Abstract
Intrinsically disordered proteins (IDPs) are abundant in nature and characterization of their potential structural propensities remains a widely pursued but challenging task. Analysis of NMR secondary chemical shifts plays an important role in such studies, but the output of such analyses depends on the accuracy of reference random coil chemical shifts. Although uniform perdeuteration of IDPs can dramatically increase spectral resolution, a feature particularly important for the poorly dispersed IDP spectra, the impact of deuterium isotope shifts on random coil values has not yet been fully characterized. Very precise 2H isotope shift measurements for 13Cα, 13Cβ, 13C’, 15N, and 1HN have been obtained by using a mixed sample of protonated and uniformly perdeuterated -synuclein, a protein with chemical shifts exceptionally close to random coil values. Decomposition of these isotope shifts into one-bond, two-bond and three-bond effects as well as intra- and sequential residue contributions shows that such an analysis, which ignores conformational dependence, is meaningful but does not fully describe the total isotope shift to within the precision of the measurements. Random coil 2H isotope shifts provide an important starting point for analysis of such shifts in structural terms in folded proteins, where they are known to depend strongly on local geometry.
Keywords: chemical shift, IDP, IUP, protein NMR, 2H, random coil, triple resonance NMR
Introduction
Intrinsically disordered proteins (IDPs) are particularly common and important in eukaryotic systems (Dyson and Wright 2005; Uversky and Dunker 2010; Wang et al. 2011), but their lack of a defined structure generally prevents their study by X-ray crystallography. Consequently, solution NMR moved to the fore as the primary method for site-specific studies of residual structural propensities of these proteins. Indeed, one of the most commonly considered features of IDPs is their secondary structure propensity, which is commonly derived from a comparison of the chemical shifts of the backbone nuclei to their literature “random coil" values (Marsh et al. 2006; Camilloni et al. 2012; Ozenne et al. 2012; Rezaei-Ghaleh et al. 2012). These reference random coil chemical shifts typically have been measured on fully protonated unstructured peptides (Richarz and Wuthrich 1978; Wishart et al. 1995; Schwarzinger et al. 2001; Kjaergaard et al. 2011), or have been derived from chemically denatured (Peti et al. 2001) or intrinsically disordered proteins (De Simone et al. 2009; Tamiola et al. 2010).
The intrinsic absence of stable secondary structure in IDPs gives rise to very poor chemical shift dispersion. Moreover, IDPs also tend to have low sequence complexity, high net charge, and low hydrophobicity (Romero et al. 2001), which further contributes to spectral congestion. The problem of spectral overlap can be alleviated by using a high field NMR spectrometer and by perdeuteration of the non-exchangeable hydrogens. Indeed, protein perdeuteration is highly beneficial for the vast majority of common NMR experiments performed on IDPs, as it improves relaxation properties, in particular for experiments utilizing TROSY-type detection schemes. Perdeuteration also removes amide proton splittings caused by 3JHN-Hα couplings, thereby further reducing spectral overlap (Maltsev et al. 2012a).
Due to the anharmonicity of zero-point C-H bond length vibrations when a hydrogen is replaced by deuterium, the average 13C-2H bond length is slightly shorter than the corresponding 13C-1H bond, thereby increasing the chemical shielding of the 13C nucleus (Jameson 1996. Because of the large fractional change in mass upon substituting 2H for 1H, the resulting one-bond isotope shift effect, 1 13C(D), often is quite substantial, and typically falls in the 0.2–0.5 ppm range. The 2H isotope shift effect extends also to other nearby nuclei, and effects of deuteration on chemical shifts can readily be seen for 1H, 13C and 15N nuclei that are up to three bonds removed from the site of substitution. Taking these isotope shift effects into account becomes particularly important when recording spectra on both protonated and deuterated forms of the same protein. Isotope shifts also give rise to unresolved splittings when fractional deuteration is used, effectively lowering attainable resolution (Nietlispach et al. 1996; Gardner et al. 1997). Taking the effects of isotope shifts into account is important when predicting protein backbone torsion angles from observed chemical shifts, using empirically optimized programs such as TALOS, PREDITOR, and TALOS+ (Cornilescu et al. 1999)(Neal et al. 2006)(Shen et al. 2009) or when identifying elements of secondary structure from chemical shifts (Wishart et al. 1991)(Gronenborn and Clore 1994)(Wishart and Sykes 1994)(Shen and Bax 2010)(Wang and Jardetzky 2002)(Eghbalnia et al. 2005)(Camilloni et al. 2012). Similarly, accounting for 2H isotope shifts is important when modeling entire protein structures on the basis of chemical shifts, through programs such as CHESHIRE, CS-Rosetta, and CS23D (Cavalli et al. 2007)(Shen et al. 2008)(Wishart et al. 2008).
When interpreting chemical shifts in structural terms, such as in the applications listed above, any 2H isotope shifts typically are either removed or accounted for by using literature 2H isotope shift values derived for the enzyme carbonic anhydrase II rich in -sheet (Venters et al. 1996). However, 1 13C(D) isotope shifts depend strongly on local structure and can vary by as much as 30%, depending on adjacent torsion angles (Lemaster 1994); Lemaster et al. 1994). Similarly, 1 15N(D) isotope shifts are quite sensitive to both local backbone torsion angles and H-bonding (Abildgaard et al. 2009).
Particularly for IDPs, where secondary structure propensities tend to be low and deviations from random coil chemical shifts are correspondingly small (Eliezer et al. 2001)(Tamiola et al. 2010; Kjaergaard et al. 2011; Camilloni et al. 2012), accurate knowledge of isotope shifts is critical when aiming to derive such structural propensities quantitatively from chemical shift data collected on perdeuterated proteins.
Here, we report highly precise, residue-specific chemical shift differences between fully protonated and perdeuterated -synuclein (aS), an IDP with chemical shifts that fall closer to random coil values than any other fully assigned IDP (Maltsev et al. 2012b). Therefore, aS is an excellent mimic for a disordered chain, despite a small, ca 20% decrease in hydrodynamic radius compared to what is expected for an ideal random coil, an effect resulting from mild electrostatic compaction (Bertoncini et al. 2005). Our measurements reflect the sum of both one-bond and longer range isotope shifts. Due to the presence of 17 different amino acid types in aS, and nearly complete chemical shift assignments for all backbone atoms, it is possible to decompose these values into their intra- and interresidue contributions (for 1HN, 15N, and 13C') and one-bond 1 13C(D), two-bond 2 13C(D), and three-bond 3 13C(D) components (for 13C and 13C). Not unexpectedly, such decompositions show a dependence on residue type, i.e. on local chemistry, and highlights the potential wealth of structural information contained in a quantitative analysis of accurately measured individual isotope shifts.
