Abstract
Osteoclasts are specialized secretory cells of the myeloid lineage important for normal skeletal homeostasis as well as pathologic conditions of bone including osteoporosis, inflammatory arthritis and cancer metastasis. Differentiation of these multinucleated giant cells from precursors is controlled by the cytokine RANKL, which through its receptor RANK initiates a signaling cascade culminating in the activation of transcriptional regulators which induce the expression of the bone degradation machinery. The transcription factor nuclear factor of activated T-cells c1 (NFATc1) is the master regulator of this process and in its absence osteoclast differentiation is aborted both in vitro and in vivo. Differential mRNA expression analysis by microarray is used to identify genes of potential physiologic relevance across nearly all biologic systems. We compared the gene expression profile of murine wild-type and NFATc1-deficient osteoclast precursors stimulated with RANKL and identified that the majority of the known genes important for osteoclastic bone resorption require NFATc1 for induction. Here, five novel RANKL-induced, NFATc1-dependent transcripts in the osteoclast are described: Nhedc2, Rhoc, Serpind1, Adcy3 and Rab38. Despite reasonable hypotheses for the importance of these molecules in the bone resorption pathway and their dramatic induction during differentiation, the analysis of mice with mutations in these genes failed to reveal a function in osteoclast biology. Compared to littermate controls, none of these mutants demonstrated a skeletal phenotype in vivo or alterations in osteoclast differentiation or function in vitro. These data highlight the need for rigorous validation studies to complement expression profiling results before functional importance can be assigned to highly regulated genes in any biologic process.
Keywords: Osteoclast, Gene array, NFATc1, Bone resorption, NHEDC2
1. Introduction
Bone is a dynamic tissue that continuously remodels through the activity of bone forming osteoblasts and bone resorbing osteoclasts [1]. An imbalance in remodeling favoring resorption over formation underlies the pathology of many common and morbid skeletal diseases such as osteoporosis, inflammatory arthritis and primary and secondary bone malignancies. Medications targeting the osteoclast such as bisphosphonates and the RANKL inhibitor, denosumab, can reverse this imbalance to improve bone quality and prevent fragility and pathologic fractures [2, 3]. However, use of these medications can be limited by side effects and cost [4–7], necessitating a better understanding of osteoclast biology to identify new treatment targets.
Osteoclasts are highly specialized, multi-nucleated giant cells, which secrete acid and enzymes to degrade the inorganic and proteinaceous components of bone, respectively. These cells differentiate from myeloid precursors under the influence of the cytokine, RANKL [8]. Osteoblast lineage cells, including osteocytes, are believed to be the source of RANKL in the skeleton [9, 10]. In pathologic circumstances other cell types, such as the synovial fibroblast, T lymphocytes and myeloma cells, produce RANKL [11]. Engagement of the receptor for RANKL, RANK, on the cell surface of the osteoclast precursor activates a series of signaling events that promotes osteoclast formation, which like many differentiation programs is driven by transcription factors regulated themselves at either the transcriptional or post-translational level or both. Important transcription factors activated early after RANK engagement include Nuclear-factor-κB and c-Fos, which in turn promote expression of nuclear factor of activated T-cells c1 (NFATc1). NFATc1 is a transcription factor that is relegated to the cytoplasm by hyperphosphorylation of a N-terminal regulatory domain. As the osteoclast precursor progresses along the differentiation program, however, intracellular calcium concentrations rise, driving activation of the phosphatase calcineurin, which dephosphorylates NFATc1 to promote nuclear localization [12, 13]. Highlighting the importance of NFATc1 to osteoclast biology, genetic deletion of Nfatc1 in mice results in profound osteoclast-poor osteopetrosis, a high bone mass state caused by a lack of osteoclast activity [14–16]. Furthermore, deletion of NFATc1 in adult mice prevents catabolic bone loss in a model of paralysis induced osteolysis [17] and systemic osteoporosis in a model of the human genetic inflammatory disease, cherubism [14].
RNA microarray profiling is a powerful technique to identify genes and pathways involved in biologic processes in an unbiased manner. However, like any discovery-based technology, hypotheses generated from microarray data must be validated through experimentation. We hypothesized that the family of NFATc1 regulated transcripts in the osteoclast would be enriched for genes associated with osteoclast function. In 2008 we reported that many of the known genes associated with osteoclast function, including Itgb3, Oscar, Calcr, Mmp9, Acp5, Ctsk, Mmp14, Car2, and Clcn7, require NFATc1 for optimal expression [14]. Accordingly, others have shown that NFATc1 is recruited to the promoters of some of these genes, often acting in concert with other osteoclast transcriptional regulators [18]. Within the family of NFATc1 regulated transcripts we also discovered an anion exchanger, Slc4a2, whose deletion in mice and cattle results in severe osteopetrosis due to dysfunctional osteoclasts incapable of secreting acid [19, 20].
Here, we report all the NFATc1-dependent transcripts discovered in a microarray comparison of highly purified wild-type (WT) and NFATc1-deficient osteoclast precursors (OCPs) stimulated with RANKL [14]. In an attempt to identify novel osteoclast regulators, five NFATc1-dependent transcripts within one of four general biological processes required for osteoclast function were selected for further study based on the availability of knockout mouse models. These processes and the chosen genes included 1) cytoskeletal reorganization and exocytosis (ras homolog gene family member C (Rhoc) and ras-related protein Rab38 (Rab38)), 2) ion exchange (Na+/H+ exchanger-like domain-containing protein 2 (Nhedc2)), 3) the generation of second messengers (adenylate cyclase 3 (Adcy3)) and 4) regulation of the proteolytic enzyme activity that degrades organic bone matrix (serpin peptidase inhibitor, clade D, member 1 (Serpind1)). Using qPCR we confirmed that these transcripts are induced by RANKL in an NFATc1-dependent manner during osteoclast differentiation. Despite reasonable hypotheses for the importance of these molecules in bone resorption and their dramatic induction during differentiation, a rigorous analysis of mice with mutations in these genes failed to reveal a function for any in osteoclast biology. These data support the critical role for NFATc1 in mediating the osteoclast transcriptional program but indicate that some NFATc1-dependent transcripts play either non-significant or redundant roles in physiologic bone resorption. Furthermore this study highlights the importance of rigorous validation approaches to complement expression profiling results.
2. Materials and Methods
2.1 Mice
Nfatc1fl/fl, Mx1-Cre, Serpind1−/−, Adcy3−/−, were previously described [14, 21–23]. Nfatc1Δ/Δ mice were generated from Nfatc1fl/fl, Mx1-Cre mice by treatment with poly I:C (GE Healthcare, Piscataway, NJ) as described [14]. Dr. Tak Mak of the University of Toronto and Dr. William J. Pavan of the National Institutes of Health kindly provided Rhoc−/− [24] and Rab38cht/cht [25] mice, respectively. Nhedc2 gene trap mice (Nhedc2GT/GT) were generated from clone CMHD-GT_108B11-3 from the Centre for Modeling Human Disease (Toronto, Canada). The vector, GepSD5, is inserted into intronic sequence 5’ to the start of exon 13. The resulting targeted transcript lacks a polyA tail and is unstable. The following PCR primers were used to genotype Nhedc2GT/GT mice NhedC2-in12-400F (AGCTGTCCTTTGACCTCCGTAG), Nhedc2-ex13-B (ACCTGAGGTACTGTTAGCAGCGTA) and SD5-501R (CGCGAAGAGTTTGTCCTCAAC). Primers NhedC2-in12-400F and Nhedc2-ex13-B generate a WT product of approximately 380 bp and primers NhedC2-in12-400F and SD5-501R generate a product of approximately 650 bp (Fig. S1A). All other strains were genotyped as described. [14, 22–25]. For all experiments each knockout mouse was paired with at least one, sex matched WT or heterozygous littermate. For the purpose of data analysis, when no differences were observed between WT and heterozygous mice, data from these two genotypes were pooled for comparison with the knockout littermates and are designated +/? where appropriate in the figures and legends. The number of mice analyzed in each experiment is indicated in the figure legends. Experimental protocols were approved by the Standing Committee on Animals at the Harvard Medical School and were designed with institutional and National Institutes of Health guidelines for the humane use of animals.
