Abstract
In this study, the population structure, incidence, and potential sources of human infection caused by the d-tartrate-fermenting variant of Salmonella enterica serovar Paratyphi B [S. Paratyphi B (dT+)] was investigated. In Germany, the serovar is frequently isolated from broilers. Therefore, a selection of 108 epidemiologically unrelated S. enterica serovar Paratyphi B (dT+) strains isolated in Germany between 2002 and 2010 especially from humans, poultry/poultry meat, and reptiles was investigated by phenotypic and genotypic methods. Strains isolated from poultry and products thereof were strongly associated with multilocus sequence type ST28 and showed antimicrobial multiresistance profiles. Pulsed-field gel electrophoresis XbaI profiles were highly homogeneous, with only a few minor XbaI profile variants. All strains isolated from reptiles, except one, were strongly associated with ST88, another distantly related type. Most of the strains were susceptible to antimicrobial agents, and XbaI profiles were heterogeneous. Strains isolated from humans yielded seven sequence types (STs) clustering in three distantly related lineages. The first lineage, comprising five STs, represented mainly strains belonging to ST43 and ST149. The other two lineages were represented only by one ST each, ST28 and ST88. The relatedness of strains based on the pathogenicity gene repertoire (102 markers tested) was mostly in agreement with the multilocus sequence type. Because ST28 was frequently isolated from poultry but rarely in humans over the 9-year period investigated, overall, this study indicates that in Germany S. enterica serovar Paratyphi B (dT+) poses a health risk preferentially by contact with reptiles and, to a less extent, by exposure to poultry or poultry meat.
INTRODUCTION
Salmonellosis is still one of the major global causes of gastroenteritis in humans and animals. The underlying bacterium Salmonella enterica is divided into more than 2,570 serovars (16). The d-tartrate-fermenting variant (dT+) of Salmonella enterica subsp. enterica (referred to as S. enterica) serovar Paratyphi B, formerly called S. enterica serovar Java, is recognized as one important cause of gastroenteritis worldwide (6, 36). According to the White-Kauffmann-Le Minor scheme, the seroformula is 4,[5],12:b:1,2, optionally expressing or not expressing the O:5 antigen (16). Since the end of the 1990s, the serovar has become established in poultry, especially in Germany, Netherlands, and Belgium (12). It was shown that this variant is multidrug resistant, carrying a class 2 integron-associated dfrA1-sat1-aadA1 gene cassette conferring resistance to trimethoprim, streptomycin, and spectinomycin (27), and that it possesses a particular pathogenicity gene repertoire (21). Outbreaks caused by S. enterica serovar Paratyphi B (dT+) have been associated with goat's milk cheese in France (10), alfalfa sprouts or aquariums in Canada (15, 32), tropical fish aquariums in Australia (25), turtle exposure in the United States (17), and poultry in several European countries (27, 33).
Multilocus sequence typing (MLST) data indicated that S. enterica serovar Paratyphi B, similar to S. enterica serovar Newport, is polyphyletic (31). Altogether, 17 distinct sequence types (STs) have been previously described, clustering in three distantly related groups (1).
The aim of this study was to get a better understanding of the clonality and genetic relatedness of S. enterica serovar Paratyphi B (dT+) in Germany and to identify the potential sources of human infection caused by this serovar and its potential transmission by poultry meat to humans. Therefore, a representative collection of 108 S. enterica serovar Paratyphi B (dT+) strains isolated in Germany from humans, the environment, reptiles, and poultry/poultry meat during the years 2002 to 2010 was investigated using phenotypic and genotypic methods. Moreover, the pathogenicity and resistance gene repertoire of selected strains were determined.
MATERIALS AND METHODS
Selection of strains.
