Abstract
This paper reports on a significant improvement of a new structural biology approach designed to probe the secondary structure of membrane proteins using the pulsed EPR technique of Electron Spin Echo Envelope Modulation (ESEEM) spectroscopy. Previously, we showed that we could characterize an α-helical secondary structure with ESEEM spectroscopy using a 2H-labeled Val side chain coupled with site-directed spin-labeling (SDSL). In order to further develop this new approach, molecular dynamic (MD) simulations were conducted on several different hydrophobic residues that are commonly found in membrane proteins. 2H-SL distance distributions from the MD results indicated that 2H-labeled Leu was a very strong candidate to significantly improve this ESEEM approach. In order to test this hypothesis, the secondary structure of the α-helical M2δ peptide of the acetylcholine receptor (AChR) incorporated into a bicelle was investigated with 2H-labeled Leu d10 at position 10 (i) and nitroxide spin labels positioned 1, 2, 3 and 4 residues away (denoted i+1 to i+4) with ESEEM spectroscopy. The ESEEM data reveal a unique pattern that is characteristic of an α-helix (3.6 residues per turn). Strong 2H modulation was detected for the i+3 and i+4 samples, but not for the i+2 sample. The 2H modulation depth observed for 2H-labeled d10 Leu was significantly enhanced (x4) when compared to previous ESEEM measurements that used 2H-labeled d8 Val. Computational studies indicate that deuterium nuclei on the Leu sidechain are closer to the spin label when compared to Val. The enhancement of 2H modulation and the corresponding Fourier Transform (FT) peak intensity for 2H-labeled Leu significantly reduces the ESEEM data acquisition time for Leu when compared to Val. This research demonstrates that a different 2H-labeled amino acid residue can be used as an efficient ESEEM probe further substantiating this important biophysical technique. Finally, this new method can provide pertinent qualitative structural information on membrane proteins in a short time (few minutes) at low sample concentrations (~50 μM).
Keywords: Electron Spin Echo Envelope Modulation, Membrane Protein, Spin Labeling, EPR
INTRODUCTION
A novel approach is being developed to characterize the secondary structure of membrane proteins utilizing Electron Spin Echo Envelope Modulation (ESEEM) spectroscopy coupled with site-directed spin-labeling (SDSL) 1. Despite the importance and large number of membrane proteins, relatively limited structural information is known about them2,3. New biophysical techniques are needed to probe their structural properties. Their hydrophobic nature and low expression yields cause difficulties for traditional structural techniques such as x-ray crystallography and solution NMR spectroscopy 3,4.
ESEEM spectroscopy indirectly observes NMR transitions through an electron spin coupled to a nearby NMR active nucleus 5-7. ESEEM can detect weak dipolar interactions between a 2H atom and a spin label out to a maximum distance of approximately 8 Å 7,8. The modulation depth for weakly coupled nuclei is proportional to 1/r6. Previously, we demonstrated using 2H-labeled Val as a probe and a strategically placed MTSL (S- (2,2,5,5-tetramethyl-2, 5-dihydro-1H-pyrrol-3-yl) methyl methanesulfonothioate), α-helical content could be detected with ESEEM spectroscopy 1. The unique structure of an α-helix consists of 3.6 residues per turn with a vertical distance of 5.4 Å separating the turn. When a Cys mutated nitroxide spin label is positioned 1, 2, 3 and 4 residues away (denoted i+1 to i+4 respectively) from a 2H-labeled residue such as the d8 Val (i) side chain, the periodicity of the α-helical structure gives rise to a unique pattern in the collective ESEEM data. The i+3 and i+4 samples reveal 2H modulation from the dipolar coupling between the 2H nuclei of the Val side chain and spin label (< 8 Å). However, in the i+1 and i+2 samples, the 2H nuclei are beyond the detection limit (> 8 Å). Due to the structure and dynamics of different amino acid side chains, a variety of patterns and signal intensities can be obtained using this approach 9,10. The aim of this approach is to provide additional tools for probing the structure of membrane proteins using SDSL and EPR spectroscopy.