2.2 Microarray analysis
Microarray analysis of Nfatc1fl/fl and Nfatc1Δ/Δ OCPs stimulated with RANKL (n=2 per genotype) was previously described in [14]. Further analysis of this data reported here was performed as follows. To generate probeset-level expression estimates, we normalized and summarized the probe-level hybridization intensities using the MAS 5 algorithm (Affymetrix, Santa Clara, CA). For identification of differentially expressed genes, we first excluded 8,453 probesets that did not exhibit sequence-specific hybridization signal as determined by their having a MAS 5 detection call of absent in all four samples. For the remaining 14,238 probesets, we identified genes significantly differentially expressed between samples from WT and KO animals by first log transforming the expression data and then employing the limma 3.10 package [26] that uses an Empirical Bayesian approach and default settings. To account for multiple-testing we calculated the FDR of the resulting p-values using the method of Benjamini and Hochberg [27]. Probeset annotations were obtained using the annaffy (version 1.26.0) and mouse4302.db (version 2.63) packages. Heatmaps were generated after z-score normalizing the log transformed expression values using a Euclidean distance metric and complete linkage clustering. All of the steps following generating probeset-level expression estimates were performed using the R language for statistical computing version 2.14.0 (R Development Core Team, 2011, http://www.R-project.org/). Microarray data has been uploaded to GEO and can be found at the following link http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37219.
The reviewer link to the data is: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zzaxpimsmoessly&acc=GSE37219
2.3 Real-time quantitative PCR
RNA was isolated by TRIzol reagent (Life Techologies, Grand Island, NY) and cDNA synthesized using kits from either Applied Biosystems (Life Technolgies) or Agilent Technolgies (Santa Clara, CA). Realtime PCR was performed using the Brilliant II SYBR Green Master Mix (Agilent Technologies) on a Mx3005P qPCR system (Agilent Technologies). Ct values for duplicate technical replicates were averaged and the amounts of mRNA relative to Hprt or Hmbs were calculated using the ΔCt method. All qRT-PCR primers used in this study are reported in Table S1 except those targeting Mmp9, Itgb3, Ctsk and Calcr, which were previously described {Aliprantis, 2008 #15835}. All qRT-PCR reactions yielded products with single dissociation peak.
2.4 Preparation and analysis of murine Osteoclasts in vitro
Cell culture experiments were performed in α-MEM (Cellgro) containing 10% fetal calf serum (Invitrogen), 100 U penicillin and 100 µg/ml streptomycin (Cellgro) at 37°C with 5% CO2. Mouse bone marrow cells were cultured for 3 days in the presence of 40 ng/mL murine recombinant M-CSF (R&D Systems) in a suspension culture dish (Corning Costar Ins) to which stromal and lymphoid cells cannot adhere. Thereafter, cells were washed with PBS once to remove nonadherent cells, harvested by pipetting with 10mM EDTA in phosphate-buffered saline (PBS) and seeded on plastic with 20 ng/mL M-CSF and 5 ng/ml RANKL (R&D Systems). After 3 days, the cytokines were replenished daily. Alternatively, osteoclasts were cultured from precursors harvested from the interface of a Histopaque 1083 (Sigma) gradient after an overnight incubation of bone marrow cells with MCSF as described in [14] using RANKL at 100–300 ng/ml that was a kind gift of Dr. Yongwon Choi from the University of Pennsylvania. To evaluate osteoclast formation, we either 1) stained for tartrate resistant acid phosphatase (TRAP) using a Leukocyte Acid Phosphatase kit (Sigma), 2) assayed for TRAP activity in the tissue culture supernatants as described [14], 3) isolated RNA to measure osteoclast associated gene expression or 4) isolated protein for western blotting. Resorption assays were performed by culturing osteoclast precursors on either bovine cortical bone, dentin or Biocoat osteologic slides (Becton Dickinson) with MCSF and RANKL. Resorption pits were identified using either toluidine blue, lectin-TRITC as described [19] or lectin-HRP, followed by 3,3'-Diaminobenzidine. Actin rings were visualized using Phalloidin-TRITC (Sigma) according to the manufacturer’s directions.
2.5 Microquantitative computed tomography
For µCT analysis, a Scanco Medical µCT 35 system with an isotropic voxel size of 7 µm was used to image the femur. Scans were conducted in 70% ethanol using an X-ray tube potential of 55 kVp, an X-ray intensity of 0.145 mA and an integration time of 600 ms. From the scans of the femur, a region beginning 0.28 millimeters proximal to the growth plate and extending 2.1 millimeters proximally was selected for trabecular bone analysis. A second region 0.6 millimeters in length and centered at the midpoint of the femur was used to calculate diaphyseal parameters. A semi-automated contouring approach was used to distinguish cortical and trabecular bone. The region of interest was thresholded using a global threshold that set the bone/marrow cut-off at 352.3 mgHA/cm3 for trabecular bone and 589.4 mgHA/cm3 for cortical bone. 3-D microstructural properties of bone, including bone volume fraction (BV/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N.), trabecular separation (Tb.Sp.), midshaft bone volume fraction (M.BV/TV) and cortical thickness (C.Th) were calculated using software supplied by the manufacturer.
2.6 Histomorphometry
Tibiae and femora were stripped of soft tissue, fixed in 10% buffered formalin, dehydrated, and embedded in methyl methacrylate before being sectioned and stained with toluidine blue [28]. To label bone mineralization fronts, mice were injected with 30 mg/kg of calcein 8 days and 24 hours prior to sacrifice. Histomorphometric measurements were performed on a fixed region just below the growth plate corresponding to the primary spongiosa [29] and analyzed by Osteomeasure software (Osteometrics, Atlanta, GA).
2.7 TRAP5b measurement
Serum TRAP5b levels were measured using a commercially available kit from Immunodiagnostic systems as described in [14].
2.8 Western blotting
Western blotting was performed using standard indirect chemiluminescence methods and antibodies targeting NHEDC2 (Biolegend), Cathepsin K (Calbiohem) and HSP90 (Santa Cruz Biotechnogies).
2.9 Intracellular cAmp Assay
Intracellular cAmp levels were determined using a kit from GE Healthcare (Buckinghamshire, UK) following the manufacturer’s directions.
2.10 Statistical analyses
Graphs show mean+/− S.D. with some graphs also depicting individual data points. When 2 or fewer samples were reported, only individual data points are shown. Sample numbers and p-values are reported in the figure legends. P values were calculated using unpaired Student's t-tests as all the data in the manuscript represent continuous variables. A p-value > 0.05 was considered non-significant and is reported in the figures by the abbreviation “n.s.” Prism version 5.0b software was used to generate all the graphs and perform all statistical analyses.
3. Results
3.1 Expression profiling of RANKL stimulated NFATc1-deficient OCPs
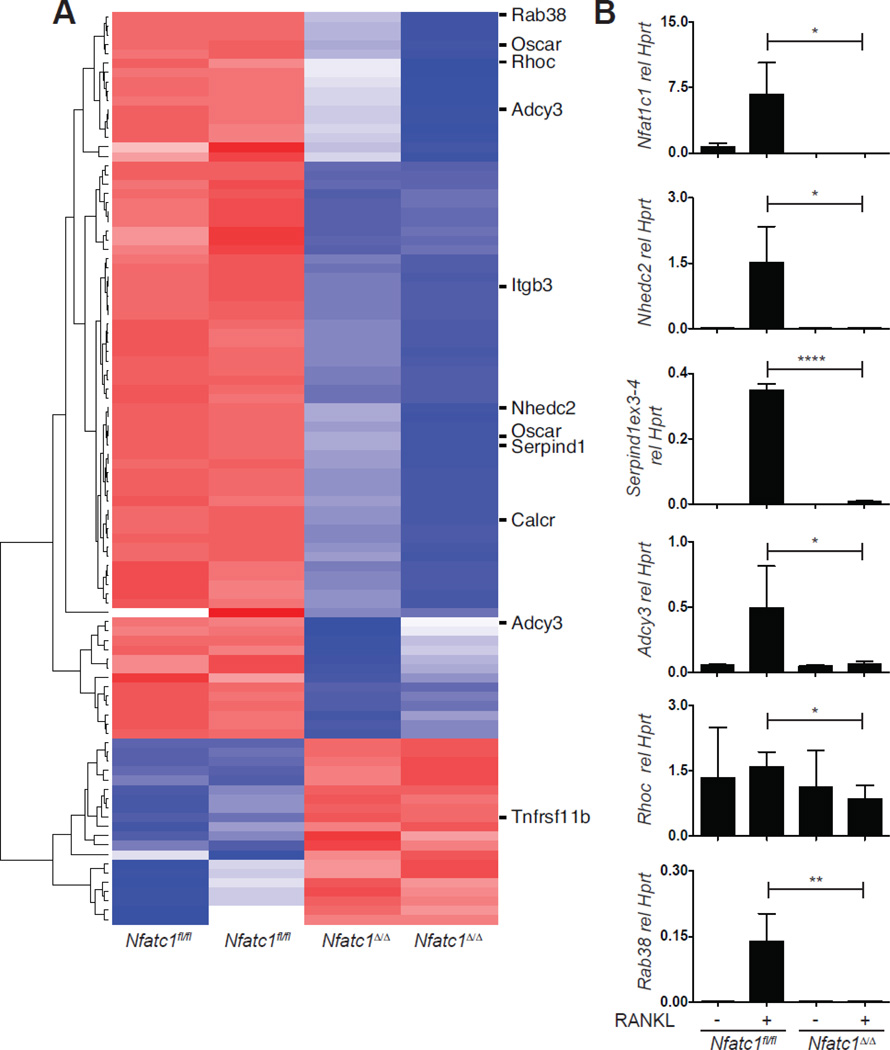
Expression profiling of Nfatc1fl/fl and Nfatc1Δ/Δ bone marrow derived osteoclasts on day 3 of culture in MCSF and RANKL was previously reported and revealed numerous NFATc1-dependent transcripts, including known regulators of osteoclast function [14]. To identify additional candidate osteoclast regulators, we reanalyzed this microarray dataset to select for transcripts highly dependent on NFATc1, with the rationale that the NFATc1-dependent transcriptome is likely to be enriched for genes important for osteoclast function. Transcripts induced or repressed at least 6-fold in the absence of NFATc1 were identified, representing 99 probe-sets and corresponding to 80 unique genes and 5 probe-sets that did not map to named loci (Fig. 1A and Table S2). Several known regulators of osteoclast function, Oscar, Itgb3, Calcr, and Tnfrsf11b (Osteoprotegerin) met these criteria, validating our approach. As outlined in each subsection below, functional information from other biological systems, as well as the availability of knockout mouse models, were used to select 5 highly NFATc1-dependent transcripts for further study: Nhedc2, Rhoc, Serpind1, Adcy3 and Rab38.
