Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that regulate expression of various target genes. miRNAs are expressed in a tissue-specific manner and play important roles in cell proliferation, apoptosis, and differentiation. Epigenetic alterations such as DNA methylation and histone modification are essential for chromatin remodeling and regulation of gene expression including miRNAs. The CCCTC-binding factor, CTCF, is known to bind insulators and exhibits an enhancer-blocking and barrier function, and more recently, it also contributes to the three-dimensional organization of the genome. CTCF can also serve as a barrier against the spread of DNA methylation and histone repressive marks over promoter regions of tumor suppressor genes. Recent studies have shown that CTCF is also involved in the regulation of miRNAs such as miR-125b1, miR-375, and the miR-290 cluster in cancer cells and stem cells. miR-125b1 is a candidate of tumor suppressor and is silenced in breast cancer cells. On the other hand, miR-375 may have oncogenic function and is overexpressed in breast cancer cells. CTCF is involved in the regulation of both miR-125b1 and miR-375, indicating that there are various patterns of CTCF-associated epigenetic regulation of miRNAs. CTCF may also play a key role in the pluripotency of cells through the regulation of miR-290 cluster. These observations suggest that CTCF-mediated regulation of miRNAs could be a novel approach for cancer therapy and regenerative medicine.
Keywords: microRNA, CTCF, cancer cell, embryonic stem cell, miR-125b1, miR-375, miR-290 cluster
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that regulate various target genes and play important roles in cell proliferation, apoptosis, and differentiation. One of the important mechanisms of miRNA expression is epigenetic alteration such as DNA methylation and histone modification. The CCCTC-binding factor, CTCF, is known to bind insulators and exhibits an enhancer-blocking and barrier function, and more recently, it also contributes to the three-dimensional organization of the genome. Although, there are a number of studies describing regulation of miRNA expression including epigenetic alterations, only a few studies have reported the association between miRNA expression and CTCF. In this report, we review recent studies regarding miRNAs and CTCF, and discuss about roles of CTCF in the regulation of miRNA expression.
miRNA
miRNAs are ~22 nucleotide (nt) non-coding RNAs that can post-transcriptionally downregulate the expression of various target genes. Currently, ~1500 human miRNAs have been identified in the human genome, and each miRNA potentially controls hundreds of target genes. In animals, miRNA genes are generally transcribed by RNA polymerase II (pol II) to form primary transcripts (pri-miRNAs). Pol II transcribed pri-miRNAs are capped with 7-methylguanosine and are polyadenylated. The nuclear RNase III enzyme Drosha and its co-factor DGCR8 process pri-miRNAs into ~60 nt precursor miRNAs (pre-miRNAs), which form an imperfect stem-loop structure. Pre-miRNAs are transported into the cytoplasm by exportin 5 and are subsequently cleaved by Dicer into mature miRNAs which are then loaded into the RNA-induced silencing complex (RISC). The miRNA/RISC complex downregulates specific gene products by translational repression via binding to partially complementary sequences in the 3′ untranslated regions of the target mRNAs or by directing mRNA degradation via binding to perfectly complementary sequences. miRNAs are expressed in a tissue-specific manner and play important roles in metabolism, proliferation, apoptosis, and differentiation. Moreover, recent studies have shown a link between aberrant expression of miRNAs and the development of cancer (Calin and Croce, 2007; Cho, 2007; Saito et al., 2009).
Epigenetic regulation of miRNA expression
Since miRNAs can have large-scale effects through regulation of a variety of genes during mammalian development and carcinogenesis, an understanding of the regulatory mechanisms controlling miRNA expression is important. There are several reports of transcription factors binding to the promoter regions of specific miRNA genes and activating the transcription of pri-miRNAs, resulting in increased expression of mature miRNAs. c-Myc binds to the regulatory region of the miR-17-92 cluster and increased expression of c-Myc leads to the activation of the miRNAs in the cluster (O'Donnell et al., 2005).