In this work, a molecular modeling study was conducted to obtain distance distributions between 2H nuclei on different amino acid side chains and a spin label. Four different 2H-labeled hydrophobic amino acids (Leu, Val, Phe, Ala) that are commonly found in membrane proteins and commercially available were compared. The Leu side chain was found to a very strong candidate for conducting the present study based on the molecular modeling studies carried on different amino acid residues in the sequence of AChR M2δ peptide. 2H-labeled d10 Leu was used to demonstrate the practicality of this method instead of 2H-labeled d8 valine. The observed experimental ESEEM data have similar patterns for α-helical segments with a significantly enhanced 2H modulation depth. The distance distributions obtained from the modeling studies between 2H nuclei on the Leu side chain and the spin label are consistent with the experimental ESEEM data. These experiments demonstrate that another 2H-labeled amino acid can be used as probe for secondary structure determination with significantly improved sensitivity; thus, making this method applicable to more proteins that contain Leu.
MATERIALS AND METHODS
The M2δ subunit of the acetylcholine receptor (AChR) was used as an α-helical model for transmembrane peptides and proteins 11-13. All peptides were synthesized using Fmoc solid-phase peptide synthesize chemistry on a CEM microwave solid phase peptide synthesizer 14. Four different peptides were designed by positioning the 2H-labeled d10 leucine at position 10 (i) and the cysteine (X) at four successive positions (i+1 to i+4). For one complete set of samples, the sequences of peptides are as follows: i+1 (EKMSTAISXLLAQAVFLLLTSQR), i+2(EKMSTAIXVLLAQAVFLLLTSQR), i+3 (EKMSTAXSVLLAQAVFLLLTSQR), and i+4 (EKMSTXISVLLAQAVFLLLTSQR), where X represents the position of the cysteine for spin labeling. Additionally, one i+3 control sample was prepared without 2H labeling at the Leu10 position.
After the peptides were cleaved from their solid support, reverse-phase HPLC was used for purification with a C4 preparation column and a gradient of 5% to 95% solvent B (90% acetonitrile) 15,16. Purified peptides were labeled with MTSL (Toronto Research Chemicals) in DMSO for 20 hours and excess MTSL was removed by HPLC 16. MALDI-TOF was utilized to confirm the molecular weight and purity of the target peptides. MTSL-labeled M2δ peptides were integrated into DMPC/DHPC (3.5/1) bicelles at 1:1000 molar ratio 16. For these experiments, bicelles were used as membrane mimic system and yielded high quality ESEEM data 17,18. Comparable ESEEM data could be obtained with liposomes.
X-band CW-EPR (~9 GHz) spectroscopy was used to measure spin concentration (~150 μM) of all bicelle samples. Three-pulse ESEEM measurements were performed on a Bruker ELEXSYS E580 with an ER 4118X MS3 resonator using a 200 ns tau value with a microwave frequency of ~9.269 GHz at 80 K 1. For all samples, a starting T of 386 ns and 512 points in 12 ns increments were used to collect the spectra 1. All ESEEM data were obtained with 40 Ol of bicelle samples and in 30 scans.
Molecular modeling and molecular dynamics studies for each sample were performed using nanoscale molecular dynamics (NAMD) 19 with the molecular graphics software VMD 20. The structure of the AChR M2δ peptide was obtained from the solution NMR coordinates (PDB entry: 1EQ8). Cysteine mutants were created at i+1, i+2, i+3, and i+4 positions using VMD, a MTSL nitroxide spin probe was attached by using CHARMM force-field topology files incorporated in NAMD. The resultant assembly of spin labeled peptides was solvated in a water box. Further equilibration and energy minimization were performed using NAMD simulations. Molecular dynamic simulations were collected out to 1 ns at room temperature using Langevin dynamics under NAMD. This timescale corresponds to the MTSL and leucine sidechain dynamics. The trajectory data were recorded in 1 ps increments. The possible distance distribution for each deuterium and SL was obtained from the analysis of the trajectory data file using VMD. All molecular dynamics simulations were run on a home-built 24-node Linux Beowulf style cluster in our lab.