Fig. 1. Identification of potential osteoclast regulators by expression profiling.
(A) Microarray analysis of Nfatc1fl/fl and Nfatc1Δ/Δ OCPs stimulated for 3 d with MCSF and RANKL. Shown is a heat map depicting genes up or down regulated 6-fold. NFATc1-dependent transcripts selected for further study, as well as representative known regulators of osteoclast function are highlighted. (B) Real-time PCR analysis for the expression of the indicated genes in cultures of Nfatc1fl/fl and Nfatc1Δ/Δ OCPs stimulated with MCSF alone or with MCSF and RANKL to stimulate osteoclast differentiation. (*, p<0.05; **, p<0.01; ****, p<0.0001).
Real-time PCR analysis of Nfatc1fl/fl and Nfatc1Δ/Δ OCPs stimulated with RANKL for 3 days confirmed that expression of Nhedc2, Rhoc, Serpind1, Adcy3 and Rab38 depends on the presence of NFATc1, and with the exception of Rhoc, is highly induced by RANKL (Fig. 1B). Analysis of Nfatc1 expression in the same samples confirmed the efficient deletion of Nfatc1 in the Nfatc1Δ/Δ cells (Fig. 1B We obtained knock-out or gene trap mice for each of these genes and examined their skeletal and osteoclast-intrinsic phenotypes using micro-computed tomography (micro-CT) and ex vivo functional assays, respectively.
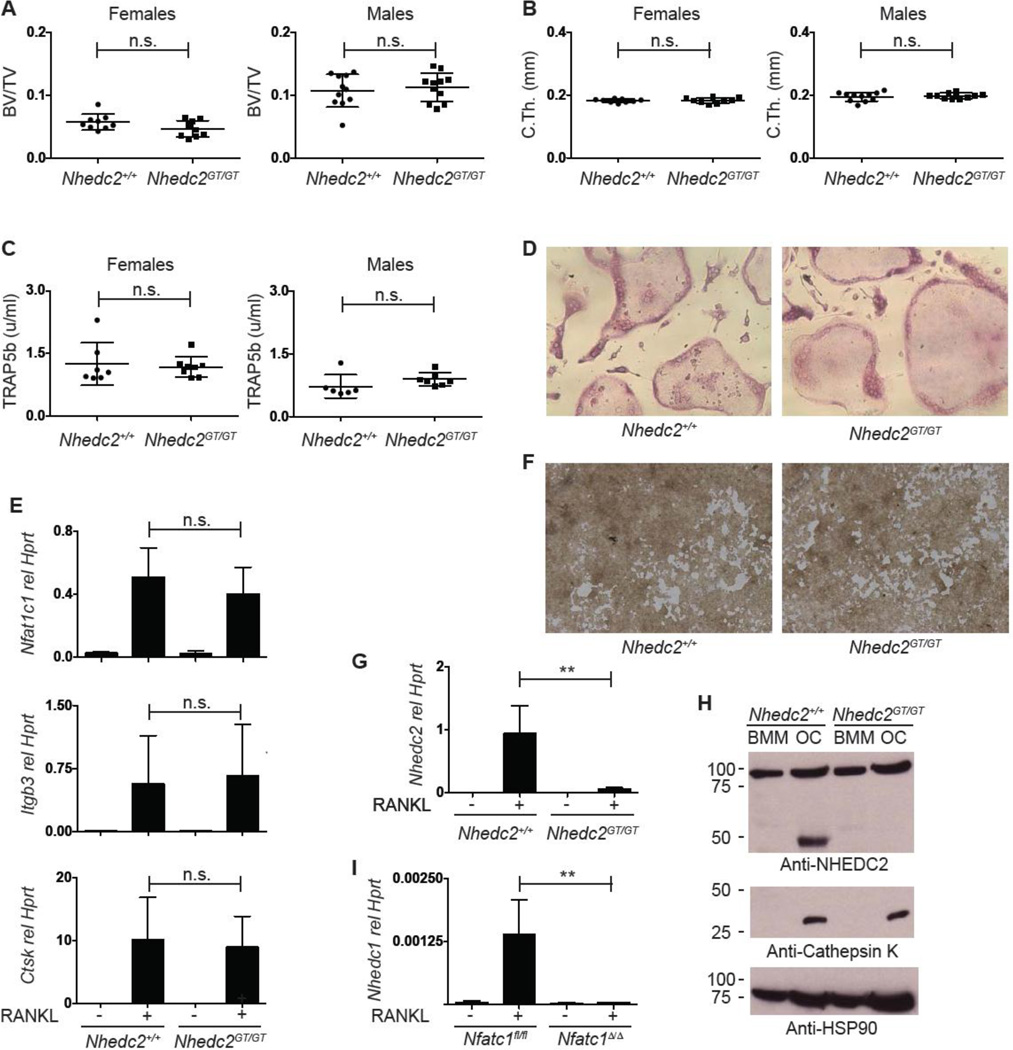
3.2 Normal bone mass and osteoclast activity in Nhedc2 gene trap mice
Previous profiling experiments showed that NHEDC2 protein and Nhedc2 message are enriched in osteoclasts compared to macrophages [30, 31]. Supporting a functional role for this molecule in the osteoclast, both amiloride, a selective inhibitor of Na+/H+ anti-porters, and shRNA knock-down of Nhedc2 in RAW264.7 cells inhibits osteoclast fusion [31]. In contrast, Hofstetter et al, reported that Nhedc2 gene trap mice lack a bone phenotype, though only a small number of mice were analyzed [30]. Given the profound RANKL- and NFATc1-dependent regulation of Nhedc2 transcription in the osteoclast, and to resolve these discrepant reports, the effect of NHEDC2 deficiency on bone mass and in vitro osteoclast function was investigated by generating gene trap mice (Nhedc2GT/GT). The ES cells used to generate these mice were targeted with a different vector (see Material and Methods) than the previous report [30]. Compared to WT littermates, no significant differences in femoral metaphyseal BV/TV, trabecular parameters or cortical bone were identified in either male or female Nhedc2GT/GT mice (Fig. 2A, B, S1B). Serum TRAP5b activity, a measure of in vivo osteoclast activity, was also similar in Nhedc2GT/GT and WT mice (Fig. 2C). Similarly, osteoclast formation in vitro as determined by formation of TRAP positive multinucleated cells and qPCR analysis of genes in the osteoclast differentiation program was also comparable (Fig. 2D, E). Lastly, no gross abnormalities in in vitro osteoclast resorption activity were observed (Fig. 2F). To confirm absence of NHEDC2 in Nhedc2GT/GT osteoclasts, expression in osteoclasts and bone marrow macrophages was examined. Nhedc2 transcripts were dramatically reduced in Nhedc2GT/GT osteoclasts (Fig. 2G). Furthermore, NHEDC2 protein was detected in WT but not Nhedc2GT/GT osteoclasts, confirming that the Nhedc2GT is a null-allele (Fig. 2H). Parallel immunoblotting for cathepsin K demonstrates that the absence of NHEDC2 was not due to a failure of osteoclast differentiation in the Nhedc2GT/GT sample. Although it is possible that NHEDC2 has a non-significant role in osteoclast function, another explanation for the lack of bone phenotype in Nhedc2GT/GT mice is functional redundancy with the mouse ortholog NHEDC1, whose mRNA interestingly is also induced by RANKL in an NFATc1-dependent manner (Fig. 2I). No compensatory increase in expression of NHE family members 1–9 was detected in Nhedc2GT/GT osteoclasts (data not shown). We conclude that despite strong suggestive evidence that NHEDC2 regulates osteoclastogenesis, an exhaustive analysis shows that Nhedc2GT/GT mice have no defect in osteoclast formation or function in vitro or in vivo.