Next to the 13C and 13C nuclei, we also report total 2H isotope shift values for backbone 15N, 1HN, and 13C' nuclei. These values, lacking one-bond contributions, are much smaller than seen for 13C and 13C, and can even change sign, i.e., result in a small downfield change in chemical shift. The intraresidue and sequential contributions to the 2H isotope shifts of the peptide bond nuclei were separated by using a uniformly perdeuterated sample with incorporation of protonated Lys residues, highlighting a downfield 2H isotope shift associated with deuteration of the preceding 13C nucleus.
The values reported in this study are for a dynamically highly disordered protein. In folded proteins, substantial deviations from these values will occur that are a function of local torsion angles. Our values therefore serve as a baseline for random coil behavior, much like random coil chemical shifts and random coil J couplings, and deviations from these values may prove useful as reporters or restraints on local geometry once sufficiently quantitative relations have been developed.
Materials and methods
Protein expression and purification
Uniformly 13C/15N-enriched aS, either fully protonated or fully perdeuterated, was expressed and purified as described previously (Bodner et al. 2009). Briefly, bacteria were grown in M9 medium at 37 °C to an OD600 of 0.6, and protein production was induced by adding 1 mM IPTG. Cells were harvested by centrifugation 3 hours later and immediately frozen at −80°C. Lysis of cells and initial protein purification were achieved by several freeze-thaw cycles followed by heat precipitation (15 min at 85°C) in 50 mM Tris pH 7.4, 500 mM NaCl. The solution was then cleared by centrifugation, and the supernatant was diluted 10-fold with 50 mM Tris pH 7.4 buffer. Anion-exchange chromatography on a Q-sepharose column was performed, and fractions containing aS were pooled together. Finally, size-exclusion chromatography was performed on a Superdex 75 HiLoad 16/60 prep grade column in 20 mM Tris pH 7.4, 50 mM NaCl. The protein was then dialyzed into water and lyophilized for storage. Protonated aS was produced in H2O-based M9 medium supplemented with 1 g/L of 1H, 13C, 15N Isogro (Sigma, St. Louis, MO). Perdeuterated aS was produced in D2O (99%), using M9 medium with 3 g/L of 2H/13C glucose as the main carbon source, supplemented with 1 g/L of 2H, 13C, 15N Isogro. Based on 1D 1H NMR, the extent of deuteration in the resulting protein was estimated to be >97%. Site-specifically labeled aS (1H/15N – Lys in 2H/13C/15N background) was prepared by growing bacteria in D2O-based 13C/15N/2H M9 medium supplemented with MEM vitamins, adding 100 mg/L of 1H/15N – Lys one hour prior to induction, and then proceeding with the regular purification procedure described above.
NMR experiments
To exclude any contributions from imprecise referencing, or small temperature or ionic strength differences, NMR experiments were performed on a mixed sample containing 150 M protonated aS and 150 M perdeuterated aS. The use of a mixed sample also eliminates any possible errors arising from imprecise pH matching, which can be significant in particular for His, Asp, and Glu residues (Kjaergaard et al. 2011). The sample was prepared in 20 mM phosphate buffer, pH 6, 0.02% sodium azide, 7% D2O, and spectra were recorded at 15 °C.
Amide 15N and 1HN differences in chemical shift for the fully protonated and perdeuterated aS molecules were measured from a highly resolved TROSY-HSQC spectrum recorded on a 900 MHz Bruker Avance III spectrometer equipped with a TCI cryogenic probe. The spectrum comprised 512*×2048* complex points for acquisition times of 246 and 206 ms in the 15N (t1) and 1H (t2) dimensions, respectively. For peak picking of the protonated aS resonances, complications resulting from sometimes poorly resolved 3JHNH splittings in the 1H (t2) dimension of the spectrum were avoided by truncating the t2 time domain to 81 ms, followed by apodization with a squared 90°-shifted sine bell window function. For peak picking of the perdeuterated components of the spectrum, the full 1H time domain was used, again with squared 90°-shifted sine bell apodization. The indirect 15N dimension was apodized with a 90°-shifted sine bell window function, truncated at 176.4°. With a recycle delay of 1s, using 8 scans per FID, the total data collection time was approximately 3.2 hours.
Deuterium isotope shifts for 13C' were derived from a TROSY-HNCO spectrum recorded on a 600 MHz Bruker Avance II spectrometer equipped with a TCI cryogenic probe. The spectrum was recorded with a mixed-time 15N evolution period (Ying et al. 2007), and regular time incrementation in the 13C' dimension, using random non-uniform sampling in both indirect dimensions, for a total sparsity of 25%. The full data matrix, including non-sampled points, corresponded to 175*×300*×900* complex points for acquisition times of 200, 225, and 120 ms in the 13C' (t1), 15N (t2), and 1H (t3) dimensions, respectively. With a recycle delay of 1.2 s and 2 scans per FID, the total recording time was approximately 44 hours. The spectrum was reconstructed in approximately 20 minutes using the IST method (Hyberts et al. 2012), using two 12-core computers (dual Xeon processors with 6 cores per CPU). A 90°-shifted sine bell window function, truncated at 176.4° was used for the two indirect dimensions, and a squared 90°-shifted sine bell window function was applied in the 1H dimension.
Total 2H isotope shifts for 13Cα and 13Cβ were measured from a constant-time TROSY-HNCACB spectrum recorded on a 600 MHz Bruker Avance II spectrometer equipped with a TCI cryogenic probe. The spectrum again was recorded with a mixed-time 15N evolution period (Ying et al. 2007), and constant-time evolution in the 13C dimension, using random non-uniform sampling in both indirect dimensions, for a total sparsity of 40%. The full data matrix comprised 212*×180*×900* complex points for acquisition times of 24.6, 135, and 120 ms in the 13C (t1), 15N (t2), and 1H (t3) dimensions, respectively. With a recycle delay of 1.2 s and 2 scans per FID, the total recording time was approximately 49 h. The spectrum was reconstructed in approximately 15 min using the IST algorithm of Hyberts et al. (2012), using a 64-core computer (4 AMD opteron processors with 16 cores per CPU). A 90°-shifted sine bell window truncated at 176.4 degree was applied for the two indirect dimensions, and a squared 90°-shifted sine bell window function was applied in the 1H dimension. All standard data processing was performed using the NMRPipe program (Delaglio et al. 1995), and peak picking was done with Sparky (Goddard and Kneller).
TROSY-HSQC and TROSY-HNCO spectra were collected on a 340 μM aS sample selectively labeled with 1H/15N – Lys in a 2H/13C/15N background. The experimental and processing parameters were the same as described above for the mixed sample.