RESULTS
Molecular modeling and molecular dynamics studies were performed on different hydrophobic amino acids (Ala, Leu, Val, and Phe) in which deuterium labeled side chains are commercially available. Distance distributions were determined between 2H nuclei on the side chain (i) and the N-O bond for the SL at the i+3 and i+4 positions. 2H-SL distance distributions were found to be within the 8 Å ESSEM detection range for all residues studied. However, 2H atoms on the Leu sidechain were found to have the shortest distances; thus, suggesting that Leu would be a strong candidate to significantly improve this ESEEM approach.
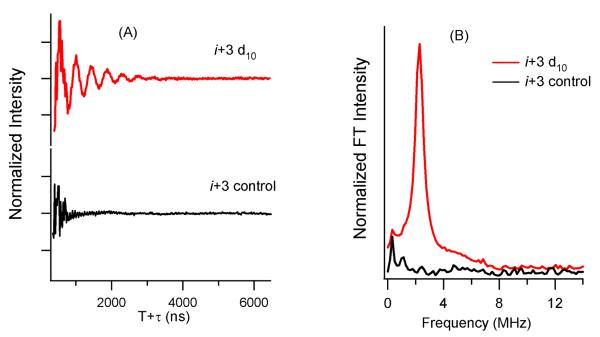
Figure 1 shows three-pulse ESEEM data for i+3 2H-labeled d10 Leu10 M2δ peptide incorporated into DMPC/DHPC lipid bicelles. Also, a non-deuterated Leu (control) sample is shown for comparison. Both low frequency 2H modulation and high frequency proton modulation appear in the time domain data (Figure 1(A)) for the 2H-labeled d10 Leu10 i+3 sample (red). However, only proton modulation appears in the control sample prepared in the absence of 2H-labeled Leu (black). FT frequency domain data for the 2H-labeled d10 Leu10 M2δ i+3 sample reveal a peak centered at the 2H Larmor frequency of 2.3 MHz. No such peak was observed for the control sample. The ESEEM spectra clearly indicate that the dipolar interaction between the SL and 2H-labeled Leu can be detected for the i+3 Leu10 M2δ sample.
Figure 1.
(A) Time domain three-pulse ESEEM spectra of Leu10 i+3 M2δ in bicelle. The top (red) is 2H-labeled d10 Leu10 i+3 and the bottom (black) is the non-deuterated Leu10 i+3 control sample. (B) Frequency domain spectra where red illustrates the 2H-labeled d10 Leu10 i+3 M2δ sample and black illustrates the control.
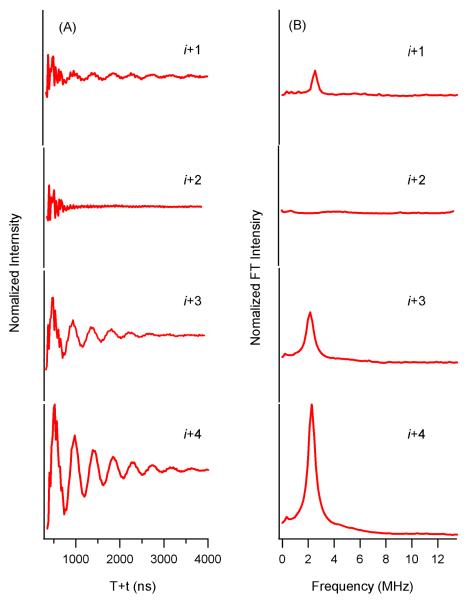
The three-pulse ESEEM data for the 2H-labeled d10 Leu10 (i+1 through i+4) M2δ peptide are shown in Figure 2, respectively. For 2H-labeled d10 Leu10 M2δ peptides, the 2H modulation is observed in the time domain for the i+1, i+3 and i+4 samples. Also, a 2H peak is clearly observed at the 2H Larmor frequency in the frequency domain. However, there is no 2H modulation for the 2H-labeled d10 Leu10 i+2 M2δ sample. To support these data, molecular modeling and molecular dynamics studies were conducted to estimate distances between the 2H nuclei on the Leu10 sidechain and the N-O bond on the SL at the i+1, i+2, i+3, and i+4 positions 1. 2H-SL distances for the i+2 (9–15 Å) were found to be outside the ESEEM detection range of 8 Å. However, 2H-SL distances for the i+1 (6–11 Å), i+3 (5–11 Å) and i+4 (3–10 Å) positions were found to be within the ESEEM detection range with different probabilities.