Epigenetic alterations such as DNA methylation and histone modification play critical roles in chromatin remodeling and regulation of gene expression in mammalian development and in human diseases. Many miRNAs are expressed in a tissue- and tumor-specific manner, implying that some miRNAs are subject to epigenetic control. We have shown that miR-127, which is embedded in a CpG island, is strongly induced by treatment with DNA methylation inhibitors and histone deacetylase inhibitors, indicating that some miRNA genes are controlled by epigenetic alterations in their promoter regions and can be activated by chromatin modifying drugs (Saito et al., 2006, 2009). Lujambio et al. (2007) compared miRNA expression profiling between the wild-type HCT116 colon cancer cell line and HCT116 after genetic disruption of both DNA methyltransferase (DNMT) 1 and DNMT3b (DKO cells). They found that 18 out of 320 miRNAs are significantly upregulated in DKO cells. In particular, miR-124a is silenced by its own CpG island hypermethylation in human tumors, but can be activated by inhibition of DNA methylation. They also demonstrated that the oncogene CDK6 is a target of miR-124a and that epigenetic silencing of miR-124a in cancer cells modulates CDK6 activity. It has been reported that miR-9-1 and miR-9-3 are potential tumor suppressor miRNAs and are inactivated by epigenetic mechanisms in human cancers (Lehmann et al., 2008; Lujambio et al., 2008). miR-34a was identified as a target of p53 and induces a G(1) cell cycle arrest, senescence and apoptosis (He et al., 2007; Tazawa et al., 2007). miR-34a expression is silenced in several types of cancer including pancreatic cancer due to aberrant CpG methylation of its promoter. Re-expression of miR-34a in a pancreatic carcinoma cell line induced senescence and cell cycle arrest at least in part by targeting CDK6, indicating that miR-34a represents a tumor suppressor gene which is inactivated by CpG methylation in pancreatic cancer (Lodygin et al., 2008). miR-34b and miR-34c are also reported to be silenced by aberrant CpG island methylation in colorectal cancer (Toyota et al., 2008). Thus, a number of miRNAs are under epigenetic control and disruption of DNA methylation patterns and histone modification in the promoter regions of miRNAs might be associated with cancer development (Esteller, 2011).
The CCCTC-binding factor, CTCF, is known to bind insulators and exhibits an enhancer-blocking function. CTCF can also serve as a barrier against the spread of DNA methylation and histone repressive marks over promoter regions of tumor suppressor genes (Recillas-Targa et al., 2011). CTCF is a highly conserved multifunctional zinc finger protein involved in transcriptional repression and activation, insulation, epigenetic events such as imprinting of the H19/IGF2 locus, and X-inactivation, and which binds preferentially to unmethylated DNA (Filippova, 2008; Phillips and Corces, 2009). Moreover, CTCF play important roles during carcinogenesis: epigenetic silencing of tumor suppressor genes such as p16 and Rb (De La Rosa-Velazquez et al., 2007; Witcher and Emerson, 2009), apoptosis of breast cancer cells (Docquier et al., 2005), and regulation of important tumor suppressor genes such as p53 (Recillas-Targa et al., 2011; Saldana-Meyer and Recillas-Targa, 2011). These findings suggest that CTCF may be involved in epigenetic regulation of non-coding RNAs including miRNAs as well as coding RNAs.
Disruption of CTCF binding at the miR-125b1 CpG island in human cancers
Recent studies have reported that expression of miR-125b is downregulated in various human cancers including glioblastoma, prostate cancer, ovarian cancer, and breast cancer (Scott et al., 2007; Zhang et al., 2011). In addition, miR-125b suppresses oncogenes such as EST1, ERBB2, ERBB3, and Bak1 as its targets, suggesting that miR-125b functions as a tumor suppressor. DNA hypermethylation at the CpG island of miR-125b was observed in cell lines and in tissue samples from patients with breast cancer (Zhang et al., 2011).