Results and discussion
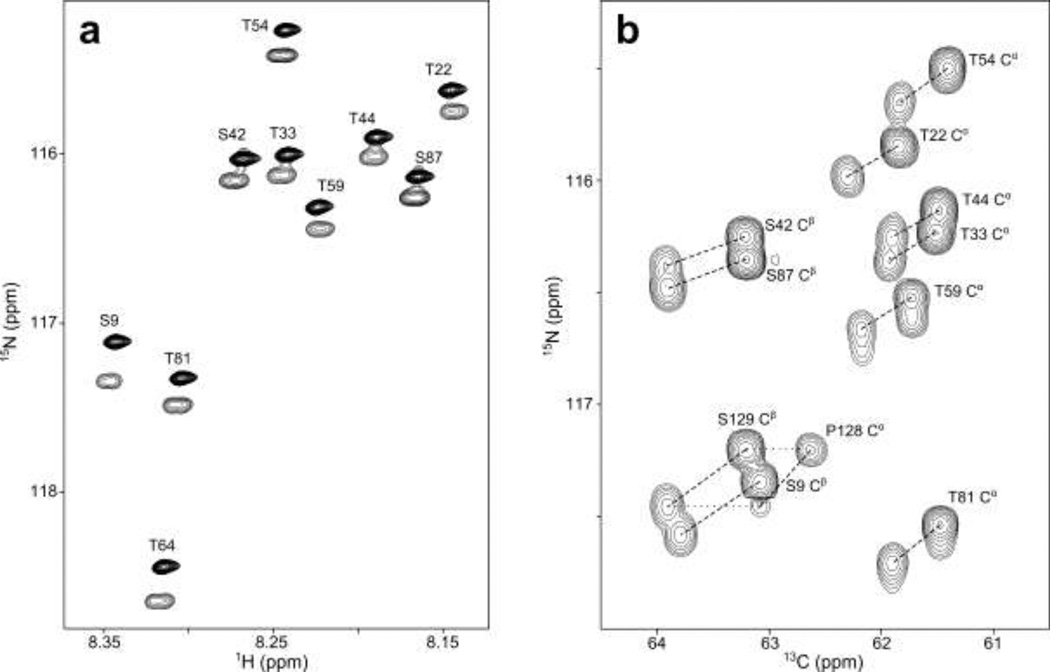
Accurate measurements of the isotope shifts were achieved by using a mixed sample of protonated and perdeuterated protein and by collecting 3D spectra at very high resolution. The latter was accomplished without requiring excessive measuring times by recording spectra with only two scans per FID, and using non-uniform, sparse sampling of the indirect time domains (Orekhov et al. 2003; Rovnyak et al. 2004)(Kazimierczuk et al. 2010)(Hyberts et al. 2012). However, to limit any possible increase in peak position uncertainty associated with sparse sampling, only a moderate 60–75% savings in measurement time compared to full sampling was employed. The resulting spectra are indeed of very high quality (Figure 1), and are nearly devoid of resonance overlap, despite the fact that the mixed sample effectively corresponds to 280 residues of an intrinsically disordered protein. In this respect, it is worth noting that the intrinsically disordered nature of aS results in favorable relaxation properties and very narrow line widths when its long transverse relaxation times are exploited by the use of non-uniform sampling, thereby allowing measurement of 2H isotope shifts for the 13Cα, 13Cβ, 13C', 15N, and 1HN nuclei at unprecedented precision.
Figure 1.
Spectra acquired on a mixture of protonated and perdeuterated aS. (a) Expanded region of the 900 MHz TROSY-HSQC spectrum. The upfield components in the 15N dimension of each pair of resonances lack the partially resolved 3JHH splitting seen on the downfield components and correspond to the perdeuterated form of the protein. With the exception of H(D) of T22 and T59, all 2H isotope shifts observed in this spectrum are negative. (b) Expanded region of a projection on the 13C/15N plane of the 600 MHz constant-time NUS TROSY-HNCACB spectrum. Dashed lines connect peaks originating from perdeuterated and protonated aS. The P128 Cα peaks are observed via residue S129. P128 and S129 peaks are connected by horizontal dotted lines. Upfield shifted components of each pair correspond to the perdeuterated form of the protein.
2H isotope shift for 13Cα and 13Cβ
The 2H isotope shifts for 13Cα and 13Cβ are particularly important considering the critical role these nuclei play when deriving structural information from secondary chemical shifts. To a very good approximation, 2H isotope shifts resulting from different substitution sites are independent of one another and are therefore additive (Jameson 1996. Isotope shifts resulting from substitutions more than three bonds away are typically very small, and consequently have been neglected in our analysis. Average values and standard deviations of the measured total 2H isotope shifts for 13Cα and 13Cβ (Table 1) show little scatter within any given residue type, confirming that 13Cα and 13Cβ 2H isotope shift corrections to a very good approximation are not impacted by their neighboring residues. However, 2H isotope shifts observed for 13C in different residue types vary considerably, ranging from −390 ppb for Asn and Asp, to −510 ppb for Val. An even larger range is observed for 13C, spanning from −575 ppb for Thr to −1.1 ppm for Leu.
Table 1.
Average 2H isotope shifts for 13Cα and 13Cβ from a single hydrogen that is one bond (1ΔC(D)), two bonds (2ΔC(D)), or three bonds (3ΔC(D)) removed, and total 2H isotope shifts, TΔC (D) and TΔC (D), extracted from spectra of protonated and perdeuterated aS.a
1ΔC(D) = −283 ± 2 ppb
2ΔC(D) = −94 ± 1 ppb
3ΔC(D) = −30 ± 1 ppb
1ΔCgly(D) = −238 ± 3 ppb
| Residue type |
Residues utilized |
TΔCα(D), ppb | TΔCβ(D), ppb | (d1, d2, d3) for Cα | (d1, d2, d3) for Cβ |
|---|---|---|---|---|---|
| Asn | 3 | −387 ± 9 | −617 ± 10 | (1, 2, 0) | (2, 1, 0) |
| Asp | 6 | −387 ± 5 | −660 ± 11 | ||
| Ser | 4 | −446 ± 9 | −706 ± 3 | ||
| His | 1 | −453 | −674 | ||
| Phe | 2 | −430 ± 6 | −846 ± 11 | ||
| Tyr | 4 | −435 ± 7 | −857 ± 7 | ||
| Gln | 6 | −480 ± 10 | −858 ± 19 | (1, 2, 2) | (2, 3, 0) |
| Glu | 15 | −489 ± 11 | −880 ± 6 | ||
| Met | 3 | −443 ± 20 | −885 ± 16 | ||
| Lys | 14 | −461 ± 15 | −1033 ± 10 | (1, 2, 2) | (2, 3, 2) |
| Pro | 5 | −447 ± 8 | −912 ± 12 | (1, 2, 4) | |
| Ala | 19 | −473 ± 12 | −876 ± 7 | (1, 3, 0) | (3, 1, 0) |
| Val | 19 | −507 ± 16 | −964 ± 10 | (1, 1, 6) | (1, 7, 0) |
| Leu | 4 | −448 ± 7 | −1104 ± 9 | (1, 2, 1) | (2, 2, 6) |
| Ile | 2 | −470 ± 9 | −1019 ± 0 | (1, 1, 5) | (1, 6, 3) |
| Thr | 10 | −431 ± 12 | −574 ± 6 | (1, 1, 3) | (1, 4, 0) |
| Gly | 17 | −475 ± 6 | ― | ― | ― |
No experimental values were obtained for Arg, Cys, and Trp residues, but values for Arg are expected to be similar to those listed for Lys, values for Cys may be approximated by Ser, and values for Trp by those of Phe and Tyr.