Between 2002 and 2010 the National Reference Laboratory for Salmonella (NRL-BfR) received for diagnostic serotyping 35,925 Salmonella strains, isolated by public or private diagnostic laboratories across Germany, from livestock, animals, food, feed, and the environment. Of these, 2.1% (751 strains) were assigned to S. enterica serovar Paratyphi B (dT+) (Table 1). Only 33 of these strains (4.4%) exhibited the O:5 antigen in addition to the 4,12 antigen. Similarly, between 2002 and 2010 the National Reference Centre for Salmonella and other Enterics (NRZ-RKI), Wernigerode, Germany, received 33,935 Salmonella strains isolated from humans with Salmonella infection in Germany. Of these, 211 strains (0.6%) were serotyped as S. enterica serovar Paratyphi B (dT+) (Table 1). Most of the S. enterica serovar Paratyphi B (dT+) strains (195 strains, 92%) expressed the O:5 antigen. Based on both collections, altogether 56 S. enterica serovar Paratyphi B (dT+) strains from humans and 52 from animals, food, and the environment were chosen for a deeper molecular typing procedure (Table 2). Within the selection, all 16 strains isolated from humans and not exhibiting the O:5 antigen were included. The remaining 40 strains (71%) isolated from humans and expressing the O:5 antigen were randomly selected. Twenty-seven of the 52 strains (52%) selected from animals, food, and the environment were O:5 antigen positive. They represent 79% of all O:5-antigen-expressing S. enterica serovar Paratyphi B (dT+) strains isolated between 2002 and 2010. The remaining 25 strains (48%) lacked the O:5 antigen and were selected with a focus on poultry and chicken meat. Two strains originated from cattle, and one strain each was from the environment (compost), ice cream, and fish.
Table 1.
Number and source of S. enterica serovar Paratyphi B (dT+) isolates in Germany received by the NRL-BfR and NRZ-RKI
| Yr of isolation | No. of isolates from: |
4,5,12:b:1,2 isolates |
4,12:b:1,2 isolates |
|||
|---|---|---|---|---|---|---|
| NRL | NRZ | Total no. | Composition of group (no. [source]) | Total no. | Composition of group (no. [source]) | |
| 2002 | 4,411 | 6,300 | 20 | 15 (human), 4 (reptile), 1 (other)a | 108 | 2 (human), 36 (poultry/meat), 1 (poultry/organ), 57 (poultry/feces), 2 (food), 3 (environment), 7 (other) |
| 2003 | 3,630 | 3,930 | 12 | 12 (human) | 79 | 4 (human), 26 (poultry/meat), 4 (poultry/organ), 26 (poultry/feces), 3 (food), 5 (environment), 11 (other) |
| 2004 | 3,604 | 3,691 | 18 | 17 (human), 1 (reptile) | 84 | 1 (human), 15 (poultry/meat), 3 (poultry/organ), 40 (poultry/feces), 1 (cattle), 7 (environment), 17 (other) |
| 2005 | 4,090 | 3,655 | 28 | 26 (human), 2 (reptile) | 56 | 2 (human), 26 (poultry/meat), 2 (poultry/organ), 21 (poultry/feces), 4 (food), 1 (other) |
| 2006 | 3,887 | 3,333 | 34 | 34 (human) | 76 | 42 (poultry/meat), 10 (poultry/organ), 5 (poultry/feces), 6 (food), 9 (environment), 4 (other) |
| 2007 | 3,955 | 3,855 | 21 | 16 (human), 2 (reptile), 3 (food) | 84 | 1 (human), 67 (poultry/meat), 8 (poultry/organ), 3 (food), 1 (environment), 4 (other) |
| 2008 | 3,606 | 3,205 | 19 | 17 (human), 2 (reptile) | 84 | 59 (poultry/meat), 9 (poultry/organ), 1 (poultry/feces), 1 (food), 2 (cattle), 5 (environment), 7 (other) |
| 2009 | 4,111 | 3,646 | 34 | 23 (human), 9 (reptile), 2 (food) | 73 | 6 (human), 34 (poultry/meat), 13 (poultry/organ), 3 (poultry/feces), 3 (food), 9 (environment), 5 (other) |
| 2010 | 4,631 | 2,320 | 42 | 35 (human), 7 (reptile) | 90 | 31 (poultry/meat), 28 (poultry/organ), 2 (poultry/feces), 11 (food), 2 (cattle), 9 (environment), 7 (other) |
| Total | 35,925 | 33,935 | 228 | 195 (human), 27 (reptile), 5 (food), 1 (other) | 734 | 16 (human), 336 (poultry/meat), 78 (poultry/organ), 155 (poultry/feces), 33 (food), 5 (cattle), 48 (environment), 63 (other) |
Other, rare isolates or source of isolation not specified.
