Abstract
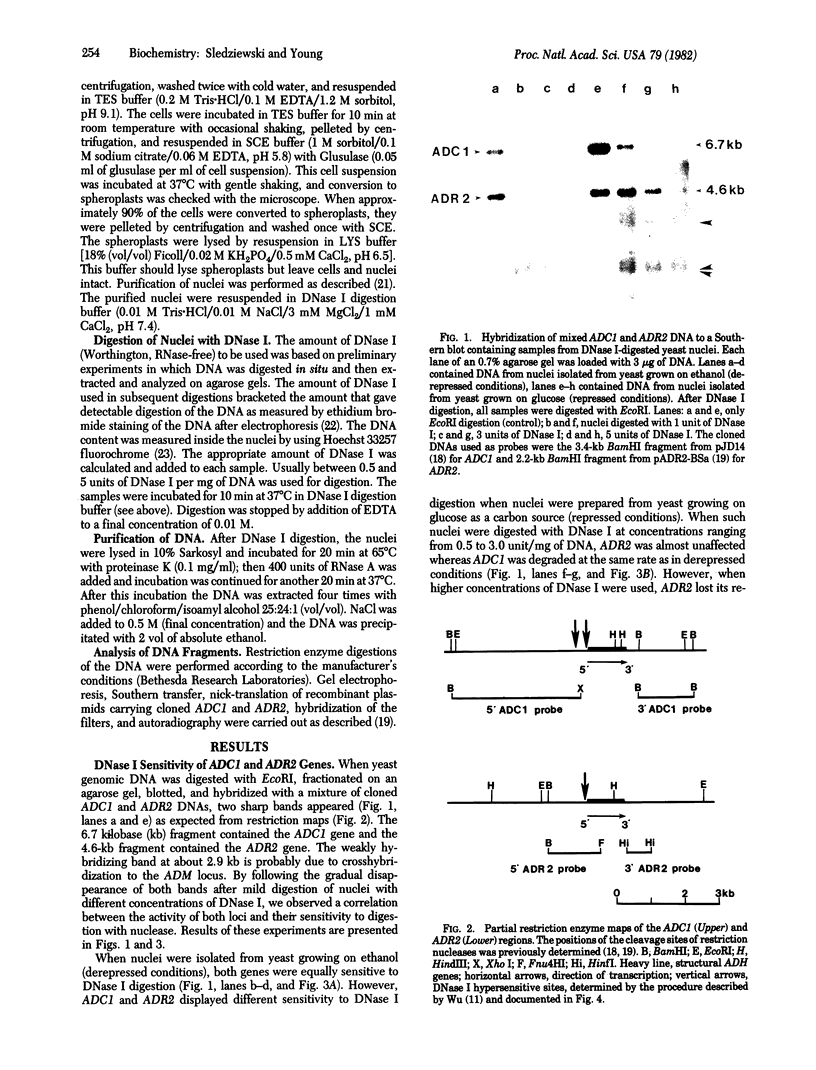
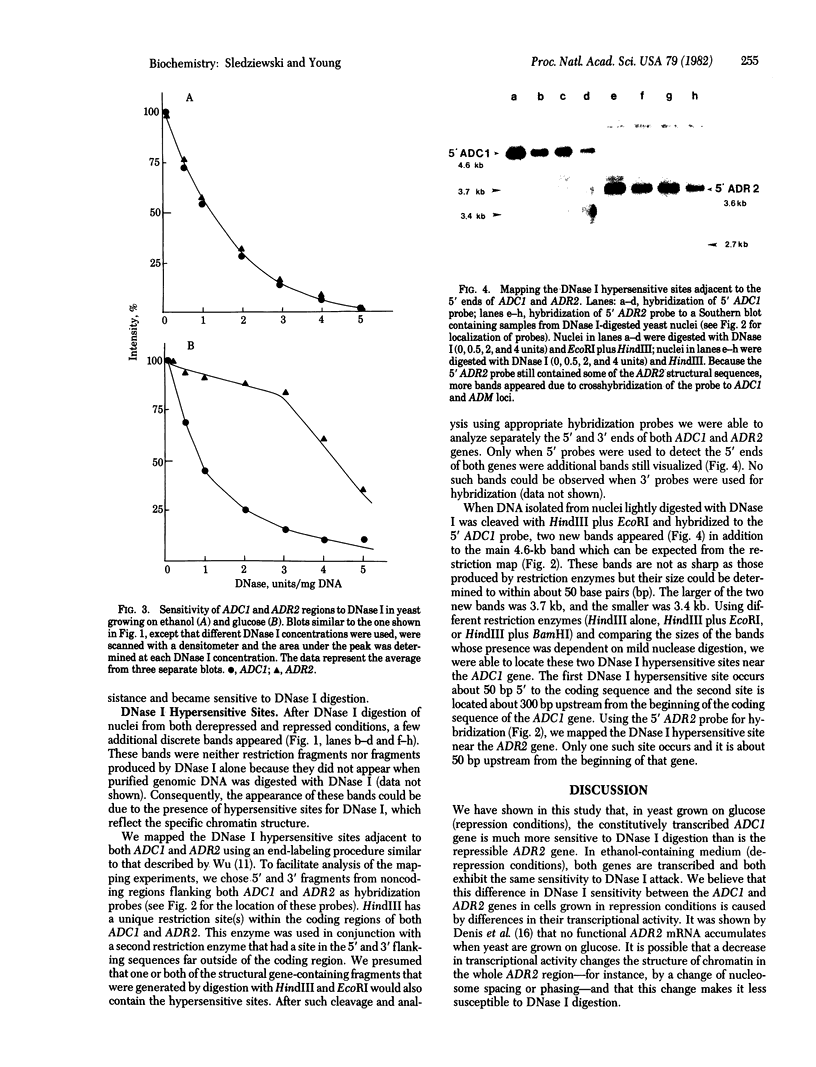
In the present study we have analyzed the kinetics of DNase I digestion of the two alcohol dehydrogenase (ADH) genes within yeast nuclei. We have found that in yeast grown on glucose the constitutively transcribed ADCI gene is much more sensitive to DNase I digestion than is the repressible ADR2 gene. In yeast grown on ethanol, both genes are transcribed and both exhibit the same sensitivity to DNase I attack. We have also found and mapped DNase I hypersensitive sites near the 5' ends of constitutive and repressible ADH genes. These sites are well correlated with the position at which transcription is initiated.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ciriacy M. Cis-dominant regulatory mutations affecting the formation of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1976 Jun 15;145(3):327–333. doi: 10.1007/BF00325831. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol Gen Genet. 1975;138(2):157–164. doi: 10.1007/BF02428119. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of further cis- and trans-acting regulatory elements involved in the synthesis of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Nov;176(3):427–431. doi: 10.1007/BF00333107. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Ciriacy M., Young E. T. A positive regulatory gene is required for accumulation of the functional messenger RNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol. 1981 Jun 5;148(4):355–368. doi: 10.1016/0022-2836(81)90181-9. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Weintraub H. M. An altered subunit configuration associated with the actively transcribed DNA of integrated adenovirus genes. Cell. 1977 Nov;12(3):783–794. doi: 10.1016/0092-8674(77)90277-x. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Das S., Neiman P., Weintraub H. Regulation of expression and chromosomal subunit conformation of avian retrovirus genomes. Cell. 1978 Aug;14(4):865–878. doi: 10.1016/0092-8674(78)90342-2. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Corces V., Lowenhaupt K., Elgin S. C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5' ends of regions of transcription. Proc Natl Acad Sci U S A. 1981 Jan;78(1):143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca C., Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980 Mar 1;102(2):344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- Lohr D., Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. doi: 10.1073/pnas.76.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. M., Turner P., Nienhuis A. W., Axelrod D. E., Gopalakrishnan T. V. Active conformation of the globin genes in uninduced and induced mouse erythroleukemia cells. Cell. 1978 Jul;14(3):511–521. doi: 10.1016/0092-8674(78)90237-4. [DOI] [PubMed] [Google Scholar]
- Samal B., Worcel A., Louis C., Schedl P. Chromatin structure of the histone genes of D. melanogaster. Cell. 1981 Feb;23(2):401–409. doi: 10.1016/0092-8674(81)90135-5. [DOI] [PubMed] [Google Scholar]
- Schultz L. D. Transcriptional role of yeast deoxyribonucleic acid dependent ribonucleic acid polymerase III. Biochemistry. 1978 Feb 21;17(4):750–758. doi: 10.1021/bi00597a031. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Stalder J., Groudine M., Dodgson J. B., Engel J. D., Weintraub H. Hb switching in chickens. Cell. 1980 Apr;19(4):973–980. doi: 10.1016/0092-8674(80)90088-4. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Bennetzen J., Young E. T., Nasmyth K., Hall B. D. Isolation of the structural gene for alcohol dehydrogenase by genetic complementation in yeast. Nature. 1980 Jan 10;283(5743):214–216. doi: 10.1038/283214a0. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Wu C., Gilbert W. Tissue-specific exposure of chromatin structure at the 5' terminus of the rat preproinsulin II gene. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1577–1580. doi: 10.1073/pnas.78.3.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]