Following Venters et al. (1996), we also decomposed the total 2H isotope shift, TΔC(D), into its individual one-bond, two-bond, and three-bond components. This decomposition was carried out using a home-written Matlab script to solve sets of linear equations of the type:
| (1) |
where TΔC(D) denotes the total observed 2H isotope shift for 13C or 13C in a given amino acid type, nΔC(D) is the n-bond isotope shift contribution per deuteron, and dn is the number of deuterons n bonds away from the given carbon nucleus (see Table 1). Note that, by convention, the sign of 2H isotope shifts is defined by the difference between the deuterated and the protonated species. The three best-fit nΔC(D) values were derived by solving this set of linear equations, which relate the experimental total isotope shifts observed for the 13Cα and 13Cβ nuclei of all residue types (except for Gly) to the corresponding sets of dn values. In this analysis, some residue types are expected to have the same 2H isotope shifts, considering their identical dn patterns (Table 1). However, when examining the experimental values observed for the largest such group, which includes Asp, Asn, Ser, His, Phe, and Tyr, it is already clear that the measured deuteration corrections differ far beyond their experimental uncertainties, indicating that a simple decomposition according to eq 1 only provides a rough estimate for the total isotope shift.
Solving the set of equations for the isotope shift values observed for aS yields 1ΔC(D) = −283 ppb, 2ΔC(D) = −94 ppb, and 3ΔC(D) = −30 ppb, values that differ substantially from those previously reported by Venters et al.(1996) (−290 ppb, −130 ppb, −70 ppb for 1ΔC(D), 2ΔC(D), and 3ΔC(D), respectively), with ours being considerably smaller in magnitude, by as much as 57% for 3ΔC(D). The reason for these differences may be related to the different types of protein studied: an IDP in our present study, and the folded carbonic anhydrase II enzyme rich in -sheet by Venters et al. The magnitude of at least the 1ΔC(D) isotope shift is known to vary considerably with structure (Lemaster et al. 1994) and substantial variations in 2H isotope shift for 13C also are known to correlate with secondary structure (Garrett et al. 1997). In this respect, it is interesting to note that the 2H isotope shift for 13C in Pro is more than 120 ppb smaller than for Lys, despite having the same numbers of hydrogens at positions one, two and three bonds away. Clearly, the isotope shift also is impacted by the restricted conformation of the Pro ring system versus the fully disordered conformation of the Lys sidechain. When excluding methyl-containing residues, for most residues in synuclein the total 2H isotope shift observed falls well within the rather large ranges observed by Gardner et al. (1997) for the corresponding type of residue in their perdeuterated, methyl-protonated SH2 domain. Presumably the substantial spread in isotope shifts observed in the folded SH2 domain reflects the impact of local structure on the magnitude of the isotope effect.
2H isotope shift for 1HN
2H isotope effects on 1HN are small, ranging from −5 to +15 ppb, but readily can be identified in Figure 1a from an E.COSY-like effect (Griesinger et al. 1986), with the much larger 2H isotope shift on 15N separating the cross peaks for the protonated and deuterated forms of the protein. We aim to separate the isotope shift into a contribution from the preceding residue, Ai−1 and an intraresidue contribution, Bi:
| (2) |
Accurate values for ΔHi(D) were measured from the TROSY-HSQC spectrum for 120 out of the 133 residues with HSQC correlations (140 minus 5 Pro residues and the two N-terminal residues). With 17 different residue types, and a sequential and intraresidue parameter for each type (except for Pro), the solution to this set of 120 equations is far less overdetermined than it was for finding the three nΔC(D) values above. In fact, the matrix form of eq 2 is singular, and an arbitrary constant could be added to all Ai−1 values and subtracted from Bi without affecting the calculated ΔHi(D) values.
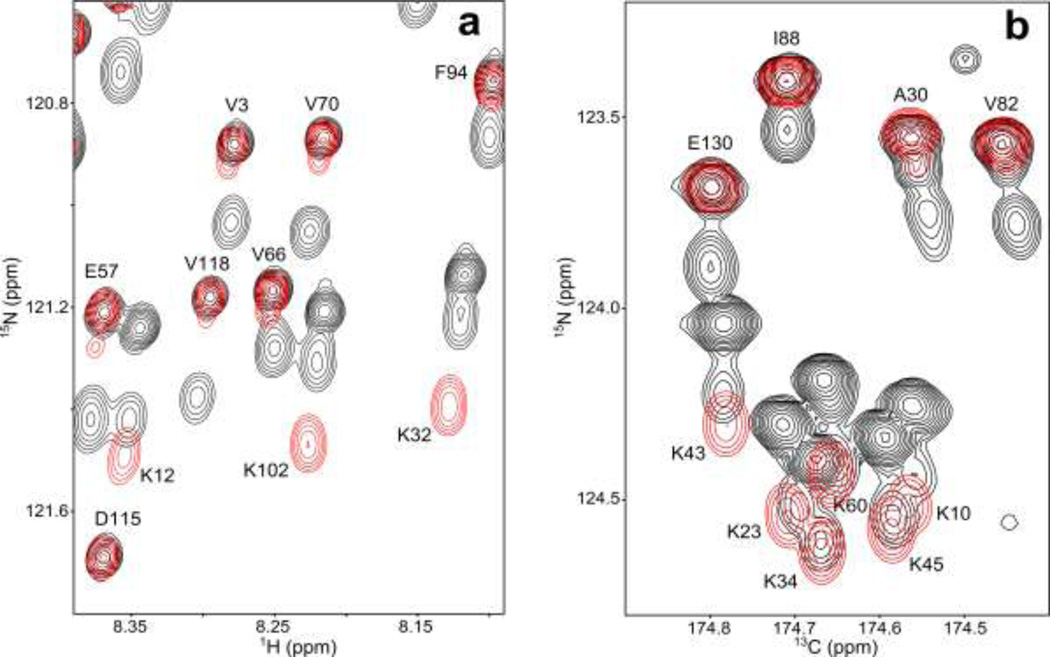
In order to resolve the above ambiguity regarding the intra- and inter-residue contributions to the observed ΔHi(D) isotope shift, we generated a uniformly 2H/13C/15N-enriched aS sample, except for Lys residues, which were 1H/15N-labeled. The effect of this selective labeling on the TROSY-HSQC spectrum is shown in Figure 2a, which presents the superposition of this 2D spectrum (red contours) on that of the mixed sample. For all residues except Lys and those following Lys, the resonance positions in the 1H/15N-Lys containing sample coincide with those in the perdeuterated protein. Correlations for Lys residues are displaced from their positions in perdeuterated aS by a nearly uniform amount. Since the protonated Lys residues lack the intraresidue 2H isotope shift, Bi, whereas the spectrum of the perdeuterated sample includes this component, the observed displacement corresponds to Bi of Lys. There are 15 Lys residues in aS and analysis of the full spectrum of Figure 2A yields an intra-residue 2H isotope effect on 1HN of Bi = −10 ± 1 ppb. This result therefore also yields the sequential 2H isotope shift contribution, Ai 1, for residues preceding Lys, thereby lifting the above noted degeneracy when solving eq 2. Independently, values for Ai-1 can be obtained by considering the peak positions of residues that are preceded by Lys, but fall outside the spectral region shown in Figure 2a. The extracted residue-specific values are listed in Table 2. Remarkably, the observation that the total isotope shift ΔHi(D) for many residues is smaller in magnitude than the intraresidue contribution, Bi, immediately indicates that the sequential isotope shift is mostly of the opposite sign, i.e., positive. The same applies for the sequential isotope shift for 15N (see below).