Table 2.
S. enterica serovar Paratyphi B (dT+) isolates used for phenotypic and molecular analysis in this study
| Strain no. | Yr of isolation | Origin | Resistancea | PFGE cluster | PFGE profile no. | MLST | sop-ST | Microarray (PAT PT)b | O antigen |
|---|---|---|---|---|---|---|---|---|---|
| 02-04534 | 2002 | Fertilizer | Susceptible | B | 44 | 149 | 12-34-25 | 9 | 4,5,12 |
| 02-00729 | 2002 | Reptile, Mississippi turtle | Susceptible | B | 32 | 43 | 11-25-25 | 5 | 4,5,12 |
| 02-04564 | 2002 | Reptile, snake | Susceptible | A | 15 | 88 | NT | NT | 4,5,12 |
| 02-04565 | 2002 | Reptile, snake | Susceptible | A | 28 | 88 | NT | NT | 4,5,12 |
| 02-04626 | 2002 | Reptile, corn snake | TET | A | 19 | 88 | NT | NT | 4,5,12 |
| 04-01793 | 2004 | Reptile, snake | TET | A | 14 | 88 | 39-28-24 | 2 | 4,5,12 |
| 05-01473 | 2005 | Reptile, snake | STR TET TMP | A | 3 | 88 | 39-28-24 | 2 | 4,5,12 |
| 05-05074 | 2005 | Reptile, snake | Susceptible | A | 28 | 88 | NT | NT | 4,5,12 |
| 07-01916-1 | 2007 | Fish, pintado | Susceptible | B | 58 | 88 | 41-28-24 | 2 | 4,5,12 |
| 07-03272 | 2007 | Reptile, snake | Susceptible | A | 1 | 88 | NT | NT | 4,5,12 |
| 07-04003 | 2007 | Reptile, unkown | Susceptible | A | 20 | 88 | NT | NT | 4,5,12 |
| 08-01440 | 2008 | Reptile, turtle | Susceptible | B | 54 | 88 | NT | NT | 4,5,12 |
| 08-04475 | 2008 | Reptile, snake | NAL CIP | A | 16 | 88 | NT | NT | 4,5,12 |
| 09-01458 | 2009 | Reptile, snake | Susceptible | A | 12 | 88 | NT | NT | 4,5,12 |
| 09-02038 | 2009 | Reptile, snake | Susceptible | A | 2 | 88 | 39-28-24 | 4 | 4,5,12 |
| 09-02329 | 2009 | Reptile, snake | TET | A | 13 | 88 | NT | NT | 4,5,12 |
| 09-02447 | 2009 | Reptile, snake | Susceptible | A | 10 | 88 | NT | NT | 4,5,12 |
| 09-02452 | 2009 | Reptile, turtle | Susceptible | A | 10 | 88 | 39-28-24 | 2 | 4,5,12 |
| 09-02772 | 2009 | Reptile, snake | Susceptible | A | 26 | 88 | NT | NT | 4,5,12 |
| 09-03579 | 2009 | Reptile, snake | Susceptible | A | 11 | 88 | NT | NT | 4,5,12 |
| 09-03667 | 2009 | Chicken, meat | Susceptible | B | 47 | 149 | 12-34-25 | 9 | 4,5,12 |
| 09-04735 | 2009 | Reptile, snake | Susceptible | A | 17 | 88 | NT | NT | 4,5,12 |
| 10-00628 | 2010 | Reptile, unkown | Susceptible | A | 18 | 88 | NT | NT | 4,5,12 |
| 10-02147 | 2010 | Reptile,snake | Susceptible | A | 22 | 88 | 39-28-24 | 2 | 4,5,12 |
| 10-03633 | 2010 | Reptile,snake | Susceptible | A | 27 | 88 | 39-28-24 | 2 | 4,5,12 |
| 10-04378 | 2010 | Reptile, snake | Susceptible | A | 8 | 88 | 39-28-24 | 3 | 4,5,12 |
| 10-04870 | 2010 | Reptile, snake | Susceptible | A | 25 | 88 | NT | NT | 4,5,12 |
| 02-04177 | 2002 | Chicken, feces | CIP NAL STR TMP | C | 60 | 28 | 38-27-0 | 1 | 4,12 |
| 03-01517 | 2003 | Fish | STR TMP | C | 60 | 28 | NT | NT | 4,12 |
| 03-02942 | 2003 | Turkey | AMP STR SMX TET TMP | C | 60 | 28 | NT | NT | 4,12 |