Soto-Reyes et al. (2012) investigated epigenetic alterations such as DNA methylation and histone modification, and association of CTCF at the locus of miR-125b1 in breast cancer cells. They found aberrant DNA methylation of the miR-125b1 CpG island and that disruption of CTCF binding correlated with incorporation of repressive histone modifications such as histone H3 lysine 9 (K9) trimethylation and histone H3 K27 trimethylation in cancer cells. In normal breast cells expressing miR-125b1, CTCF might prevent the recruitment of epigenetic silencing components, such as DNA methylation and repressive histone modifications, and also favors an open chromatin structure. In breast cancer cells, the loss of CTCF is associated with CpG island methylation and the gain of repressive histone modifications such as histone H3 K9 trimethylation and histone H3 K27 trimethylation. Disruption of CTCF binding at CpG island induces silencing of miR-125b1 expression (Figure 1). These findings suggest that CTCF plays an important role in the regulation of the tumor suppressor miR-125b1 in cooperation with DNA methylation and histone modification in breast cancer cells. A recent study has also demonstrated that miR-125b1 can be silenced by DNA methylation, which may lead to activation of the ETS1 proto-oncogene and a worse prognosis in breast cancer patients (Zhang et al., 2011). Reactivation of the tumor suppressor miR-125b1 by epigenetic therapy using DNA methylation inhibitors may have clinical promise for the treatment of breast cancer patients.
Figure 1.

Disruption of CTCF binding at the miR-125b1 CpG island in human cancers. In normal breast cells, CTCF might prevent the recruitment of epigenetic silencing components, such as DNA methylation and repressive histone modifications, and also favors an open chromatin structure. Meanwhile, in breast cancer cells, the loss of CTCF is associated with CpG island methylation and the gain of repressive histone modifications such as histone H3 K9 trimethylation and histone H3 K27 trimethylation. Open circle, unmethylated DNA; filled circle, methylated DNA; H3K9Me, histone H3 K9 methylation; H3K27Me, histone H3 K27 methylation.
Role of CTCF in the regulation of miR-375 expression in breast cancer cells
Breast cancer is the leading cause of cancer death in women worldwide. Estrogen receptor α (ER α) upregulation causes abnormal cell proliferation in approximately 70% of breast cancers (Shoker et al., 1999; Vargo-Gogola and Rosen, 2007). A recent study has reported that miR-375 is overexpressed in ERα-positive breast cancer cell lines and plays an important role in cell proliferation (de Souza Rocha Simonini et al., 2010). There are CpG islands in the upstream region of the miR-375 gene. DNA hypermethylation is observed in the CpG island of ERα-positive breast cancer cells showing high expression of miR-375, whereas DNA hypomethylation and histone H3 K9 dimethylation are observed in the CpG islands of ERα-negative breast cancer cells. CTCF binds to unmethylated DNA in the CpG islands of ERα-negative cells and induces silencing of miR-375 expression. These findings suggest that overexpression of miR-375 is caused by dissociation of CTCF from the CpG island of miR-375 gene via loss of epigenetic marks including local DNA hypomethylation and histone H3 K9 dimethylation (de Souza Rocha Simonini et al., 2010) (Figure 2). It has been shown that miR-375 suppresses Ras dexamethasone-induced 1 (RASD1) as its potential target, and RASD1 can suppress the growth of breast cancer cells and down-regulate ERα expression (Vaidyanathan et al., 2004; de Souza Rocha Simonini et al., 2010). Thus the modulation of ERα expression by miR-375 is achieved through the repression of RASD1. These observations provide a possibility that inhibition of miR-375 could be a novel clinical approach for the treatment of ERα-positive breast cancer.
Figure 2.