Fig. 2. Analysis of Nhedc2GT/GT mice and osteoclasts.
(A, B) Micro-CT quantification of femoral (A) metaphyseal trabecular BV/TV and (B) midshaft cortical thickness of 12–14 week old female and male Nhedc2+/+ (n=9, female; n=11, male) and Nhedc2GT/GT (n=9, female; n=11, male) mice. (C) Serum TRAP5b levels in 10-week old female and male Nhedc2+/+ (n=7, female; n=6, male) and Nhedc2GT/GT (n=8, female; n=7, male) mice. (D) TRAP stain of Nhedc2+/+ and Nhedc2GT/GT osteoclast cultures. (E) Real time PCR analysis for osteoclast marker genes in samples from Nhedc2+/+ (n=5) and Nhedc2GT/GT (n=4) OCPs stimulated with MCSF alone or with MCSF and RANKL to induce osteoclast differentiation. (F) resorption of calcium phosphate coated tissue culture wells incubated with Nhedc2+/+ and Nhedc2GT/GT osteoclasts. (G) Real time PCR analysis for Nhedc2 expression in Nhedc2+/+ (n=5) and Nhedc2GT/GT (n=4) OCPs stimulated with MCSF alone or with MCSF and RANKL to induce osteoclast differentiation. (H) Western blot for NHEDC2 protein on samples from Nhedc2+/+ and Nhedc2GT/GT OCPs stimulated with MCSF alone or with MCSF and RANKL to induce osteoclast differentiation. Immunoblots for Cathepsin K and HSP90 are shown as positive controls for osteoclast differentiation and protein loading, respectively. (I) Real time PCR analysis for Nhedc1 expression in Nfatc1fl/fl and Nfatc1Δ/Δ OCPs stimulated with MCSF alone or with MCSF and RANKL to induce osteoclast differentiation. (n.s., not significant; **, p<0.01.)
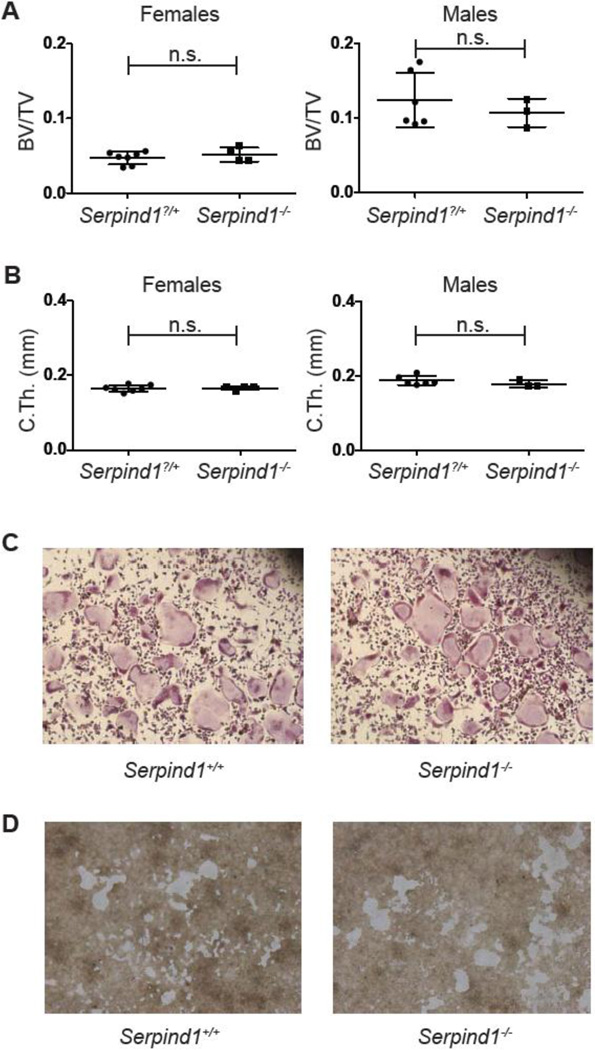
3.3 Serpind1−/− mice lack abnormalities in bone mass and osteoclast differentiation and function
SERPIND1, also known as heparin cofactor II, is a member of a large family of serine protease inhibitors. This plasma protein is primarily produced in the liver, demonstrates heparin-or dermatan sulfate-dependent inhibition of thrombin, and circulates free (~1 µM) or in a complex with thrombin (trace) (reviewed in [32]). Serpind1−/− mice are phenotypically indistinguishable from WT mice at baseline, but demonstrate increased rates of thrombotic carotid artery occlusion after endothelial cell injury [22]. SERPIND1 also has inhibitory activity against chymotrypsin and cathepsin G [33, 34]. As proteases, including cathepsins, are critical for resorption of the bone matrix by osteoclasts, we hypothesized that RANKL induced expression of SERPIND1 might serve an intrinsic role in the osteoclast to mitigate resorptive function. However, micro-CT analysis of Serpind1−/− males and females showed no difference in femoral BV/TV, trabecular parameters or cortical thickness (Fig. 3A, B, S2A) compared to WT, sex-matched littermates. Consistent with the normal bone phenotype in Serpind1−/− mice, in vitro assays for bone marrow derived osteoclast differentiation, expression of markers of osteoclast differentiation and resorptive function were not affected by SERPIND1 deficiency (Fig. 3C, S2B, 3D). Deletion of Serpind1 mRNA in Serpind1−/− mice was confirmed by qPCR on liver (Fig. S2C).
Fig. 3. Analysis of Serpind1−/− mice and osteoclasts.
(A, B) Micro-CT quantification of femoral (A) metaphyseal trabecular BV/TV and (B) midshaft cortical thickness of 12 week old female and male Serpind1?/+ (female, n=7; male, n=6) and Serpind1−/− (female, n=4; male, n=3) mice. (C) TRAP stain of Serpind1+/+ and Serpind1−/− osteoclast cultures. (D) resorption of calcium phosphate coated tissue culture wells incubated with Serpind1+/+ and Serpind1−/− osteoclasts. (n.s., not significant; **, p<0.01.)
Our data suggest two possible explanations for the lack of a bone phenotype in Serpind1−/− mice. While a transcript containing exon 3 of Serpind1 is induced in osteoclasts by RANKL through an NFATc1 pathway (Fig. 1B), primers spanning the exon 2–3 junction do not amplify a product in osteoclasts (Fig. S2D). These Serpind1 exon 2–3 primers do detect robust and specific expression in WT liver (Fig. S2C), indicating that the lack of detection of exon 2 in osteoclasts is not an artifact of the choice of primers. Since Serpind1−/− mice were generated through targeted deletion of exon 2 [22], an alternative hypothesis for the lack of a bone phenotype in these mice is that the functionally relevant Serpind1 transcript in osteoclasts is a splice variant lacking exon 2, which is unaffected in the knockout mice. Alternatively, SERPIND1 may have a redundant regulatory role in osteoclast function, as our microarray data suggests that Serpinb1a, Serpinb2, Serpind1, Serpine2 and Serpinh1 are also expressed in osteoclasts in a manner dependent on NFATc1 (Table S2). By qPCR using validated primers, we find that Serpinb1a and Serpinb2, but not Serpine2 or Serpinh1, are expressed in bone marrow derived osteoclasts, though there is no compensatory increase in expression in the absence of Serpind1 (Fig. S2E and data not shown).