| 04-03840 | 2004 | Cattle, organ | CIP KAN NAL STR SMX TET TMP | C | 60 | 28 | NT | NT | 4,12 |
| 05-01581 | 2005 | Compost | AMP CIP NAL STR SMX TMP | C | 60 | 28 | NT | NT | 4,12 |
| 06-02092 | 2006 | Ice cream | CIP NAL STR TMP | C | 60 | 28 | NT | NT | 4,12 |
| 06-05076 | 2006 | Chicken, meat | AMP CHL STR SMX TET TMP | C | 60 | 28 | NT | NT | 4,12 |
| 08-00451 | 2008 | Chicken, meat | AMP CIP NAL STR TMP | C | 60 | 28 | NT | NT | 4,12 |
| 08-01554 | 2008 | Chicken, meat | SMX STR TET TMP | C | 62 | 28 | NT | NT | 4,12 |
| 08-02848 | 2008 | Cattle, organ | CIP NAL TMP | C | 65 | 28 | 38-27-0 | 1 | 4,12 |
| 08-04157 | 2008 | Chicken, meat | AMP CIP GEN NAL SMX TMP | C | 68 | 28 | NT | NT | 4,12 |
| 08-04806 | 2008 | Chicken, meat | CIP NAL SMX TMP | C | 60 | 28 | NT | NT | 4,12 |
| 09-02946 | 2009 | Chicken, meat | AMP CHL CIP GEN KAN NAL SMX STR TET TMP | C | 73 | 28 | 38-27-0 | 1 | 4,12 |
| 09-03610 | 2009 | Chicken, meat | CHL CIP NAL SMX STR TET TMP | C | 69 | 28 | NT | NT | 4,12 |
| 09-04217 | 2009 | Chicken, meat | AMP FOT TAZ TMP | C | 60 | 28 | 38-27-0 | 1 | 4,12 |
| 10-00908 | 2010 | Turkey | CIP KAN NAL SMX TET TMP | C | 60 | 28 | NT | NT | 4,12 |
| 10-01119 | 2010 | Chicken, meat | AMP CHL CIP NAL SMX STR TET TMP | C | 66 | 28 | NT | NT | 4,12 |
| 10-01673 | 2010 | Chicken, meat | AMP CIP NAL SMX TET TMP | C | 60 | 28 | NT | NT | 4,12 |
| 10-01782 | 2010 | Chicken, meat | CIP NAL TMP | C | 70 | 28 | 38-27-0 | 1 | 4,12 |
| 10-03460 | 2010 | Cattle, organ | AMP CHL CIP NAL SMX TET TMP | C | 60 | 28 | NT | NT | 4,12 |
| 10-04808 | 2010 | Turkey | AMP CHL CIP GEN KAN NAL SMX STR TET TMP | C | 60 | 28 | 38-27-0 | 1 | 4,12 |
| 10-05176 | 2010 | Chicken | CIP NAL STR TMP | C | 71 | 28 | NT | NT | 4,12 |
| 11-01472 | 2002 | Human | AMP CHL FFN SMX STR TET | B | 34 | 43 | NT | NT | 4,5,12 |
| 11-01474 | 2003 | Human | STR TET | A | 5 | 88 | NT | NT | 4,5,12 |
| 11-01475 | 2003 | Human | SMX STR | B | 33 | 43 | NT | NT | 4,5,12 |
| 11-01476 | 2004 | Human | Susceptible | B | 45 | 149 | NT | NT | 4,5,12 |
| 11-01478 | 2004 | Human | Susceptible | B | 52 | 43 | NT | NT | 4,5,12 |
| 11-01479 | 2005 | Human | STR | B | 46 | 149 | NT | NT | 4,5,12 |
| 11-01480 | 2005 | Human | COL | B | 40 | 110 | NT | NT | 4,5,12 |
| 11-01481 | 2005 | Human | Susceptible | B | 50 | 43 | 11-25-25 | 5 | 4,5,12 |
| 11-01482 | 2006 | Human | Susceptible | B | 42 | 149 | NT | NT | 4,5,12 |
| 11-01483 | 2007 | Human | Susceptible | B | 58 | 88 | NT | NT | 4,5,12 |
| 11-01484 | 2007 | Human | Susceptible | A | 6 | 88 | 39-28-24 | 3 | 4,5,12 |
| 11-01485 | 2009 | Human | Susceptible | A | 23 | 88 | NT | NT | 4,5,12 |
| 11-01486 | 2009 | Human | Susceptible | B | 55 | 88 | 41-28-24 | 2 | 4,5,12 |
| 11-01487 | 2009 | Human | Susceptible | B | 59 | 88 | NT | NT | 4,5,12 |