Figure 2.
Three-pulse ESEEM experimental data with a τ=200ns of i+1 to i+4 2H-labeled d10 Leu10 in lipid bicelle samples. (A) Time domain spectra, (B) Frequency domain spectra.
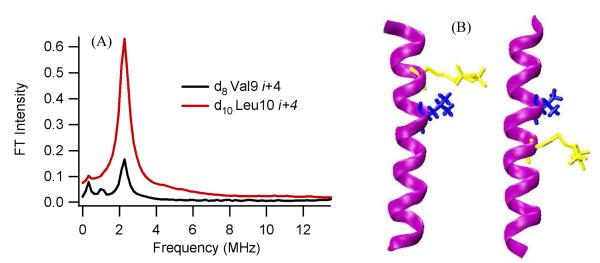
Figure 3(A) shows the comparison of the ESEEM frequency domain data between 2H-labeled d10 Leu10 and 2H-labeled d8 Val9 peptides at the i+4 position. These frequency domain data reveal a dramatic signal enhancement when 2H-labeled d10 Leu is used instead of 2H-labeled d8 Val. Distances and conformations provided by molecular dynamic simulation supported this result. Figure 3(B) displays the minimal energy conformations of the AChR M2δ peptides with 2H-labeled d10 Leu10 (left) and 2H-labeled d8 Val9 (right) with MTSL at the i+4 positions. The figure indicates that the distance from the N-O bond on the nitroxide to the nearest deuteron is much smaller in AChR M2δ peptides with 2H-labeled d10 Leu10 (4.4 Å) than that with 2H-labeled d Val9 (7.6 Å) 1
Figure 3.
Comparison between 2H-labeled d8 Val9 and 2H-labeled d10Leu10 M2δ bicelle samples. (A) Normalized FT frequency domain modulation data for 2H-labeled d10Leu10 i+4 (blue) and 2H-labeled d8 Val9 i+4 (red). (B) Likely conformation of M2δ with and 2H-labeled d10Leu10 (left) and M2δ with 2H-labeled d8 Val9 (right) both with MTSL at i+4 from MD simulation
DISCUSSION
In previous 2H-labeled d8 Val9 experiments, three-pulse ESEEM data showed 2H peaks for both i+3 and i+4, but not for i+1 and i+2 for the AChR M2δ peptide 1. This unique pattern is indicative of an α-helical secondary structure. MD simulations suggest that 2H-labeled Leu d10 atoms on the side chain will be closer in distance to the SL for i+3 and i+4 positions when compared to Val, Ala, Phe. 2H-labeled Leu d10 is a strong candidate for this new methodology method due to its longer chain and larger number of available deuterium atoms within the ESEEM ~8 Å detection limit.
For the 2H-labeled d10 Leu10 ESEEM experiments conducted, similar data were obtained with significantly enhanced 2H modulation as shown in Figure 3(B). MD simulations reveal a significant population of 2H-SL distances below the 8 Å ESEEM detection limit for 2H-labeled d10 Leu10 when compared to 2H-labeled d8 Val9. In the case of Leu, there are two torsion angle rotations about χ1 and χ2 and two free rotation modes about the Cγ and Cδ bonds, which correspond to two (CD3) methyl groups 9,21,22. As shown in Figure 3(B), the additional C-C bonds in the Leu sidechain bring the deuterons closer to the N-O nitroxide bond when compared to the Val sidechain. Therefore, the MTSL has a higher probability of being able to detect the 2H nuclei at a closer distance and resulting in a significant increase in 2H modulation depth for Leu at this position. However, different conformations of Val and Leu might be favored due to unique sidechain or tertiary interactions. For MTSL, there are three torsional angle rotations about χ1, χ2 and χ3 and two additional free torsion angle rotations about χ4 and χ5 23-25. Thus, various orientations could be favored due to the interaction of MTSL and peptide backbone or amino acid side chains. All of these factors play a role in the 2H modulation depth and can alter the corresponding FT intensity. Thus, the observed 2H modulation depth for 2H-labeled Leu or Val can vary depending upon the biological system studied.