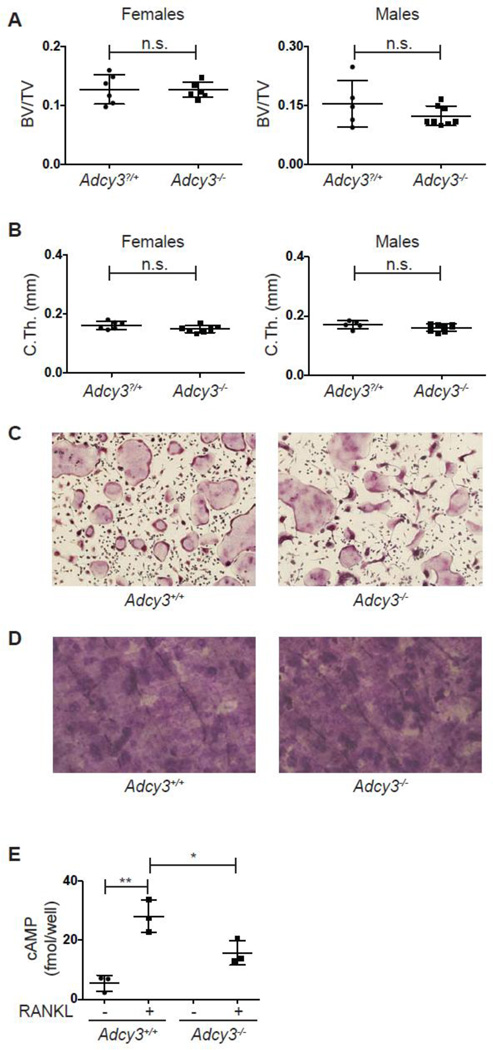
3.4 Normal bone mass and osteoclast function in Adcy3−/− mice
Levels of cAMP are regulated by the balance between adenylate cyclases, which catalyze cAMP formation from ATP, and phosphodiesterases that degrade this signaling intermediate. Of nine isoforms of adenylate cyclase tested, only Adcy3 is induced during osteoclastogenesis [35]. cAMP affects osteoclastogenesis both directly and indirectly. Indirectly, cAMP promotes osteoclastogenesis via induction of RANKL on osteoblasts [36, 37]. Directly, increased cAMP levels induced by treatment with forskolin inhibit osteoclastogenesis in vitro and in vivo [35]. Conversely, knockdown of Adcy3 expression significantly increases osteoclast differentiation in bone marrow macrophages [35]. This report suggested that ADCY3 mediated increases in cAMP levels attenuate osteoclastogenesis by promoting inhibitory phosphorylation of NFATc1 by the cAMP activated kinase PKA [35, 38] thereby interrupting NFATc1 autoamplification. We confirmed the induction of Adcy3 expression by RANKL (Fig. 1B). Given the previous findings indicating that intracellular cAMP inhibits osteoclastogenesis we predicted that Adcy3−/− mice would have low bone mass.
Adcy3−/− mice are runted in the early perinatal period but paradoxically obese in adulthood because of reduced locomotor activity, hyperphagia, and leptin insensitivity [39]. Thus the skeletal phenotype of Adcy3−/− mice was analyzed 8 weeks after birth, a time point when the mutant mice display similar body weight to their WT littermates. Deficiency of Adcy3 did not result in a bone phenotype, as demonstrated by similar BV/TV, trabecular parameters and cortical thickness in littermate male and female WT and Adcy3−/− mice (Fig. 4A, B, S3A). As predicted based on a previous report [39], body weight was similar between the two groups at this time point (Fig. S3B). Consistent with the lack of a skeletal phenotype, osteoclast formation, expression of markers of osteoclast maturation and resorption activity was similar between WT and Adcy3−/− bone marrow derived osteoclast cultures (Fig. 4C, S3C, 4D). Absence of Adcy3 mRNA (Fig. S3D) and a significant reduction in RANKL-induced cAMP was demonstrated for Adcy3−/− osteoclasts (Fig. 4E). These data suggest that while ADCY3 is responsible for a share of the cAMP in osteoclasts, the reduction of cAMP levels in the absence of this gene is insufficient to augment differentiation or effect changes in bone mass in vivo.
Fig. 4. Analysis of Adcy3−/− mice and osteoclasts.
(A, B) Micro-CT quantification of femoral (A) metaphyseal trabecular BV/TV and (B) midshaft cortical thickness of 8-week old female and male Adcy3?/+ (female, n=6; male, n=5) and Adcy3−/− (female, n=7; male, n=8) mice. (C) TRAP stain of Adcy3+/+ and Adcy3−/− osteoclast cultures. (D) Toluidine blue stained bovine cortical bone slices cultured with Adcy3+/+ and Adcy3−/− osteoclasts. (E) cAMP levels in cultures of in Adcy3+/+ and Adcy3−/− OCPs stimulated with MCSF alone or MCSF and RANKL to induce osteoclast differentiation (n=3 per group). (n.s., not significant; *, p<0.05; **, p<0.01.)
3.5 The small GTPases, RHOC and RAB38, are dispensable for osteoclast function
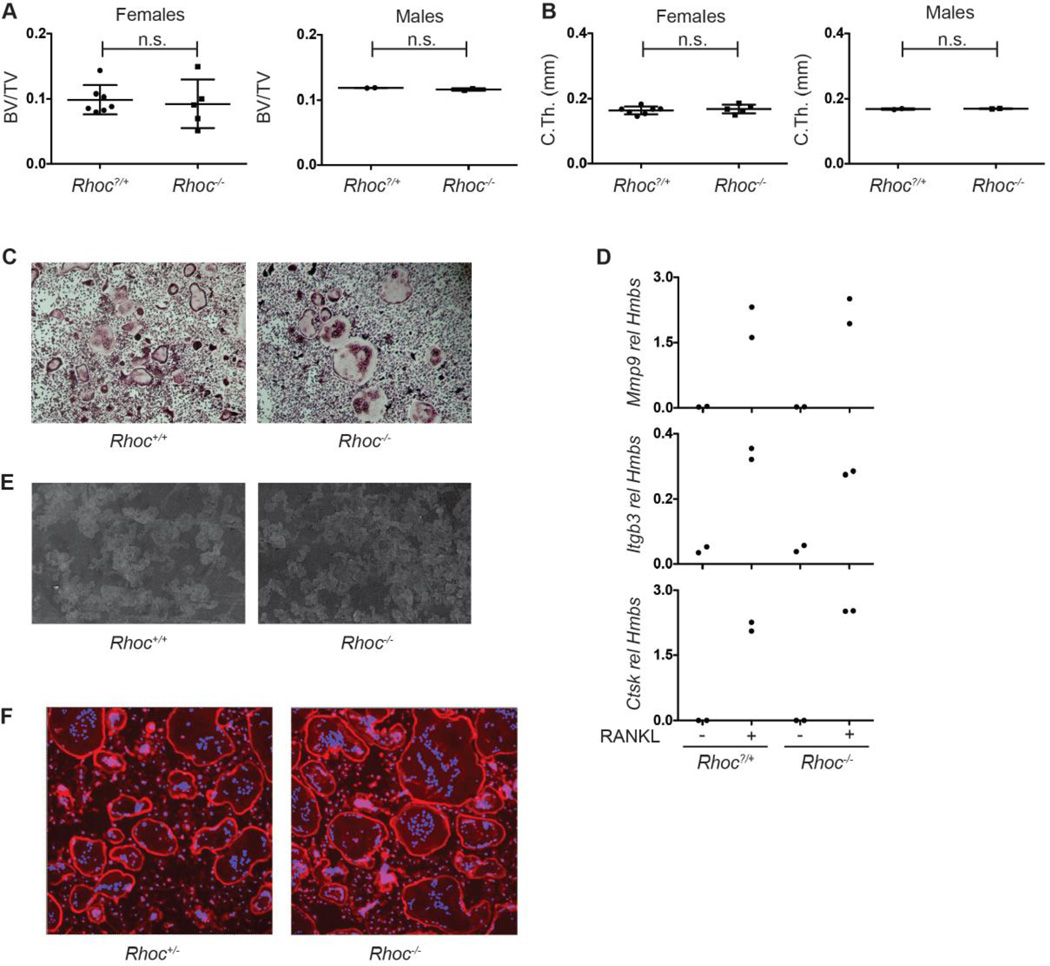
The Ras GTPases include the Ras, Rho, and Rab subfamilies. Ras family proteins regulate many cellular processes including organization of the actin cytoskeleton and vesicular transport, two cell biologic processes required by osteoclasts to polarize, spread on the bone surface and secrete acid and proteolytic enzymes. The importance of small GTPases for osteoclast function is demonstrated by the discovery that nitrogen-containing bisphosphonates inhibit osteoclast resorptive function by blocking prenylation, the protein modification required for correct subcellular localization of these proteins [40]. Several Rho subfamily members, including RHOA, CDC42 and RAC are essential for osteoclast function [40–43]. Our microarray analysis strategy identified Rhoc as an NFATc1-dependent transcript in osteoclasts, and thus a candidate regulator of these cells. To determine if RHOC exhibits a non-redundant function in osteoclasts, the bone phenotype of Rhoc−/− mice was examined. Trabecular BV/TV, trabecular parameters and cortical thickness was equivalent in RHOC-deficient mice and their WT sex-matched littermates (Fig. 5A, B and S4A). Consistent with this result, bone histomorphometric parameters, including measures of osteoblast and osteoclast numbers and activity, were similar between female WT and Rhoc−/− mice (Table S3). Ex vivo WT and Rhoc−/− bone marrow derived osteoclasts were comparable, as assessed by the formation of TRAP+ multinucleated cells (Fig. 5C), the expression of key genes in the osteoclast differentiation program (Fig. 5D), and pit formation on dentin (Fig. 5E). As generalized inhibition of Rho proteins interferes with actin podosome function [44], actin ring formation was examined in Rhoc−/− osteoclasts and no alterations were identified (Fig. 5F). The absence of Rhoc mRNA in Rhoc−/− osteoclasts was confirmed by qPCR to ensure that the lack of an observed phenotype was not due to residual transcripts (Fig. S4B). Thus, unlike other Rho subfamily GTPases, RHOC does not appear to be a significant regulator of osteoclast formation or function.