| 11-01488 | 2009 | Human | Susceptible | B | 55 | 88 | NT | NT | 4,5,12 |
| 11-01489 | 2009 | Human | Susceptible | A | 24 | 88 | NT | NT | 4,5,12 |
| 11-01490 | 2009 | Human | Susceptible | A | 7 | 88 | NT | NT | 4,5,12 |
| 11-01491 | 2009 | Human | Susceptible | A | 7 | 88 | NT | NT | 4,5,12 |
| 11-01492 | 2009 | Human | AMP CHL FFN SMX STR TET | B | 34 | 43 | 11-25-25 | 6 | 4,5,12 |
| 11-01493 | 2009 | Human | Susceptible | B | 42 | 149 | NT | NT | 4,5,12 |
| 11-01494 | 2009 | Human | Susceptible | B | 42 | 149 | 12-34-25 | 10 | 4,5,12 |
| 11-01495 | 2009 | Human | Susceptible | B | 49 | 149 | NT | NT | 4,5,12 |
| 11-01496 | 2009 | Human | Susceptible | B | 31 | 896 | 12-24-25 | 8 | 4,5,12 |
| 11-01497 | 2009 | Human | Susceptible | B | 42 | 149 | NT | NT | 4,5,12 |
| 11-01498 | 2009 | Human | Susceptible | B | 42 | 149 | NT | NT | 4,5,12 |
| 11-01499 | 2009 | Human | Susceptible | B | 58 | 88 | NT | NT | 4,5,12 |
| 11-01500 | 2009 | Human | Susceptible | B | 38 | 43 | NT | NT | 4,5,12 |
| 11-01501 | 2009 | Human | AMP CHL FFN SMX STR TET | B | 34 | 43 | NT | NT | 4,5,12 |
| 11-01502 | 2009 | Human | Susceptible | B | 36 | 307 | 11-25-25 | 11 | 4,5,12 |
| 11-01503 | 2010 | Human | Susceptible | B | 51 | 43 | NT | NT | 4,5,12 |
| 11-01504 | 2010 | Human | Susceptible | B | 29 | 307 | 11-25-25 | 5 | 4,5,12 |
| 11-01505 | 2010 | Human | Susceptible | B | 41 | 43 | NT | NT | 4,5,12 |
| 11-01506 | 2010 | Human | Susceptible | B | 35 | 43 | NT | NT | 4,5,12 |
| 11-01507 | 2010 | Human | AMP SMX | B | 34 | 43 | 11-25-25 | 6 | 4,5,12 |
| 11-01508 | 2010 | Human | Susceptible | B | 37 | 43 | NT | NT | 4,5,12 |
| 11-01509 | 2010 | Human | Susceptible | B | 56 | 88 | NT | NT | 4,5,12 |
| 11-01510 | 2010 | Human | STR | B | 43 | 149 | NT | NT | 4,5,12 |
| 11-01511 | 2010 | Human | Susceptible | A | 9 | 88 | NT | NT | 4,5,12 |
| 11-01512 | 2010 | Human | Susceptible | A | 4 | 88 | 39-28-24 | 2 | 4,5,12 |
| 11-01513 | 2010 | Human | Susceptible | A | 4 | 88 | NT | NT | 4,5,12 |
| 11-01515 | 2002 | Human | AMP CIP NAL SMX STR TET TMP | C | 60 | 28 | NT | NT | 4,12 |
| 11-01516 | 2002 | Human | Susceptible | B | 30 | 110 | 12-24-25 | 8 | 4,12 (7-bp deletion) |
| 11-01517 | 2003 | Human | CIP NAL SMX STR TMP | C | 63 | 28 | NT | NT | 4,12 |
| 11-01518 | 2003 | Human | AMP CIP NAL SMX STR TMP | C | 60 | 28 | NT | NT | 4,12 |
| 11-01519 | 2003 | Human | STR TMP | C | 60 | 28 | NT | NT | 4,12 |
| 11-01520 | 2003 | Human | AMP CIP NAL SMX STR TMP | C | 60 | 28 | NT | NT | 4,12 |
| 11-01521 | 2004 | Human | AMP FOT STR TAZ TMP | C | 60 | 28 | NT | NT | 4,12 |
| 11-01522 | 2005 | Human | Susceptible | B | 48 | 149 | 46-34-25 | 9 | 4,12 (7-bp deletion) |
| 11-01523 | 2005 | Human | Susceptible | A | 21 | 88 | 39-28-24 | 2 | 4,12 (7-bp deletion) |
| 11-01524 | 2007 | Human | CHL CIP NAL SMX STR TET TMP | C | 67 | 28 | NT | NT | 4,12 |