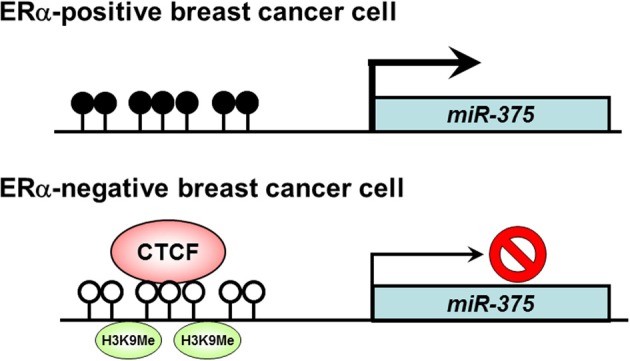
Role of CTCF in the regulation of miR-375 expression in breast cancer cells. In ERα-positive breast cancer cells, DNA hypermethylation is observed in the CpG island of the miR-375 gene, and miR-375 expression is activated. On the other hand, in ERα-negative breast cancer cells, DNA hypomethylation and histone H3 K9 dimethylation are observed, resulting in silencing of miR-375 expression by binding of CTCF in the CpG island. Open circle, unmethylated DNA; filled circle, methylated DNA; H3K9Me, histone H3 K9 methylation.
CTCF modulates expression of the early embryonic miRNA cluster
Human embryonic stem cells (ESCs) are derived from the inner cell mass of the human blastocyte and can be kept in an undifferentiated, self-renewing state indefinitely. ESCs have the advantage of being pluripotent, which endows them with the ability to differentiate into virtually every cell type in the human body. Thus, ESCs have gained popularity as a potentially ideal cell candidate for regenerative medicine. The early embryonic miRNA cluster (EEmiRC) has been identified in ESCs of mammals, and shows a remarkable cross-eutherian species conservation at the levels of both pre-miRNA hairpins and the core-promoter region (Houbaviy et al., 2003, 2005). EEmiRC encodes 7 miRNAs (miR-290, -291a, -292, -291b, -293, -294 and -295), which have been labeled as ESC-specific/pluripotency-associated miRNAs controlling cell-cycle progression, proliferation, and DNA methylation in undifferentiated/pluripotent cells. Therefore, understanding the biology of ESCs requires detailed knowledge of the mechanisms regulating EEmRC expression.
Little is known about the molecular mechanisms underlying the regulation of the EEmiRC expression. Recent studies have showed that the sequences upstream to the EEmiRC promoter contains active binding sites for Nanog, Oct3/4, Sox2, Tcf3, c-Myc, and 4n-Myc. Histone H3 K4 trimethylation and histone H3 K27 trimethylation were observed in ESCs and in differentiated cells, respectively (Chen et al., 2008; Judson et al., 2009). However, attempts to activate EEmiRC expression by ectopic expression of these individual transcriptional factors in fibroblasts were unsuccessful, suggesting that EEmiRC expression is under epigenetic control (Judson et al., 2009). Tata et al. (2011) identified a 332-bp intragenic enhancer (IE) region within the EEmiRC, which is able to modulate the transcription of the mouse EEmiRC locus. These miRNAs involve pluripotency factors and epigenetic mechanisms in pluripotent and differentiated cells. The results of chromatin immunoprecipitation (ChIP) assays demonstrated that the level of occupancy of Oct3/4, Sox2, and CTCF in this region gradually and dramatically decreased during ESC differentiation, suggesting a functional role for these transcription factors in regulating EEmiRC expression. This IE also contains a CpG island showing a differential pattern of DNA and histone methylation marks during differentiation of ESCs. Since, miR-290 cluster miRNAs have been shown to suppress Rbl2 as their target and Rbl2 modulates DNMTs (Benetti et al., 2008; Sinkkonen et al., 2008), EEmiRC may comprise a feedback loop with DNMTs. These findings indicate that this region plays a critical role in the regulation of EEmiRC expression, presumably through binding of transcription modulators such as Oct3/4, Sox2, and CTCF. Cohesin is a DNA-binding protein complex that is essential for sister chromatid cohesion and facilitates the repair of damaged DNA. Recent experiments have revealed that cohesin binds to the same sites in mammalian genomes as CTCF and cooperates with CTCF in regulating gene expression (Herold et al., 2012). Epigenetic effectors including CTCF and cohesin may modulate the pluripotency of cells through the regulation of miR-290 cluster.