Figure 2.
Overlays of two spectra acquired on 1H/15N-Lys, perdeuterated aS (red) and on a mixture of protonated and perdeuterated aS (black). (a) Expanded region of the 900 MHz TROSY-HSQC spectrum, where the spectrum of the mixed sample presents a different region of the same spectrum shown in Figure 1a. Small downfield shoulders on the red correlations observed for Val residues originate from incomplete perdeuteration of their sidechains. Resonances for Lys residues in the 1H/15N-Lys, perdeuterated aS (red) spectrum are more downfield than those of the fully protonated component of the mixed sample spectrum (black), indicating that perdeuteration of the residue preceding Lys causes a downfield 2H isotope shift for both 15N and 1HN. In the 1H/15N-Lys, perdeuterated aS spectrum the Lys peaks are displaced from their positions in perdeuterated aS by a nearly uniform amount. However, the displacement of Lys peaks from their positions in fully protonated aS is very different for K12 compared to K32 and K102. This results from the fact that K12 is preceded by Ala, while K32 and K102 are preceded by Gly, and the neighboring-residue contribution of Gly on 15N is about double that of Ala. (b) Expanded region of a projection of the constant-time 600 MHz NUS TROSY-HNCO spectrum on the 13C/15N plane. Resonances are labeled by the observed amide pair following the 13C'. The small, mostly downfield change in chemical shift relative to the broad component in the mixed sample (black) corresponds to the intraresidue 2H isotope shift effect on the 13C' of the residue preceding Lys.
Table 2.
Contributions of the preceding and intraresidue type to the total 2H isotope shift observed for 1HN in perdeuterated proteins, as described by the coefficients of Eq 2.a
| Residue type |
Ai−1, ppb | Bi, ppb |
|---|---|---|
| Ala | 2 ± 1 | −9 ± 1 |
| Asp | 9 ± 2 | −8 ± 2 |
| Glu | 6 ± 1 | −11 ± 1 |
| Phe | 3 ± 2 | −7 ± 2 |
| Gly | 7 ± 1 | −15 ± 1 |
| His | 4 ± 2 | −8 ± 2 |
| Ile | 0 ± 2 | −8 ± 2 |
| Lys | 3 ± 1 | −10 ± 1 |
| Leu | 4 ± 2 | −8 ± 2 |
| Met | 1 ± 2 | −8 ± 2 |
| Asn | 6 ± 2 | −9 ± 2 |
| Pro | 0 ± 2 | ― |
| Gln | 4 ± 1 | −9 ± 2 |
| Ser | 2 ± 2 | −9 ± 2 |
| Thr | 3 ± 1 | −3 ± 1 |
| Val | −2 ± 1 | −8 ± 1 |
| Tyr | 7 ± 2 | −5 ± 2 |
Uncertainties are based on Monte-Carlo simulations assuming 2 ppb variability of measured isotope shifts (see Text).
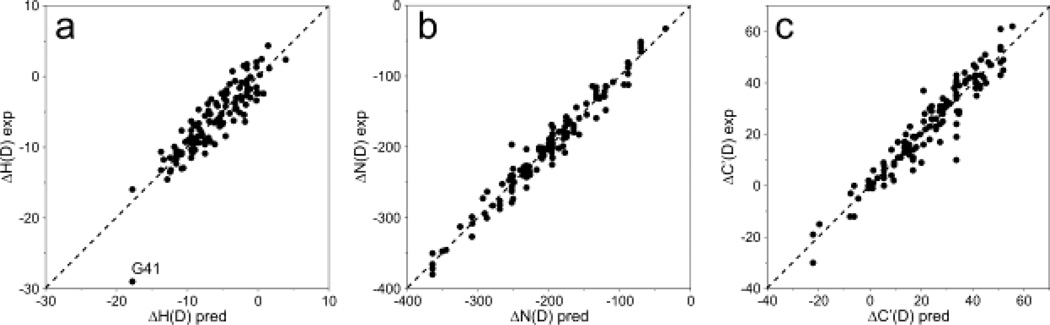
The system of equations (2) was solved using a home-written Matlab script. The measured value of Lys Bi was incorporated into the system as one additional equation. In order to establish uncertainties on the obtained Ai−1 and Bi solutions, a Monte-Carlo analysis was performed by adding normally distributed noise to the experimental values of 2H isotope shifts. The amplitude of the added noise was 2 ppb, matching the rmsd between the experimental and predicted values of 2H isotope shifts (Figure 3a). The noise in this analysis does not correspond to measurement uncertainties but rather reflects on the larger contributions to 2H isotope shifts from factors not accounted for by our simple analysis. The value of Lys Bi was also varied in the Monte-Carlo analysis, but with a standard deviation of 1 ppb, corresponding to its measurement uncertainty. The Monte-Carlo analysis involved solving the system of equations 5000 times, after which the average values and standard deviations of all Ai−1 and Bi solutions were calculated.