| 11-01525 | 2009 | Human | AMP CHL CIP COL GEN KAN NAL SMX STR TET TMP | C | 72 | 28 | NT | NT | 4,12 |
| 11-01526 | 2009 | Human | AMP CIP FOT NAL SMX STR TAZ TET TMP | C | 64 | 28 | 38-27-0 | 1 | 4,12 |
| 11-01527 | 2009 | Human | Susceptible | B | 57 | 88 | 41-28-24 | 2 | 4,12 (7-bp deletion) |
| 11-01528 | 2009 | Human | Susceptible | B | 57 | 88 | NT | NT | 4,12 (7-bp deletion) |
| 11-01529 | 2009 | Human | STR | B | 58 | 88 | 41-28-24 | 2 | 4,12 (7-bp deletion) |
| 11-01530 | 2009 | Human | CIP NAL STR TMP | C | 61 | 28 | NT | NT | 4,12 |
| 11-01532 | 2006 | Food, spice | Susceptible | B | 53 | 43 | 11-25-25 | 5 | 4,5,12 |
| 11-01533 | 2008 | Sludge | AMP CIP SMX | B | 39 | 43 | 11-25-25 | 7 | 4,5,12 |
| 11-01534 | 2005 | Compost | STR TMP | C | 60 | 28 | NT | NT | 4,12 |
See Materials and Methods for abbreviations.
NT, not tested.
The strains selected have no epidemiological link (i.e., they were not isolated at the same place or time or from the same animals or foods). They cover various geographical origins and potential sources in Germany isolated between 2002 and 2010, with an emphasis on the years 2009 and 2010.
To study the pathogenicity gene repertoire using DNA microarrays and to compare sop gene sequence typing (sop-ST) (18) with MLST (23), a subset of 35 S. Paratyphi B (dT+) strains was further chosen to reflect the diversity of pulsed-field gel electrophoresis (PFGE) XbaI profiles within the set of 108 epidemiologically unrelated strains (Table 2).
Serotyping.
Serotyping was performed according to the White-Kauffmann-Le Minor scheme (16) by slide agglutination with O- and H-antigen-specific sera (Sifin Diagnostics, Berlin, Germany).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility of strains was tested against 14 antimicrobials or antimicrobial combinations by determining the MICs using the CLSI broth microdilution method (8) in combination with the semiautomatic Sensititre system (TREK Diagnostic Systems, Cleveland, OH). Cutoff values to be used to determine susceptibility to 10 antimicrobials were applied as described in the Commission Decision on a harmonized monitoring of antimicrobial resistance in poultry and pigs [(EG) 2007/407], as follows (mg/liter): cefotaxime (FOT), >0.5; nalidixic acid (NAL), >16; ciprofloxacin (CIP), >0.06; ampicillin (AMP), >4; tetracycline (TET) >8; chloramphenicol (CHL) >16; gentamicin (GEN) >2; streptomycin (STR) >32; trimethoprim (TMP) >2; and sulfamethoxazole (SMX) >256. Cutoff values for the remaining four antimicrobials were adopted from the European Committee on Antimicrobial Susceptibility Testing ([EUCAST] 2011 breakpoints [http://www.eucast.org/clinical_breakpoints/]), as follows (mg/liter): colistin (COL), >2; florfenicol (FFN), >16; kanamycin (KAN) >32; and ceftazidime (TAZ), >2.