Perspectives and conclusion
Table 1 shows a summary of the association between miRNAs and CTCF. These findings indicate that the insulator protein CTCF plays various roles in the regulation of miRNAs such as miR-125b1, miR-375, and the miR-290 cluster during mammalian development and carcinogenesis. miR-125b1 is a candidate of tumor suppressor and is silenced in breast cancer cells. On the other hand, miR-375 may have oncogenic function and is overexpressed in breast cancer cells. CTCF is involved in the regulation of both miR-125b1 and miR-375, indicating that there are various patterns of CTCF-associated epigenetic regulation of miRNAs. CTCF-mediated regulation of these miRNAs may provide a novel therapeutic approach for breast cancer. CTCF may also play a key role in the pluripotency of cells through the regulation of miR-290 cluster. Since, the link between miRNAs and CTCF has only just begun to be understood, other miRNA genes regulated by CTCF will be identified. Further studies are necessary to investigate whether CTCF-mediated regulation of miRNAs could be a novel approach for cancer therapy and regenerative medicine.
Table 1.
miRNAs associated with CTCF.
| miRNA | Expression | Target genes | Association with CTCF | References |
|---|---|---|---|---|
| miR-125b1 | Decreased in human cancers including glioblastoma, prostate cancer, ovarian cancer and breast cancer | EST1, ERBB2, ERBB3, Bak1 | In breast cancer cells, disruption of CTCF binding at the miR-125b1 CpG island correlated with DNA methylation and methylation of histone H3K9 and K27 induces silencing of miR-125b1 expression | Scott et al., 2007; Zhang et al., 2011; Soto-Reyes et al., 2012 |
| miR-375 | Overexpressed in ERα-positive breast cancer cells | RASD1 | In ERα-positive breast cancer cells, miR-375 overexpression was caused by dissociation of CTCF from the miR-375 promoter via loss of epigenetic marks including local DNA hypomethylation and histone H3 K9 methylation | de Souza Rocha Simonini et al., 2010 |
| miR-290 cluster | miR-290 cluster (miR-290, -291a, -292, -291b, -293, -294 and -295) have been identified as ESC-specific/pluripotency-associated miRNAs | Rbl2 | CTCF binds to intragenic enhancer region within the early embryonic miRNA cluster (EEmiRC) and modulates the expression of the EEmiRC | Houbaviy et al., 2003, 2005; Benetti et al., 2008; Chen et al., 2008; Sinkkonen et al., 2008; Judson et al., 2009; Tata et al., 2011 |
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Grant-in-Aid for Young Scientists A (23680090 to Yoshimasa Saito) and a Grant-in-Aid for Scientific Research C (24590993 to Hidetsugu Saito) from the Japan Society for the Promotion of Science (JSPS), Takeda Science Foundation (to Yoshimasa Saito), and Inaida Foundation (to Hidetsugu Saito).
Glossary
Abbreviations
- miRNA
microRNA
- RISC
RNA-induced silencing complex
- ERα
estrogen receptor α
- ESC
embryonic stem cell
- EEmiRC
early embryonic miRNA cluster
- IE
intragenic enhancer.