Fig. 5. Analysis of Rhoc−/− mice and osteoclasts.
(A, B) Micro-CT quantification of femoral (A) metaphyseal trabecular BV/TV and (B) midshaft cortical thickness of 8–10 week old female and male Rhoc?/+ (female, n=7; male, n=2) and Rhoc−/− (female, n=5; male, n=2) mice. (C) TRAP stain of Rhoc+/+ and Rhoc−/− osteoclast cultures. (D) Real time PCR analysis for osteoclast marker genes in mRNA samples from Rhoc?/+ and Rhoc−/− OCPs stimulated with MCSF alone or with MCSF and RANKL to induce osteoclast differentiation (n=2 per group). Lack of differences in gene expression between Rhoc+/+ and Rhoc−/− osteoclasts is representative of 3 independent experiments. (E) Lectin-TRITC stained dentin slices cultured with Rhoc+/+ and Rhoc−/− osteoclasts. (F) Phalloidin stain of Rhoc+/+ and Rhoc−/− osteoclast cultures. (n.s., not significant). (n.s., not significant)
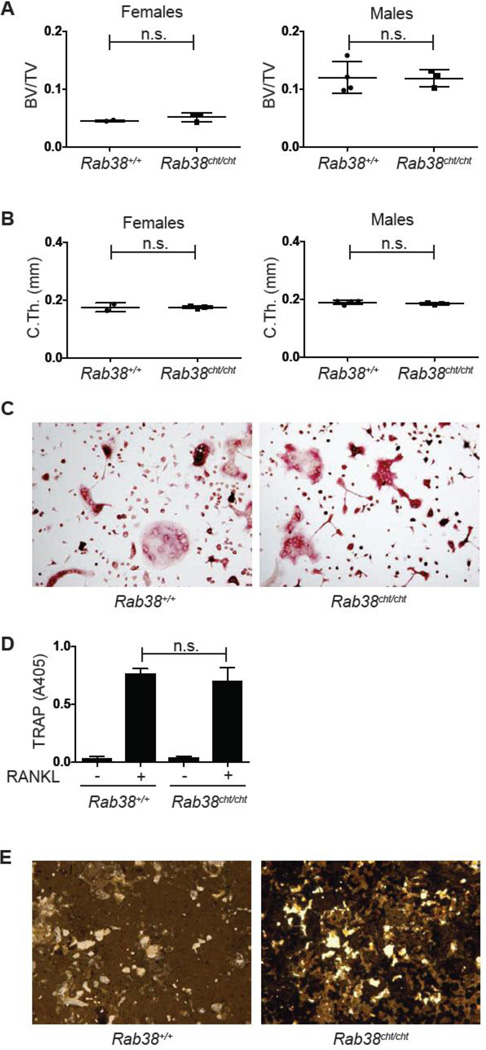
RAB subfamily proteins are essential regulators of vesicular trafficking in osteoclasts, with RAB7 and RAB3D regulating vesicular trafficking to the ruffled border [45, 46]. Although a number of additional RAB subfamily members are expressed in osteoclasts, their specific function is unknown [40]. RAB38, a member of the RAB subfamily of small GTPases, is strongly induced by RANKL in an NFATc1 dependent manner (Fig. 1B). RAB38 is essential for protein sorting to the melanosome [25], and plays a role in phagosome and lamellar body maturation [47, 48], but its expression and function in osteoclasts has not previously been reported. Rab38cht/cht mice (chocolate mice) harbor a spontaneously occurring G146T mutation in Rab38, resulting in a Gly to Val substitution in the GTP-binding pocket predicted to disrupt function based on the effect of analogous mutations in RAS and RAB5 [25]. Examination of Rab38cht/cht mice and WT sex-matched littermates revealed no differences in BV/TV, trabecular parameters, or cortical thickness (Fig. 6A, B and S5A). Similarly, the Rab38cht/cht mutation did not affect in vitro bone marrow derived osteoclasts as demonstrated by the formation of TRAP+ multinucleated osteoclasts (Fig. 6C), tissue culture supernatant TRAP activity (Fig. 6D), the expression of markers of osteoclast maturation (Fig. S5B), or resorptive function (Fig. 6E). Thus, these data indicate that RAB38 is not a significant regulator of osteoclast formation or function and that the multiple RAB subfamily members expressed in osteoclasts may have redundant roles.
Fig. 6. Analysis of Rab38cht/cht mice and osteoclasts.
(A, B) Micro-CT quantification of femoral (A) metaphyseal trabecular BV/TV and (B) midshaft cortical thickness of 16 week old female and male Rab38+/+ (female, n=2; male, n=4) and Rab38cht/cht (female, n=3; male, n=3) mice. (C) TRAP stain of Rab38+/+ and Rab38cht/cht osteoclast cultures. (D) TRAP assay on the culture supernatants of Rab38+/+ and Rab38cht/cht (n=6 per genotype) OCPs stimulated with MCSF along or MCSF and RANKL to induce osteoclast differentiation. (E) Von kossa stain of calcium phosphate coated tissue culture wells incubated with Rab38+/+ and Rab38cht/cht osteoclasts. (n.s., not significant)
4. Discussion
Commercially available RNA microarrays have made the rapid analysis of the complete transcriptome of cells under defined conditions affordable. Microarray profiling provides an unbiased method to detect genes that are differentially expressed between two or more conditions and identify genes and pathways relevant to biologic processes. We set out to identify regulators of osteoclast function utilizing an RNA microarray profiling approach. As NFATc1 is a key regulator of the osteoclast transcriptional program, subsets of genes associated with osteoclast function are likely to be NFATc1-dependent. We compared the transcriptome of WT and NFATc1-deficient OCPs differentiated with MCSF and RANKL and detected a number of known regulators of osteoclast differentiation and function ([14] and Fig. 1A and Table S2). This approach also previously identified Slc4a2, which encodes an anion exchanger required for osteoclast acid secretion, thus validating our focus on NFATc1-regulated transcripts [19]. To identify additional regulators of osteoclast function, we further analyzed this data set, now selecting for transcripts highly (6-fold or more) induced in the presence of NFATc1. This approach identified 80 unique genes, including 4 known osteoclast regulators. Utilizing existing knowledge about general biologic processes in osteoclasts to filter the list of 76 NFATc1-dependent genes without previously identified roles in osteoclast differentiation, we identified five candidate osteoclast regulators. A reasonable biologic rationale for a role for each of these candidates in bone biology existed, and the regulated expression of each during osteoclastogenesis was confirmed. Despite this, a rigorous analysis utilizing knockout mouse models did not detect a role for any of these 5 candidates in osteoclast function or maintenance of normal adult bone mass.
Biologic or functional significance is often inferred from large differences in gene transcription between two conditions, but as our results demonstrate, this information should be considered hypothesis generating rather than definitive evidence for the functional relevance of a given gene product. The inability to detect functional significance for the highly differentially expressed genes examined here could have several biologic explanations including redundancy of function and compensatory changes in transcription other genes in the same pathway.
A great strength of RNA microarray analysis is its capacity to analyze large numbers of transcripts in parallel. However, multiple comparison testing and the intrinsic variability of microarray data require that results from microarray analysis be validated by qPCR or Northern blotting. Moreover, microarray probes are designed on the basis of inferences from genomic sequence data and probe choice may influence the detection of differentially expressed genes, particularly if a transcript has multiple splice variants. The example of Serpind1 demonstrates that whether or not a gene is detected as differentially expressed by microarray can be highly dependent on the transcript region targeted by the probeset. Serpind1 probset 1418680_at detected the exon 5 and 3’UTR region of the Serpind1 mRNA. To validate this finding we used primers targeting exons 3 and 4 of Serpind1 and identified a RANKL- and NFATc1-dependent transcript in the osteoclast. Regulation of Serpind1 expression however was revisited after discovering that Serpind1−/− mice lack an osteoclast phenotype. Interestingly, no qPCR product was detected in osteoclasts using primers spanning the exon 2–3 junction. These data suggest that the relevant mRNA generated from the Serpind1 locus in osteoclasts lacks exon 2 and could explain the absence of a skeletal or osteoclast phenotype in Serpind1−/− mice in which exon 2 was deleted. Identification of the relevant Serpind1 transcript in osteoclasts is being pursued.