Figure 3.
Correlation plots of the total deuterium isotope shifts for (A) 1HN, (B) 15N, and (C) 13C’. Experimental values (vertical axis) are plotted against values predicted by using eqs 2–4 with the coefficients of Tables 2–4. Pairwise root mean sqare differences between measured (exp) and predicted (pred) H(D), N(D), and C'(D) isotope shifts are 2, 14, and 5 ppb, respectively.
The isotope shift contributions of Table 2 show trends that roughly correlate with amino acid type. For example, a preceding bulky hydrophobic residue such as Val, Ile, and Pro yields a nearly zero Ai−1 contribution to ΔHi(D), whereas a preceding small polar residue such as Asp or Asn yields the largest sequential isotope shifts (ca +7 ppb) effect. The intraresidue effect of perdeuteration on the 1HN chemical shift is largest for Gly (−15 ppb), which has two 1H contributors that are separated by three bonds from 1HN, and becomes smallest (−3 ppb) for Thr. Even within identical pairs of amino acids, however, some variation can be seen. For example, T22, T33, T44 T59 and T81 are all preceded by a Lys residue, but nevertheless significant variation in the 2H isotope shift for the 1HN resonances of these Thr residues is observed in Figure 1a. The origin of this variation, even in a highly disordered structure such as aS, remains not understood.
2H isotope shift for 15N
As can be seen in Figure 1a, the total 2H isotope effect on 15N chemical shifts is substantial and gives rise to well-resolved resonances in the TROSY-HSQC spectrum. Interestingly, whereas the T22, T33 and T44 residues (all preceded by Lys) show comparable ΔNi(D) shifts, T64 preceded by Val shows a much larger ΔNi(D). Clearly, the effect from deuteration of the preceding residue must be substantial. Again, the total isotope shifts can be decomposed into a contribution from the preceding residue, Ci−1, and an intraresidue component, Di:
| (3) |
This system of equations is underdetermined in the same way as eq 2, discussed above. Again, to resolve this problem we measured the intraresidue contribution for Lys residues from the spectra shown in Figure 2a, yielding Di = −253 ± 8 ppb.
With a total of 120 experimental ΔNi(D) values available, and 33 independent variables to be extracted for 17 residues (no intraresidue Pro 15N shifts were measured), best-fit solutions resulted in the values listed in Table 3. The uncertainties in the solutions were established using the same Monte-Carlo procedure described above for 1HN, with the root-mean-square amplitude of the added Gaussian noise set to 14 ppb (Figure 3b). As can be seen from this Table, 2H isotope effects on 15N are relatively large and show considerable variation. Intra-residue effects are smallest for the -branched residues Thr, Val and Ile, and largest for Ala. Not unexpectedly, Gly residues which have two H atoms separated by three bonds from the sequential 15N induce the largest sequential 2H isotope shift on the 15N (164 ppb) and, together with sequential isotope shifts from most other residues, have a positive sign. Exceptions are Ile and Val, which yield small negative values (Table 3).
Table 3.
Contributions of the preceding and intraresidue type to the total 2H isotope shift observed for 15N in perdeuterated proteins, as described by the coefficients of Eq 3.a
| Residue type |
Ci−1, ppb | Di, ppb |
|---|---|---|
| Ala | 77 ± 10 | −330 ± 10 |
| Asp | 88 ± 12 | −290 ± 11 |
| Glu | 79 ± 10 | −273 ± 10 |
| Phe | 59 ± 15 | −252 ± 14 |
| Gly | 164 ± 10 | −274 ± 10 |
| His | 77 ± 17 | −311 ± 17 |
| Ile | −21 ± 14 | −200 ± 14 |
| Lys | 63 ± 11 | −253 ± 8 |
| Leu | 45 ± 12 | −258 ± 13 |
| Met | 73 ± 16 | −259 ± 13 |
| Asn | 96 ± 13 | −295 ± 13 |
| Pro | 36 ± 12 | ― |
| Gln | 73 ± 11 | −263 ± 15 |
| Ser | 71 ± 12 | −286 ± 12 |
| Thr | 39 ± 10 | −197 ± 11 |
| Val | −35 ± 10 | −235 ± 10 |
| Tyr | 87 ± 13 | −253 ± 12 |
Uncertainties are based on Monte-Carlo simulations assuming 14 ppb variability of measured isotope shifts (see Text).
2H isotope shift for 13C'
Total 2H isotope shift values for 13C' are remarkably small and, remarkably, mostly in the downfield direction. Again, the total 2H isotope shift can be split into an intraresidue contribution, and a component resulting from the next residue, i+1:
| (4) |
This system of equations is underdetermined in the same way as discussed above for equations 2 and 3. The TROSY-HNCO spectrum of the 1H/15N-Lys, perdeuterated aS sample lacks correlations to Lys-13C'. However, the correlations to 13C' nuclei of residues preceding Lys are observed (Figure 2b). Comparison of these chemical shifts with corresponding shifts measured for fully deuterated aS yields the sequential residue contribution Fi+1 = 4 ± 4 ppb of Lys to the total 2H isotope effect on 13C'. Again, using this value leads to a fully determined system of equations, and allows residue-specific values for the Ei and Fi+1 terms to be determined (Table 4).
Table 4.
Contributions of the preceding and intraresidue type to the total 2H isotope shift observed for 13C' in perdeuterated proteins, as described by the coefficients of Eq. 4.a
| Residue type |
Ei, ppb | Fi+1, ppb |
|---|---|---|
| Ala | 18 ± 4 | 16 ± 4 |
| Asp | −14 ± 5 | 10 ± 5 |
| Glu | 11 ± 4 | 6 ± 4 |
| Phe | 6 ± 6 | 8 ± 5 |
| Gly | −3 ± 4 | 17 ± 4 |
| His | −3 ± 7 | 8 ± 6 |
| Ile | 39 ± 6 | 6 ± 5 |
| Lys | 27 ± 4 | 4 ± 4 |
| Leu | 29 ± 5 | 2 ± 5 |
| Met | 7 ± 6 | 10 ± 5 |
| Asn | −28 ± 5 | 7 ± 5 |
| Pro | 35 ± 5 | ― |
| Gln | 10 ± 4 | 7 ± 5 |
| Ser | −5 ± 5 | 12 ± 5 |
| Thr | 13 ± 4 | 6 ± 4 |
| Val | 35 ± 4 | 8 ± 4 |
| Tyr | 3 ± 5 | 8 ± 5 |
Uncertainties are based on Monte-Carlo simulations assuming 5 ppb variability of measured isotope shifts (see Text).