Genomic DNA purification.
DNA for PCRs and DNA microarray experiments was isolated from strains grown in Luria-Bertani broth (Merck, Darmstadt, Germany) at 37°C for 16 to 18 h. A 1.6-ml aliquot was used for purification using an RTP Bacteria DNA Mini Kit (Stratec Molecular GmbH, Berlin, Germany) according to the manufacturer's protocol with one additional step. After the cell lysis step at 95°C for 5 to 10 min, 5 μl of 100 mg/ml RNase (Qiagen GmbH, Hilden, Germany) was added, and the sample was incubated at room temperature for 30 min. The quality and quantity determinations of DNA were performed spectrophotometrically. For DNA labeling with fluorophores, a minimum of 4 μg of DNA was used, and for PCRs 1-ng/μl DNA dilutions in Tris-EDTA (TE) buffer were used.
Multilocus sequence typing (MLST) and sop-ST typing.
MLST was carried out as previously described including partial sequences of the seven housekeeping genes: aroC, dnaN, hemD, hisD, purE, sucA, and thrA (23). Alleles and sequence types were assigned according to the MLST scheme (available at http://mlst.ucc.ie/mlst/dbs/Senterica). Unknown alleles were submitted to the website and newly named. The analysis was carried out in BioNumerics, version 6.5 (Applied Maths, Sint-Martens-Latem, Belgium). The comparisons were made by advanced cluster analysis using the analysis template maximum spanning tree (MST) for categorical data on merged sequences of the seven genes. The sop gene sequence typing (sop-ST) was carried out as described previously (18) including the genes sopA, sopB, and sopD. Laboratory-internal allele numbers were assigned to new alleles, resulting in a sop-ST allele pattern (sopA-sopB-sopD) for a given strain. Based on a multiple alignment of merged sop sequences, similarities were analyzed by the unweighted pair group method with arithmetic averages (UPGMA) in BioNumerics, version 6.5 (Applied Maths, Sint-Martens-Latem, Belgium).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed according to the Pulse-Net protocol (30) using the restriction enzyme XbaI for digestion of genomic DNA. The analyses of the gel images were carried out in BioNumerics, version 6.5. The comparisons were made by cluster analysis using the Dice coefficient and UPGMA with a position tolerance of 1.5% and optimization of 1.0%. Fragments that were smaller than 25 kb were not considered for cluster analysis.
DNA microarray analysis.
The DNA microarray used in this study was applied as previously described (20). Altogether, 80 pathogenic markers, 22 fimbrial gene markers, and 49 resistance gene markers were analyzed from 35 strains representing the diversity of PFGE profiles. Analysis of raw data was performed as previously described (20). After normalization (presence/absence of gene), the data for each strain were imported in BioNumerics, version 6.5, as character values. For comparison, a cluster analysis with the simple matching binary coefficient, using the UPGMA dendrogram type, was applied on the basis of the 80 pathogenicity and 22 fimbriae markers.
oafA and d-tartrate PCRs.
Presence of the oafA gene responsible for O:5 antigen expression in Salmonella was performed according to Hauser et al. (19) using primers P-439 and P-440 amplifying a 433-bp PCR product. To detect a 7-bp deletion within the open reading frame, primers P-439 and P-1072 were used, resulting in a PCR product of 170 bp. In case of the 7-bp deletion, no PCR product was obtained. The ability of S. enterica serovar Paratyphi B (dT+) strains to ferment d-tartrate was analyzed according to the PCR protocol described by Malorny et al. (26).
Statistical methods.