The 2H-labeled d10 Leu10 ESEEM spectra reveal a 2H peak for the i+1 sample, indicating that 2H nuclei are weakly coupled to the MTSL. However, no coupling was detected for the i+2 sample. Since α-helices have 3.6 amino acids per turn, the Leu residue in the i+2 sample lies on the opposite side of the helix as the MTSL. It is expected that distances between 2H nuclei on the Leu10 sidechain and the i+1 SL are closer than that of i+2. Also, MD simulations indicated 2H-SL distances ranged from 9–15 Å for the i+2 and 6-11 Å for i+1 sample which matches well with the ESEEM data of the i+1 and i+2 samples. MD simulations show similar 2H-SL distance ranges for the i+1and i+3 samples. However, the population of distance distributions within the 8 Å limit for i+3 is significantly greater than i+1 (data not shown), which is consistent with the i+3 sample having larger 2H modulation and a bigger FT 2H peak in experimental ESEEM data (Figure 2).
Theoretically, the distance between a SL and a 2H nucleus can be determined by simulation of the ESEEM spectra. For this three-pulse ESEEM (π/2-τ-π /2-T- π /2), the modulation depth produced by the dipolar-coupled nucleus is proportional to 1/r6,7. However, due to the variation of conformations and dynamics of the leucine side chain and the MTSL, it is difficult to get quantitative distance information from this method so far 21,26-28. Since the three-pulse ESEEM measurements were conducted at 80K, the leucine side chain and MTSL adopt a variety of conformations with respect to each other, yielding a range of distances between the MTSL and multiple 2H nuclei. In order to improve this approach to get more quantitative distance information, different 2H-labeled amino acids need to be investigated. Amino acids with only one 2H atom or magnetically equivalent 2H nuclei, such as 2H-labeled d3 Ala can be employed with this approach to provide quantitative distance measurements via ESEEM spectral simulations.
This ESEEM structural approach is comparable to rotational echo double resonance (REDOR) solid-state NMR spectroscopy. Solid State NMR spectroscopy is a very powerful technique for studying the structure of membrane proteins 12. REDOR NMR can be used to probe the alpha helical component when coupled with spectral simulation by measuring dipolar couplings between NMR active nuclei, such as 13C and 15N 29,30. However, this technique suffers from low signal sensitivity. As a result, REDOR experiments usually require milligram quantities of isotopically labeled protein or peptide and abundant instrumental time. By using ESEEM and SDSL, secondary structure determination for membrane peptides and proteins can be performed on the μmol scale and with minimal data acquisition time (5 to 10 minutes).
CONCLUSION
2H-labeled d10 Leu was shown to be a very powerful secondary structure probe with enhanced 2H modulation by a factor of 4 when compared to Val. This modulation enhancement leads to a significant increase in sensitivity and corresponding decrease in data acquisition time. Thus, the amount of time required to achieve the same signal-to-noise using 2H-labeled d10 leucine as a probe instead of 2H-labeled d8 valine decreases by a factor of ~16. The Leu ESEEM data further validates this structural biology approach and provide researchers with another amino acid to probe secondary structure for proteins and peptides. This ESEEM method uses SDSL and selective deuterium labels, both of which can be incorporated into standard expression systems using routine molecular biology techniques for applications to larger protein systems (not just peptides) 31,32. 2H and 15N residue specific isotopic labeling schemes are used in a variety of NMR structural biology experiments33,34.