References
- Benetti R., Gonzalo S., Jaco I., Munoz P., Gonzalez S., Schoeftner S., Murchison E., Andl T., Chen T., Klatt P., Li E., Serrano M., Millar S., Hannon G., Blasco M. A. (2008). A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 15, 268–279 10.1038/nsmb.1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A., Croce C. M. (2007). Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. Clin. Invest. J. 117, 2059–2066 10.1172/JCI32577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 10.1016/j.cell.2008.04.043 [DOI] [PubMed] [Google Scholar]
- Cho W. C. (2007). OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer 6, 60 10.1186/1476-4598-6-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Rosa-Velazquez I. A., Rincon-Arano H., Benitez-Bribiesca L., Recillas-Targa F. (2007). Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTC. Cancer Res. 67, 2577–2585 10.1158/0008-5472.CAN-06-2024 [DOI] [PubMed] [Google Scholar]
- de Souza Rocha Simonini P., Breiling A., Gupta N., Malekpour M., Youns M., Omranipour R., Malekpour F., Volinia S., Croce C. M., Najmabadi H., Diederichs S., Sahin O., Mayer D., Lyko F., Hoheisel J. D., Riazalhosseini Y. (2010). Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 70, 9175–9184 10.1158/0008-5472.CAN-10-1318 [DOI] [PubMed] [Google Scholar]
- Docquier F., Farrar D., D'Arcy V., Chernukhin I., Robinson A. F., Loukinov D., Vatolin S., Pack S., Mackay A., Harris R. A., Dorricott H., O'Hare M. J., Lobanenkov V., Klenova E. (2005). Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res. 65, 5112–5122 10.1158/0008-5472.CAN-03-3498 [DOI] [PubMed] [Google Scholar]
- Esteller M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Filippova G. N. (2008). Genetics and epigenetics of the multifunctional protein CTC. Curr. Top. Dev. Biol. 80, 337–360 10.1016/S0070-2153(07)80009-3 [DOI] [PubMed] [Google Scholar]
- He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007). A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 10.1038/nature05939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold M., Bartkuhn M., Renkawitz R. (2012). CTCF: insights into insulator function during development. Development 139, 1045–1057 10.1242/dev.065268 [DOI] [PubMed] [Google Scholar]
- Houbaviy H. B., Dennis L., Jaenisch R., Sharp P. A. (2005). Characterization of a highly variable eutherian microRNA gene. RNA 11, 1245–1257 10.1261/rna.2890305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy H. B., Murray M. F., Sharp P. A. (2003). Embryonic stem cell-specific MicroRNAs. Dev. Cell 5, 351–358. 10.1016/S1534-5807(03)00227-2 [DOI] [PubMed] [Google Scholar]
- Judson R. L., Babiarz J. E., Venere M., Blelloch R. (2009). Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 27, 459–461 10.1038/nbt.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U., Hasemeier B., Christgen M., Muller M., Romermann D., Langer F., Kreipe H. (2008). Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. Pathol. J. 214, 17–24 10.1002/path.2251 [DOI] [PubMed] [Google Scholar]
- Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Korner H., Knyazev P., Diebold J., Hermeking H. (2008). Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7, 2591–2600 10.4161/cc.7.16.6533 [DOI] [PubMed] [Google Scholar]
- Lujambio A., Calin G. A., Villanueva A., Ropero S., Sanchez-Cespedes M., Blanco D., Montuenga L. M., Rossi S., Nicoloso M. S., Faller W. J., Gallagher W. M., Eccles S. A., Croce C. M., Esteller M. (2008). A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. U.S.A. 105, 13556–13561 10.1073/pnas.0803055105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A., Ropero S., Ballestar E., Fraga M. F., Cerrato C., Setien F., Casado S., Suarez-Gauthier A., Sanchez-Cespedes M., Git A., Spiteri I., Das P. P., Caldas C., Miska E., Esteller M. (2007). Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 67, 1424–1429 10.1158/0008-5472.CAN-06-4218 [DOI] [PubMed] [Google Scholar]
