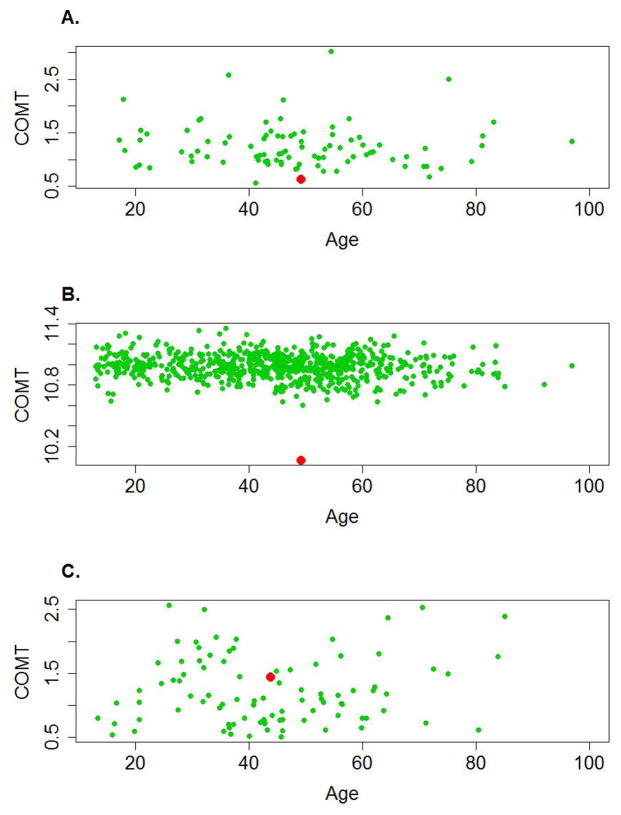
Figure 1.
Expression of COMT mRNA in the CNV region 22q11.21. A, B Sample 1, a patient with schizophrenia (red dot) with a deletion in 22q11.21, and controls without the CNV (green dots) measured by qPCR (A) and Illumina BeadArrays (B) in the postmortem dorsolateral prefrontal cortex. There is good agreement between qPCR and microarray data. C. Sample 9, a patient with major depression (red dot), with a duplication in 22q11.21, and controls without the CNV (green dots) measured by qPCR. There are no array data for this subject. Y axes represent expression of COMT mRNA normalized as described in the Methods. X axes represent age of subjects at death. Every dot represents a subject.
Discussion
We have performed a study of CNVs in DNA from a relatively large sample of human brains and explored effects on gene expression. Because of the rarity of recurrent CNVs previously associated with schizophrenia, we expected to find few in this sample. Our observation of four cases diagnosed with schizophrenia having CNVs in regions previously associated with this diagnosis and other developmental disorders is consistent with the expected rates of discovery of such CNVs in association with these diagnoses (2–3%). We found no evidence, however, for an overall greater gene-centric CNV burden across the genome in patients with schizophrenia compared with controls and no difference in genome-wide CNV burden between patients with the diagnosis of schizophrenia, major depression or bipolar disorder. These negative results must be viewed with caution as the increase in genome wide CNV burden reported in some earlier studies was very small and our study is underpowered to make similar observations. Nevertheless, the possibility that some of the previously reported private CNVs in single patients with schizophrenia were not of germ line origin (8) is not excluded by our data.
Perhaps of greater interest, gene expression in the CNV cases is not as distinctly extreme as might have been expected. While this may represent methodological noise, it also raises the possibility that compensation is made on the intact chromosome, limiting the pathogenicity of some of these structural genetic deviations. This possibility requires further study. It is also possible that the effects of duplications are less detectable because of the gene dosage effect, which is smaller in case of duplications than deletions. Moreover, we would add the further cautions in interpreting our data that in addition to limitations of our sample size, SNP arrays are limited in CNV detection and we have adopted a conservative approach to CNV detection. These caveats notwithstanding, we confirm in brain DNA the presence of recurrent CNVs in several patients with neuropsychiatric disorders and show evidence suggestive of incomplete penetrance of some CNVs on expression of genes in their respective affected regions in human brain.
We have concentrated in this study on CNVs in regions previously reported in large clinical case-control studies to be statistically associated with schizophrenia and with autism. In all instances, these CNVs are rare, occurring at most in 0.5 to 1.0% of cases for a given CNV. This catalogue of recurrent CNVs is not likely to represent the full spectrum of CNVs that are found in DNA of psychiatric patient samples, and indeed, at least one CNV >100kb is typically found in any given human being. CNVs found in only one case in a sample, even if not found in controls, is difficult to interpret in pathogenic terms. Thus, we have not addressed the possibility that any of the rare novel CNVs found in this study might be a relevant illness factor in an individual case. All of the CNVs called in our study, both in control and patient samples, will be available in a public database at the “BrainCloud” website (http://libd.org/BrainCloud [17]) for further analysis. As additional brain and clinical samples become available, some of the novel CNVs found in our cases may be found also in other case samples and not in controls, and thus achieve potential pathogenic association.
Supplementary Material
Acknowledgments
The authors wish to express their appreciation to Amy Deep-Soboslay, M.Ed. and Llewellyn B. Bigelow, M.D. of the Clinical Brain Disorders Branch, GCAP, IRP, and NIMH for their efforts in clinical diagnosis and demographic characterization, and Vesna Imamovic, Yeva Snitkovsky, Jewell King, and Jonathan Sirovatka for their excellent technical assistance.
Footnotes
Financial Disclosure Statement
All authors reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 2.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 5.Raychaudhuri S, Korn JM, McCarroll SA, Altshuler D, Sklar P, et al. International Schizophrenia Consortium. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6:e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahti S, Kumar K, Castellani C, O’Riley R, Singh S. Ontogenetic de novo copy number variations (CNVs) as a source of genetic individuality: studies on two families with MZD twins for schizophrenia. PLoS One. 2011;6:e17125. doi: 10.1371/journal.pone.0017125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, Kleinman JE. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant S, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Research. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 13.Johnson WE, Rabinovic A, Li C. Adjusting batch effects in microarray expression data using Empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 14.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burnside RD, Pasion R, Mikhail FM, Carroll AJ, Robin NH, Youngs EL, et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: a susceptibility region for neurological dysfunction including developmental and language delay. Hum Genet. 2011;130:517–528. doi: 10.1007/s00439-011-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Autism Genome Project Consortium. Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of the human neocortical transcriptome across the lifespan. Nature. 2011;478:519–524. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.