The uncertainties in the solutions were established using the same Monte-Carlo procedure described above for 1HN, with the root-mean-square amplitude of the added Gaussian noise set to 5 ppb (Figure 3c). As can be seen from Table 4, 2H isotope shifts measured for the 13C' of residue i originating from perdeuteration of residue i+1 are very small. By contrast, effects from intraresidue perdeuteration show a substantial range, from ca. +39 ppb for the hydrophobic residues Ile, Val, and Pro to negative values of −14 and −28 ppb for Asp and Asn, respectively. It is important to note that the values reported here apply for the highly disordered protein aS, and that effects in folded proteins will depend on local conformation. Indeed, considerably larger variations were observed for a perdeuterated/methyl-protonated form of an SH2 domain (Gardner et al. 1997). Also, it is worth noting that the eq 4 decomposition into an intra- and sequential residue contribution is not quite adequate to fully describe the isotope shift, even in a highly disordered protein such as aS. For example, there are five occurrences of the dipeptide KT in the protein, but with total ΔC'(D) ranging from +10 ppb for K58-T59 to +40 ppb for K21-T22 (Supplementary Material), with the measurement uncertainty of the 13C' chemical shifts being at most 3 ppb.
Concluding remarks
The highly favorable relaxation properties of aS resulted in intrinsically very narrow line widths and therefore in very high precision at which the chemical shift positions were measured. Using non-uniform sampling in a very conservative manner, sampling between 25–40% of the total number of indirect time domain data points, reconstructed 3D spectra were essentially free of artifacts and distortion of line shape or peak position. Indeed, comparison of the total 2H isotope shifts for 1H and 15N extracted from 900-MHz fully sampled 2D spectra and sparsely sampled 3D spectra, collected at 600 MHz, showed a pairwise rmsd of less than 2 ppb for 1H and 7 ppb for 15N.
-Synuclein contains 17 different amino acid types, and our study therefore has yielded a nearly complete set of residue-specific 2H isotope shifts. The three residues not represented in aS are Arg, Cys, and Trp. However, considering the close similarity of these residues to Lys, Ser, and Phe/Tyr, respectively, the backbone and 13C isotope shifts of these latter residues should be representative for these missing residues.
2H isotope shifts for 13C', 15N, and 1HN nuclei are much smaller than those for 13Cα and 13Cβ. Indeed, values for 13C' and 1HN previously were considered too small for adequate characterization (Gardner et al. 1997; Garrett et al. 1997), while corrections for 15N were differentiated only by their intra-residue type (Gardner et al. 1997). Using very high resolution spectra recorded on an IDP, we have shown that 2H isotope shifts for 13C', 15N, and 1HN also are impacted by the nearest neighboring residue. This conclusion is also easily validated by the fact that aS contains multiple pseudo-repeats, meaning that many pairs of consecutive amino acids are repeated multiple times in the sequence. Examples of such pairs include: EG (7 occurrences), GV (6 occurrences), KT (5 occurrences), AA (5 occurrences), and VA (5 occurrences). As shown in Supplementary Table 1, isotope shifts for such repeated pairs agree much closer with each other than when simply considering the intra-residue type alone, yielding root-mean-square deviations from their average values of ~10, ~10, and ~2 ppb for 13C', 15N, and 1HN, respectively, when considering amino acid pairs, versus ~11, ~55, and ~4 ppb when ignoring the adjacent amino acid type. Including the effect of the nearest neighbor through eqs 2–4 yields a good fit (Figure 3) between observed isotope shifts and values predicted using these equations with coefficients of Tables 2–4. Indeed the match between the experimentally measured deuteration corrections and those predicted by the equations then becomes 5, 14, and 2.1 ppb, for 13C', 15N, and 1HN, respectively, comparable to values seen in the pseudo repeats. Interestingly, however, these residual rmsd values remain considerably larger than their measurement uncertainties, in particular for 13C' and 15N. This observation highlights the dependence of the 2H isotope shift on more than just residue type, even in an IDP.
Accurate knowledge of 2H isotope shifts is important for at least three reasons: First, analysis of larger proteins frequently involves collection of spectra on both protonated and perdeuterated forms of such a protein and knowledge of the expected isotope shift is important when ensuring that resonance assignments are fully consistent between the different samples. Second, when analyzing chemical shifts in terms of structure, it is important to remove 2H isotope shift contributions. Although isotope shifts observed for 1HN, 15N, and 13C' are too small to impact such analyses, isotope shifts for 13C and 13C are rather large and need to be accounted for. Prior 2H isotope shifts reported by Venters et al. for human carbonic anhydrase II tend to be systematically larger in magntide than we found for intrinsically disordered aS, despite the higher level of deuteration (>97%) used in our study. Values reported for a perdeuterated SH2 domain (Gardner et al. 1997) agree closer with our aS values, although fractional methyl protonation of Ala, Val, Ile, and Leu in that study resulted in smaller magnitudes of the 2H isotope shifts observed for 13C and 13C. Third, and perhaps most important, 2H isotope shifts report on local structure (Lemaster et al. 1994; Abildgaard et al. 2009) and may provide useful experimental parameters to describe structural propensities in a manner that complements the use of secondary chemical shifts and 3JHH couplings for this purpose (Ottiger and Bax 1997). To further exploit 2H isotope shifts for this purpose, we are currently studying the dependence of such isotope shifts on local geometry, both by quantum chemical calculations, and by validating such calculations in a protein of accurately known structure.
Supplementary Material
ACKNOWLEDGMENT
We thank James L. Baber for experimental support. This work was funded by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH) and the Intramural AIDS-Targeted Antiviral Program of the Office of the Director, NIH.