This new ESEEM secondary structure approach can be applied to a wide variety of different protein systems that are not amiable to x-ray crystallography or solution NMR including membrane proteins. Several other biophysical techniques such as CD, NMR, CW-EPR, FRET, and 2D IR can provide protein secondary structural information. This ESEEM approach is advantageous because it has no size limitations, is straightforward, uses small spectroscopic labels, minimal sample requirements, and provides selective secondary structural information on specific protein segments. For SDSL EPR researchers, this ESEEM approach will allow researchers to probe not only the dynamics of a specific spin-labeled site with CW-EPR lineshape analysis, but also the secondary structure. Traditional EPR experiments that probe secondary structure typically require multiple SDSLs in a row to detect differences in lineshape or relaxation that follow a pattern that depends upon the environment and local secondary structure of the protein. Sometimes the EPR data can be ambiguous or challenging to interpret. Certain biophysical techniques such as CD provide global secondary structural information, whereas this ESEEM approach can probe the secondary structure of specific segments of a protein like the solid-state NMR REDOR technique. Direct identification of site-specific secondary structure can be obtained using this ESEEM approach with small amounts of sample and minimal instrumentation time when compared to NMR.
ACKNOWLEDGMENTS
We greatly appreciate the National Science Foundation Award CHE-1011909 (to GAL) and National Institute of Health Grants RO1 GM60259-01 (to GAL) for supporting this work generously.
Funding Sources:
This work was generously supported by the National Science Foundation Award CHE-1011909 (to GAL). Funding was also provided by the National Institute of Health Grants RO1 GM60259-01 (to GAL).
ABBREVIATIONS
- ESEEM
Electron Spine Echo Envelope Modulation
- AChR
Acetylcholine Receptor
- MTSL
(S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate)
- SDSL
Site-Directed Spin-Labeling
- DMSO
Dimethyl sulfoxide
- MALDI
Matrix-Assisted Laser Desorption/Ionization
- TOF
Time of Flight
- DMPC
1,2-Dimyristoyl-sn-glycero-3-phosphorylcholine
- DHPC
1,2- dihexanoyl-sn-glycero-3- phosphorylcholine
- CW
Continuous Wave
- EPR
Electron Paramagnetic Resonance
- MD
Molecular Dynamic
- REDOR
Rotational Echo Double Resonance
REFERENCES
- (1).Mayo D, Zhou A, Sahu I, McCarrick R, Walton P, Ring A, Troxel K, Coey A, Hawn J, Emwas A-H, Lorigan GA. Protein Science. 2011;20:1100. doi: 10.1002/pro.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).McLuskey K, Roszak AW, Zhu Y, Isaacs NW. European Biophysics Journal with Biophysics Letters. 2010;39:723. doi: 10.1007/s00249-009-0546-6. [DOI] [PubMed] [Google Scholar]
- (3).Bordag N, Keller S. Chemistry and Physics of Lipids. 2010;163:1. doi: 10.1016/j.chemphyslip.2009.07.009. [DOI] [PubMed] [Google Scholar]
- (4).Huang C, Mohanty S. Journal of the American Chemical Society. 2010;132:3662. doi: 10.1021/ja100078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lorigan GA, Britt RD, Kim JH, Hille R. Biochimica Et Biophysica Acta-Bioenergetics. 1994;1185:284. doi: 10.1016/0005-2728(94)90243-7. [DOI] [PubMed] [Google Scholar]
- (6).Force DA, Randall DW, Lorigan GA, Clemens KL, Britt RD. Journal of the American Chemical Society. 1998;120:13321. [Google Scholar]
- (7).Mims WB. Physical Review B-Solid State. 1972;5:2409. [Google Scholar]
- (8).Milov AD, Samoilova RI, Shubin AA, Gorbunova EY, Mustaeva LG, Ovchinnikova TV, Raap J, Tsvetkov YD. Applied Magnetic Resonance. 2010;38:75. [Google Scholar]