In the case of Nhedc2, as well as Serpind1, redundancy is a possible explanation for the lack of a bone phenotype in the knockout mice, as homologs of both are expressed in osteoclasts in an NFATc1-dependent manner. Resolution of roles of these protein families in bone biology in vivo will require the generation of compound mutant mice. This will be particularly challenging for the Nhedc homologs, Nhedc2 and Nhedc1, as these genes are located adjacent to one another on mouse chromosome 3. Thus, any targeting strategy would need to generate null alleles with a single vector, as opposed to generating knockout mice in parallel and crossing the resultant mutant lines.
For RHOC and RAB38, redundancy is also a likely explanation for the normal bone phenotype of Rhoc−/− and Rab38cht/cht mice. Several Rho subfamily members, including RHOA, CDC42 and RAC are essential for osteoclast function. CDC42 regulates BMM proliferation, osteoclast differentiation and survival. Mice with a conditional deletion of Cdc42 in osteoclasts are osteopetrotic, while deletion of Cdc42Gap, whose gene product negatively regulates CDC42 function, results in osteopenia [42]. Deficiency of both Rac1 and Rac2 in osteoclasts decreases osteoclast formation and abrogates bone resorption, resulting in osteopetrosis [41, 43]. Rho GTPases are essential for formation and organization of the osteoclast podosomes, though the experimental method did not distinguish between RHOA, RHOB and RHOC function [44]. RHO A, B and C share 85% sequence identity, and while most studies focus on RHOA function, differential expression patterns, as well as differences in binding regulators and effectors suggest that these proteins may have non-redundant functions in some systems [49]. Our data however, indicate that RHOC is not essential for osteoclast function. Likewise although other RAB family members have been implicated in vesicular trafficking in osteoclasts [40, 45, 46], RAB38 appears dispensable. However, it is also possible that the Rab38cht mutation does not affect RAB38 function in the osteoclast and that a true null allele would reveal a phenotype.
It is more difficult to argue the case for redundancy as an explanation for the observation that Adcy3−/− osteoclasts differentiate and resorb normally despite solid evidence for a negative regulatory role for cAMP in osteoclast differentiation. Although transcripts from Adcy2, 5, 6, 7 and 9 are also detected in osteoclasts, measurement of cAMP levels in cultured osteoclasts demonstrates that cAMP levels are significantly diminished in the absence of ADCY3. Thus, the absence of an in vitro osteoclast phenotype in Adcy3−/− cultures suggests that the lower levels of cAMP induced by RANKL are insufficient to affect osteoclastogenesis. However, cAMP does rise in response to RANKL in Adcy3−/− osteoclasts, suggesting either that other adenylate cyclases or reductions in phosphodiesterase activity contribute to elevations in cAMP during differentiation.
Strengths of our study include the 1) independent validation of our microarray results with qPCR, 2) rigorous use of WT sex-matched littermates as controls for our micro-CT studies, 3) analysis of both male and female WT and mutant mice by micro-CT and 4) complementation of the in vivo studies with ex vivo osteoclast differentiation and activity assays. The lack of histomorphometric data for 4 of the 5 mutants we analyzed is a weakness of our study, but the absence of a significant bone mass phenotype by micro-CT and normal osteoclast function ex vivo strongly suggests this data would not have provided additional insight. Given the cost of bone histomorphometry, a decision was made not to pursue these studies. An additional weakness is that mutant mice were only analyzed at a single age at baseline. Thus, it is possible that a function for some or all of these genes in the osteoclast in vivo would be revealed at a different age or in models of pathologic bone resorption. Lastly, we have not determined whether the genes examined here (Nhedc2, Adcy3, Serpind1, Rhoc, Rab38) are direct NFATc1 target genes. Perhaps had the list of NFATc1-dependent transcripts reported in Table S2 been refined for direct regulation by NFATc1, by chromatin immunoprecipitation for example, our list of genes for follow-up in vivo studies would have been enriched for functional importance.
5. Conclusion
Gene expression profiling of murine wild-type and NFATc1-deficient osteoclast precursors stimulated with RANKL identified 80 unique transcripts that are highly NFATc1-dependent, including 4 known osteoclast regulators. Analysis of additional candidate osteoclast regulators whose transcription was validated to be RANKL-induced and NFATc1-dependent did not identify genes required for osteoclast function. Our findings underscore the difficulty of extrapolating functional significance from differential expression, highlight the need for careful validation studies of expression, and exemplify the need for experimental validation before functional importance can be attributed to differentially expressed genes in a given biologic process. These data also suggest that NFATc1 ensures the fidelity of the bone resorption pathway by inducing the expression of a large number of functionally redundant genes in the osteoclast.
Supplementary Material
Highlights.
Microarray profiling was used to identify NFATc1 dependent transcripts in the osteoclast
These NFATc1 dependent transcripts encompass 80 genes, including 4 known osteoclast regulators
Five candidate regulators were selected for study: Nhedc2, Serpind1, Adcy3, Rhoc and Rab38
Knock out mice in each of these genes exhibited normal bone mass and osteoclast function
Acknowledgements
The authors acknowledge Dr. Marc Lenburg, for assistance with microarray analysis. Histomorphometric analysis was performed by the Yale Core Center for Musculoskeletal Disorders, supported by NIH P30 AR046032. Funding for this work was provided by the American College of Rheumatology Research and Education Foundation Rheumatology Investigator Award (JFC), International Bone and Mineral Society, Société Française de Rhumatologie, Association pour la Recherche sur le Cancer and the Philippe Foundation (FC), Howard Hughes Fellowship (RS) and NIH grants K08AR054859 (AOA) and R01AR060363 (AOA), DC04156 (DRS), and HL55520 (DMT). A.O.A. also holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Abbreviations
- Nfatc1
Nuclear factor of activated T-cells c1
- Rhoc
ras homolog gene family member C
- Rab38
ras-related protein
- Nhedc2
Na+/H+ exchanger-like domain-containing protein 2
- Adcy3
adenylate cyclase 3
- Serpind1
serpin peptidase inhibitor, clade D, member 1
- OCPs
osteoclast precursors
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sims NA, Gooi JH. Bone remodeling: Multiple cellular interactions required for coupling of bone formation and resorption. Semin Cell Dev Biol. 2008;19:444–451. doi: 10.1016/j.semcdb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Miller PD. Denosumab: anti-RANKL antibody. Curr Osteoporos Rep. 2009;7:18–22. doi: 10.1007/s11914-009-0004-5. [DOI] [PubMed] [Google Scholar]
- 3.Kendler DL, Roux C, Benhamou CL, Brown JP, Lillestol M, Siddhanti S, Man HS, San Martin J, Bone HG. Effects of denosumab on bone mineral density and bone turnover in postmenopausal women transitioning from alendronate therapy. J Bone Miner Res. 2010;25:72–81. doi: 10.1359/jbmr.090716. [DOI] [PubMed] [Google Scholar]
- 4.Rizzoli R, Reginster JY, Boonen S, Breart G, Diez-Perez A, Felsenberg D, Kaufman JM, Kanis JA, Cooper C. Adverse reactions and drug-drug interactions in the management of women with postmenopausal osteoporosis. Calcif Tissue Int. 89:91–104. doi: 10.1007/s00223-011-9499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo SB, Hellstein JW, Kalmar JR. Narrative [corrected] review: bisphosphonates and osteonecrosis of the jaws. Ann Intern Med. 2006;144:753–761. doi: 10.7326/0003-4819-144-10-200605160-00009. [DOI] [PubMed] [Google Scholar]
- 6.Sellmeyer DE. Atypical fractures as a potential complication of long-term bisphosphonate therapy. JAMA. 2010;304:1480–1484. doi: 10.1001/jama.2010.1360. [DOI] [PubMed] [Google Scholar]
- 7.Jonsson B, Strom O, Eisman JA, Papaioannou A, Siris ES, Tosteson A, Kanis JA. Cost-effectiveness of Denosumab for the treatment of postmenopausal osteoporosis. Osteoporos Int. 22:967–982. doi: 10.1007/s00198-010-1424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takayanagi H. Osteoimmunology and the effects of the immune system on bone. Nat Rev Rheumatol. 2009;5:667–676. doi: 10.1038/nrrheum.2009.217. [DOI] [PubMed] [Google Scholar]