References
- Abildgaard J, Hansen PE, Manalo MN, LiWang A. Deuterium isotope effects on N-15 backbone chemical shifts in proteins. J. Biomol. NMR. 2009;44:119–126. doi: 10.1007/s10858-009-9316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodner CR, Dobson CM, Bax A. Multiple Tight Phospholipid-Binding Modes of alpha-Synuclein Revealed by Solution NMR Spectroscopy. J. Mol. Biol. 2009;390:775–790. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilloni C, De Simone A, Vranken WF, Vendruscolo M. Determination of Secondary Structure Populations in Disordered States of Proteins Using Nuclear Magnetic Resonance Chemical Shifts. Biochemistry. 2012;51:2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc. Natl. Acad. Sci. U.S.A. 2007;>104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- De Simone A, Cavalli A, Hsu STD, Vranken W, Vendruscolo M. Accurate Random Coil Chemical Shifts from an Analysis of Loop Regions in Native States of Proteins. J. Am. Chem. Soc. 2009;131:16332–16333. doi: 10.1021/ja904937a. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe - a multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Eghbalnia HR, Wang LY, Bahrami A, Assadi A, Markley JL. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J. Biomol. NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- Eliezer D, Kutluay E, Bussell R, Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- Gardner KH, Rosen MK, Kay LE. Global folds of highly deuterated, methyl-protonated proteins by multidimensional NMR. Biochemistry. 1997;36:1389–1401. doi: 10.1021/bi9624806. [DOI] [PubMed] [Google Scholar]
- Garrett DS, Seok YJ, Liao DI, Peterkofsky A, Gronenborn AM, Clore GM. Solution structure of the 30 kDa N-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system by multidimensional NMR. Biochemistry. 1997;36:2517–2530. doi: 10.1021/bi962924y. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; 2008. [Google Scholar]
- Griesinger C, Sørensen OW, Ernst RR. Correlation of connected transitions by two-dimensional NMR spectroscopy. J. Chem. Phys. 1986;85:6837–6852. [Google Scholar]
- Gronenborn AM, Clore GM. Identification of N-terminal helix capping boxes by means of 13C chemical shifts. J. Biomol. NMR. 1994;4:455–458. doi: 10.1007/BF00179351. [DOI] [PubMed] [Google Scholar]
- Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson CJ. Isotope effects on chemical shifts & coupling constants. In: Grant DM, Harris RK, editors. Encyclopedia of Nuclear Magnetic Resonance. Vol. 4. New York: Wiley; 1996. pp. 2638–2655. [Google Scholar]
- Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W. Random sampling in multidimensional NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2010;57:420–434. doi: 10.1016/j.pnmrs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Kjaergaard M, Brander S, Poulsen FM. Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J. Biomol. NMR. 2011;49:139–149. doi: 10.1007/s10858-011-9472-x. [DOI] [PubMed] [Google Scholar]
- Lemaster DM. Isotope labeling in solution protein assignment and structural analysis. Prog. Nucl. Magn. Reson. Spectrosc. 1994;26:371–419. [Google Scholar]
- Lemaster DM, Laiuppa JC, Kushlan DM. Differential deuterium isotope shifts and one-bond 1H-13C scalar couplings in the conformational analysis of protein glycine residues. J. Biomol. NMR. 1994;4:863–870. doi: 10.1007/BF00398415. [DOI] [PubMed] [Google Scholar]
- Maltsev AS, Grishaev A, Bax A. Monomeric alpha-Synuclein Binds Congo Red Micelles in a Disordered Manner. Biochemistry. 2012a;51:631–642. doi: 10.1021/bi201435d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltsev AS, Ying JF, Bax A. Impact of N-Terminal Acetylation of α-Synuclein on Its Random Coil and Lipid Binding Properties. Biochemistry. 2012b;51:5004–5013. doi: 10.1021/bi300642h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia ZC, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal S, Berjanskii M, Zhang HY, Wishart DS. Accurate prediction of protein torsion angles using chemical shifts and sequence homology. Magn. Reson. Chem. 2006;44:S158–S167. doi: 10.1002/mrc.1832. [DOI] [PubMed] [Google Scholar]
- Nietlispach D, Clowes RT, Broadhurst RW, Ito Y, Keeler J, Kelly M, Ashurst J, Oschkinat H, Domaille PJ, Laue ED. An approach to the structure determination of larger proteins using triple resonance NMR experiments in conjunction with random fractional deuteration. J. Am. Chem. Soc. 1996;118:407–415. [Google Scholar]
- Orekhov VY, Ibraghimov I, Billeter M. Optimizing resolution in multidimensional NMR by three-way decomposition. J. Biomol. NMR. 2003;27:165–173. doi: 10.1023/a:1024944720653. [DOI] [PubMed] [Google Scholar]
- Ottiger M, Bax A. An empirical correlation between amide deuterium isotope effects on C-13(alpha) chemical shifts and protein backbone conformation. J. Am. Chem. Soc. 1997;119:8070–8075. [Google Scholar]
- Ozenne V, Bauer F, Salmon L, Huang J-r, Jensen MR, Segard S, Bernado P, Charavay C, Blackledge M. Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics. 2012;28:1463–1470. doi: 10.1093/bioinformatics/bts172. [DOI] [PubMed] [Google Scholar]
- Peti W, Smith LJ, Redfield C, Schwalbe H. Chemical shifts in denatured proteins: Resonance assignments for denatured ubiquitin and comparisons with other denatured proteins. J. Biomol. NMR. 2001;19:153–165. doi: 10.1023/a:1008307323283. [DOI] [PubMed] [Google Scholar]
- Rezaei-Ghaleh N, Blackledge M, Zweckstetter M. Intrinsically Disordered Proteins: From Sequence and Conformational Properties toward Drug Discovery. ChemBioChem. 2012;13:930–950. doi: 10.1002/cbic.201200093. [DOI] [PubMed] [Google Scholar]
- Richarz R, Wuthrich K. 13C NMR chemical shifts of common amino acid residues measured in aqueous solutions of linear tetrapeptides H-GLY-GLY-X-L-ALA-OH. Biopolymers. 1978;17:2133–2141. [Google Scholar]
- Romero P, Obradovic Z, Li XH, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins-Structure Function and Genetics. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Rovnyak D, Frueh DP, Sastry M, Sun ZYJ, Stern AS, Hoch JC, Wagner G. Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J. Magn. Reson. 2004;170:15–21. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J. Biomol. NMR. 2010;46:199–204. doi: 10.1007/s10858-009-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu GH, Eletsky A, Wu YB, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiola K, Acar B, Mulder FAA. Sequence-Specific Random Coil Chemical Shifts of Intrinsically Disordered Proteins. J. Am. Chem. Soc. 2010;132:18000–18003. doi: 10.1021/ja105656t. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Understanding protein non-folding. BBA-Proteins Proteomics. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters RA, Farmer BT, Fierke CA, Spicer LD. Characterizing the use of perdeuteration in NMR studies of large proteins C-13, N-15 and H-1 assignments of human carbonic anhydrase II. J. Mol. Biol. 1996;264:1101–1116. doi: 10.1006/jmbi.1996.0699. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fisher JC, Mathew R, Ou L, Otieno S, Sublet J, Xiao L, Chen J, Roussel MF, Kriwacki RW. Intrinsic disorder mediates the diverse regulatory functions of the Cdk inhibitor p21. Nat. Chem. Biol. 2011;7:214–221. doi: 10.1038/nchembio.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G. CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36:496–502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H-13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The C-13 chemical shift index - a simple method for the identification of protein secondary structure using C-13 chemical shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Ying JF, Chill JH, Louis JM, Bax A. Mixed-time parallel evolution in multiple quantum NMR experiments: sensitivity and resolution enhancement in heteronuclear NMR. J. Biomol. NMR. 2007;37:195–204. doi: 10.1007/s10858-006-9120-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.