- (9).Mulder FAA. Chembiochem. 2009;10:1477. doi: 10.1002/cbic.200900086. [DOI] [PubMed] [Google Scholar]
- (10).Liu W, Crocker E, Siminovitch DJ, Smith SO. Biophysical Journal. 2003;84:1263. doi: 10.1016/S0006-3495(03)74941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kim J, McNamee MG. Biochemistry. 1998;37:4680. doi: 10.1021/bi972666k. [DOI] [PubMed] [Google Scholar]
- (12).Inbaraj JJ, Cardon TB, Laryukhin M, Grosser SM, Lorigan GA. Journal of the American Chemical Society. 2006;128:9549. doi: 10.1021/ja0622204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Sankararamakrishnan R, Sansom MSP. Biophysical Chemistry. 1995;55:215. doi: 10.1016/0301-4622(95)00006-j. [DOI] [PubMed] [Google Scholar]
- (14).Oblattmontal M, Buhler LK, Iwamoto T, Tomich JM, Montal M. J. of Biol. Chem. 1993;268:14601. [PubMed] [Google Scholar]
- (15).Bhargava K, Feix JB. Biophys. J. 2004;86:329. doi: 10.1016/S0006-3495(04)74108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mayo DJ, Inbaraj JJ, Subbaraman N, Grosser SM, Chan CA, Lorigan GA. J. Am. Chem. Soc. 2008;130:9656. doi: 10.1021/ja803590w. [DOI] [PubMed] [Google Scholar]
- (17).Marcotte I, Auger M. Concepts Magn. Reson., Part A. 2005;24A:17. [Google Scholar]
- (18).Jesorka A, Orwar O. Liposomes: Technologies and analytical applications. Annual Review of Analytical Chemistry. 2008;1:801. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- (19).Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. of Comput. Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Humphrey W, Dalke A, Schulten K. J. of Mol. Graphics & Modell. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- (21).Batchelder LS, Sullivan CE, Jelinski LW, Torchia DA. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1982;79:386. doi: 10.1073/pnas.79.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wand A. J. Nat. Struct. Biol. 2001;8:926. doi: 10.1038/nsb1101-926. [DOI] [PubMed] [Google Scholar]
- (23).Sezer D, Freed JH, Roux B. J. of Phys. Chem. B. 2008;112:5755. doi: 10.1021/jp711375x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Beier C, Steinhoff H-J. Biophys. J. 2006;91:2647. doi: 10.1529/biophysj.105.080051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Columbus L, Hubbell WL. Trends in Biochem. Sci. 2002;27:288. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- (26).Fanucci GE, Cafiso DS. Curr. Opin. Struct. Biol. 2006;16:644. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- (27).McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Biochemistry. 1996;35:7692. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- (28).Lee AL, Kinnear SA, Wand A. J. Nat. Struct. Biol. 2000;7:72. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- (29).Howell SC, Mesleh MF, Opella S. J. Biochemistry. 2005;44:5196. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- (30).Chu S, Coey AT, Lorigan GA. Biochimica Et Biophysica Acta-Biomembranes. 2010;1798:210. doi: 10.1016/j.bbamem.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Klare JP, Steinhoff H. J. Photosynth Res. 2009;102:377. doi: 10.1007/s11120-009-9490-7. [DOI] [PubMed] [Google Scholar]
- (32).Cheng H, Westler WM, Xia B, Oh BH, Markley JL. Archives of Biochemistry and Biophysics. 1995;316:619. doi: 10.1006/abbi.1995.1082. [DOI] [PubMed] [Google Scholar]
- (33).Simplaceanu V, Lukin JA, Fang TY, Zou M, Ho NT, Ho C. Biophys J. 2000;79:1146. doi: 10.1016/S0006-3495(00)76368-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Sailer M, Helms GL, Henkel T, Niemczura WP, Stiles ME, Vederas JC. Biochemistry. 1993;32:310. doi: 10.1021/bi00052a039. [DOI] [PubMed] [Google Scholar]