- O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- Phillips J. E., Corces V. G. (2009). CTCF: master weaver of the genome. Cell 137, 1194–1211 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F., de la Rosa-Velazquez I. A., Soto-Reyes E. (2011). Insulation of tumor suppressor genes by the nuclear factor CTC. Biochem. Cell Biol. 89, 479–488 10.1139/o11-031 [DOI] [PubMed] [Google Scholar]
- Saito Y., Liang G., Egger G., Friedman J. M., Chuang J. C., Coetzee G. A., Jones P. A. (2006). Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9, 435–443 10.1016/j.ccr.2006.04.020 [DOI] [PubMed] [Google Scholar]
- Saito Y., Suzuki H., Hibi T. (2009). The role of microRNAs in gastrointestinal cancers. Gastroenterol. J. 44Suppl. 19, 18–22 10.1007/s00535-008-2285-3 [DOI] [PubMed] [Google Scholar]
- Saito Y., Suzuki H., Tsugawa H., Nakagawa I., Matsuzaki J., Kanai Y., Hibi T. (2009). Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene 28, 2738–2744 10.1038/onc.2009.140 [DOI] [PubMed] [Google Scholar]
- Saldana-Meyer R., Recillas-Targa F. (2011). Transcriptional and epigenetic regulation of the p53 tumor suppressor gene. Epigenetics 6, 1068–1077 10.4161/epi.6.9.16683 [DOI] [PubMed] [Google Scholar]
- Scott G. K., Goga A., Bhaumik D., Berger C. E., Sullivan C. S., Benz C. C. (2007). Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. Biol. Chem. J. 282, 1479–1486 10.1074/jbc.M609383200 [DOI] [PubMed] [Google Scholar]
- Shoker B. S., Jarvis C., Clarke R. B., Anderson E., Hewlett J., Davies M. P., Sibson D. R., Sloane J. P. (1999). Estrogen receptor-positive proliferating cells in the normal and precancerous breast. Am. J. Pathol. 155, 1811–1815 10.1016/S0002-9440(10)65498-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L., Hugenschmidt T., Berninger P., Gaidatzis D., Mohn F., Artus-Revel C. G., Zavolan M., Svoboda P., Filipowicz W. (2008). MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 15, 259–267 10.1038/nsmb.1391 [DOI] [PubMed] [Google Scholar]
- Soto-Reyes E., Gonzalez-Barrios R., Cisneros-Soberanis F., Herrera-Goepfert R., Perez V., Cantu D., Prada D., Castro C., Recillas-Targa F., Herrera L. A. (2012). Disruption of CTCF at the miR-125b1 locus in gynecological cancers. BMC Cancer 12, 40 10.1186/1471-2407-12-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata P. R., Tata N. R., Kuhl M., Sirbu I. O. (2011). Identification of a novel epigenetic regulatory region within the pluripotency associated microcluster, RNA, EEmiRC. Nucleic Acids Res. 39, 3574–3581 10.1093/nar/gkq1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. (2007). Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 15472–15477 10.1073/pnas.0707351104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M., Suzuki H., Sasaki Y., Maruyama R., Imai K., Shinomura Y., Tokino T. (2008). Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 68, 4123–4132 10.1158/0008-5472.CAN-08-0325 [DOI] [PubMed] [Google Scholar]
- Vaidyanathan G., Cismowski M. J., Wang G., Vincent T. S., Brown K. D., Lanier S. M. (2004). The Ras-related protein AGS1/RASD1 suppresses cell growth. Oncogene 23, 5858–5863 10.1038/sj.onc.1207774 [DOI] [PubMed] [Google Scholar]
- Vargo-Gogola T., Rosen J. M. (2007). Modelling breast cancer: one size does not fit all. Nat. Rev. Cancer 7, 659–672 10.1038/nrc2193 [DOI] [PubMed] [Google Scholar]
- Witcher M., Emerson B. M. (2009). Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell 34, 271–284 10.1016/j.molcel.2009.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yan L. X., Wu Q. N., Du Z. M., Chen J., Liao D. Z., Huang M. Y., Hou J. H., Wu Q. L., Zeng M. S., Huang W. L., Zeng Y. X., Shao J. Y. (2011). miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 71, 3552–3562 10.1158/0008-5472.CAN-10-2435 [DOI] [PubMed] [Google Scholar]