- 9.Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, Feng JQ, Bonewald LF, Kodama T, Wutz A, Wagner EF, Penninger JM, Takayanagi H. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat Med. 2011;17:1231–1234. doi: 10.1038/nm.2452. [DOI] [PubMed] [Google Scholar]
- 10.Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O'Brien CA. Matrixembedded cells control osteoclast formation. Nat Med. 2011;17:1235–1241. doi: 10.1038/nm.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones D, Glimcher LH, Aliprantis AO. Osteoimmunology at the nexus of arthritis, osteoporosis, cancer, and infection. J Clin Invest. 2011;121:2534–2542. doi: 10.1172/JCI46262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 13.Takayanagi H. The role of NFAT in osteoclast formation. Ann N Y Acad Sci. 2007;1116:227–237. doi: 10.1196/annals.1402.071. [DOI] [PubMed] [Google Scholar]
- 14.Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, Ostrowski MC, Olsen BR, Glimcher LH. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118:3775–3789. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, Crabtree GR. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell. 2006;10:771–782. doi: 10.1016/j.devcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, Morita I, Wagner EF, Mak TW, Serfling E, Takayanagi H. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202:1261–1269. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aliprantis AO, Stolina M, Kostenuik PJ, Poliachik SL, Warner SE, Bain SD, Gross TS. Transient muscle paralysis degrades bone via rapid osteoclastogenesis. FASEB J. 2012;26:1110–1118. doi: 10.1096/fj.11-196642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sitara D, Aliprantis AO. Transcriptional regulation of bone and joint remodeling by NFAT. Immunol Rev. 2010;233:286–300. doi: 10.1111/j.0105-2896.2009.00849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Glimcher LH, Aliprantis AO. HCO3-/Cl- anion exchanger SLC4A2 is required for proper osteoclast differentiation and function. Proc Natl Acad Sci U S A. 2008;105:16934–16939. doi: 10.1073/pnas.0808763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyers SN, McDaneld TG, Swist SL, Marron BM, Steffen DJ, O'Toole D, O'Connell JR, Beever JE, Sonstegard TS, Smith TP. A deletion mutation in bovine SLC4A2 is associated with osteopetrosis in Red Angus cattle. BMC Genomics. 2010;11:337. doi: 10.1186/1471-2164-11-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 22.He L, Vicente CP, Westrick RJ, Eitzman DT, Tollefsen DM. Heparin cofactor II inhibits arterial thrombosis after endothelial injury. J Clin Invest. 2002;109:213–219. doi: 10.1172/JCI13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong ST, Trinh K, Hacker B, Chan GC, Lowe G, Gaggar A, Xia Z, Gold GH, Storm DR. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27:487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 24.Hakem A, Sanchez-Sweatman O, You-Ten A, Duncan G, Wakeham A, Khokha R, Mak TW. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 2005;19:1974–1979. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loftus SK, Larson DM, Baxter LL, Antonellis A, Chen Y, Wu X, Jiang Y, Bittner M, Hammer JA, 3rd, Pavan WJ. Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci U S A. 2002;99:4471–4476. doi: 10.1073/pnas.072087599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the False Discover Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 28.Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, et al. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development. 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 29.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 30.Hofstetter W, Siegrist M, Simonin A, Bonny O, Fuster DG. Sodium/hydrogen exchanger NHA2 in osteoclasts: subcellular localization and role in vitro and in vivo. Bone. 2010;47:331–340. doi: 10.1016/j.bone.2010.04.605. [DOI] [PubMed] [Google Scholar]
- 31.Ha BG, Hong JM, Park JY, Ha MH, Kim TH, Cho JY, Ryoo HM, Choi JY, Shin HI, Chun SY, Kim SY, Park EK. Proteomic profile of osteoclast membrane proteins: identification of Na+/H+ exchanger domain containing 2 and its role in osteoclast fusion. Proteomics. 2008;8:2625–2639. doi: 10.1002/pmic.200701192. [DOI] [PubMed] [Google Scholar]
- 32.Tollefsen DM. Heparin cofactor II modulates the response to vascular injury. Arterioscler Thromb Vasc Biol. 2007;27:454–460. doi: 10.1161/01.ATV.0000256471.22437.88. [DOI] [PubMed] [Google Scholar]
- 33.Church FC, Noyes CM, Griffith MJ. Inhibition of chymotrypsin by heparin cofactor II. Proc Natl Acad Sci U S A. 1985;82:6431–644. doi: 10.1073/pnas.82.19.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratt CW, Tobin RB, Church FC. Interaction of heparin cofactor II with neutrophil elastase and cathepsin G. J Biol Chem. 1990;265:6092–6097. [PubMed] [Google Scholar]
- 35.Yoon SH, Ryu J, Lee Y, Lee ZH, Kim HH. Adenylate cyclase and calmodulindependent kinase have opposite effects on osteoclastogenesis by regulating the PKA-NFATc1 pathway. J Bone Miner Res. 2011;26:1217–1229. doi: 10.1002/jbmr.310. [DOI] [PubMed] [Google Scholar]
- 36.Raisz LG, Vanderhoek JY, Simmons HA, Kream BE, Nicolaou KC. Prostaglandin synthesis by fetal rat bone in vitro: evidence for a role of prostacyclin. Prostaglandins. 1979;17:905–914. doi: 10.1016/0090-6980(79)90061-3. [DOI] [PubMed] [Google Scholar]
- 37.Takami M, Cho ES, Lee SY, Kamijo R, Yim M. Phosphodiesterase inhibitors stimulate osteoclast formation via TRANCE/RANKL expression in osteoblasts: possible involvement of ERK and p38 MAPK pathways. FEBS Lett. 2005;579:832–838. doi: 10.1016/j.febslet.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 38.Sheridan CM, Heist EK, Beals CR, Crabtree GR, Gardner P. Protein kinase A negatively modulates the nuclear accumulation of NF-ATc1 by priming for subsequent phosphorylation by glycogen synthase kinase-3. J Biol Chem. 2002;277:48664–48676. doi: 10.1074/jbc.M207029200. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Li V, Chan GC, Phan T, Nudelman AS, Xia Z, Storm DR. Adult type 3 adenylyl cyclase-deficient mice are obese. PLoS One. 2009;4:e6979. doi: 10.1371/journal.pone.0006979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itzstein C, Coxon FP, Rogers MJ. The regulation of osteoclast function and bone resorption by small GTPases. Small Gtpases. 2011;2:117–130. doi: 10.4161/sgtp.2.3.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croke M, Ross FP, Korhonen M, Williams DA, Zou W, Teitelbaum SL. Rac deletion in osteoclasts causes severe osteopetrosis. J Cell Sci. 2011;124:3811–3821. doi: 10.1242/jcs.086280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito Y, Teitelbaum SL, Zou W, Zheng Y, Johnson JF, Chappel J, Ross FP, Zhao H. Cdc42 regulates bone modeling and remodeling in mice by modulating RANKL/M-CSF signaling and osteoclast polarization. J Clin Invest. 2010;120:1981–1993. doi: 10.1172/JCI39650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Lebowitz D, Sun C, Thang H, Grynpas MD, Glogauer M. Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. J Bone Miner Res. 2008;23:260–270. doi: 10.1359/jbmr.071013. [DOI] [PubMed] [Google Scholar]
- 44.Zhang D, Udagawa N, Nakamura I, Murakami H, Saito S, Yamasaki K, Shibasaki Y, Morii N, Narumiya S, Takahashi N, et al. The small GTP-binding protein, rho p21, is involved in bone resorption by regulating cytoskeletal organization in osteoclasts. J Cell Sci. 1995;108(Pt 6):2285–2292. doi: 10.1242/jcs.108.6.2285. [DOI] [PubMed] [Google Scholar]
- 45.Zhao H, Laitala-Leinonen T, Parikka V, Vaananen HK. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J Biol Chem. 2001;276:39295–39302. doi: 10.1074/jbc.M010999200. [DOI] [PubMed] [Google Scholar]
- 46.Pavlos NJ, Xu J, Riedel D, Yeoh JS, Teitelbaum SL, Papadimitriou JM, Jahn R, Ross FP, Zheng MH. Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption. Mol Cell Biol. 2005;25:5253–5269. doi: 10.1128/MCB.25.12.5253-5269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seto S, Tsujimura K, Koide Y. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic. 2011;12:407–420. doi: 10.1111/j.1600-0854.2011.01165.x. [DOI] [PubMed] [Google Scholar]
- 48.Osanai K, Higuchi J, Oikawa R, Kobayashi M, Tsuchihara K, Iguchi M, Huang J, Voelker DR, Toga H. Altered lung surfactant system in a Rab38-deficient rat model of Hermansky-Pudlak syndrome. Am J Physiol Lung Cell Mol Physiol. 2010;298:L243–L251. doi: 10.1152/ajplung.00242.2009. [DOI] [PubMed] [Google Scholar]
- 49.Wheeler AP, Ridley AJ. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp Cell Res. 2004;301:43–49. doi: 10.1016/j.yexcr.2004.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.