To assess the discrimination index of PFGE, Simpson's index of diversity (ID) and 95% confidence intervals (CI) were calculated using software at the Comparing Partitions website(http://darwin.phyloviz.net/ComparingPartitions/index.php?link = Tool)
Nucleotide sequence accession numbers.
New alleles detected by sop-ST were deposited in GenBank under the following accession numbers: JQ045575 to JQ045580 for sopA11, sopA12, sopA38, sopA39, sopA41, and sopA46; JQ067612 to JQ067615 for sopB28, sopB27, sopB25, and sopB24; JQ067610 for sopB34; JQ067622 for sopD24; and JQ067623 for sopD25.
RESULTS
O:5 antigen distribution in S. enterica serovar Paratyphi B (dT+).
A minority of S. enterica serovar Paratyphi B (dT+) strains collected between 2002 and 2010 at the National Reference Laboratory for Salmonella (NRL-BfR) (33 strains, 4.4%) expressed the O:5 antigen in addition to the 4,12 antigen (O:4,5,12). These were isolated mostly from reptiles (27 strains, 82%) and only sporadically from food (5 strains, 15%) and the environment (1 strain, 3%) (Table 1). In contrast, O:5-antigen-negative strains were isolated mainly from poultry and products thereof (569 strains, 79%) and sporadically from pigs, cattle, and diverse food products but not from reptiles. The National Reference Centre for Salmonella and other Enterics (NRZ-RKI), Wernigerode, Germany, received between 2002 and 2010 a total of 211 S. enterica serovar Paratyphi B (dT+) strains isolated from humans in Germany. Most of them (195 strains, 92%) expressed the O:5 antigen. It is obvious that data show a contrast with respect to expression of the O:5 antigen between strains isolated from humans and those isolated from livestock, especially poultry.
For the investigation of clonality and genetic relatedness, 108 epidemiologically unrelated S. enterica serovar Paratyphi B (dT+) strains from the collection of the NRL-BfR and NRZ-RKI were selected (Table 2) (for selection criteria, see Materials and Methods).
Antimicrobial resistance.
Fifty-nine of the 108 S. Paratyphi B (dT+) strains (55%) tested were susceptible to all 14 antimicrobials. Three strains were monoresistant to tetracycline, one strain was monoresistant to streptomycin, and one was monoresistant to colistin. Forty-two strains (39%) were multidrug resistant to two or more antimicrobials. All strains isolated from livestock or poultry meat were multidrug resistant to up to 10 antimicrobials (Table 2) and did not express the O:5 antigen. Strains isolated from humans and lacking the O:5 antigen were mainly multidrug resistant, but five strains were susceptible. A PCR screening of the presence of the oafA gene encoding the O:5 antigen revealed the absence of the gene in multidrug-resistant strains. Susceptible O:5-antigen-negative strains harbored the oafA gene, but a 7-bp deletion within the open reading frame obviously caused a frameshift leading to the loss of O:5 antigen expression, as previously described by Hauser et al. (19) for S. enterica serovar Typhimurium. Human strains expressing factor 5 were mainly susceptible. Three strains exhibited multidrug resistance to AMP, CHL, FFN, SMX, STR, and TET, indicating the existence of a Salmonella genomic island 1 (SGI1), a cluster of genes encoding multidrug resistance (35). Responsible genes for antimicrobial resistance were determined by DNA microarray. Poultry-associated multidrug-resistant strains harbored a class 2 integrase gene. In combination with dfrA1, sat1 (Tn7), and aadA1, this indicated that a class 2 integron was carried by these strains, as previously described by Miko et al. (28). Additionally, six strains had a class 1 integrase gene in combination with sul1. Multidrug-resistant strains expressing the O:5 antigen showed the typical resistance genes usually found in SGI1, namely, floR, aadA2, blaPSE1, tet(G), and sul1. Two strains isolated from humans and one strain isolated from chicken meat were resistant to extended-spectrum β-lactams TAZ and FOT. Responsible antimicrobial resistance gene families could be assigned by DNA microarray to blaCMY-2-like (09-04217, chicken), blaCTX-M2-like (11-01526, human), and blaTEM1-like (11-01521, human) genes.
Typing by PFGE.
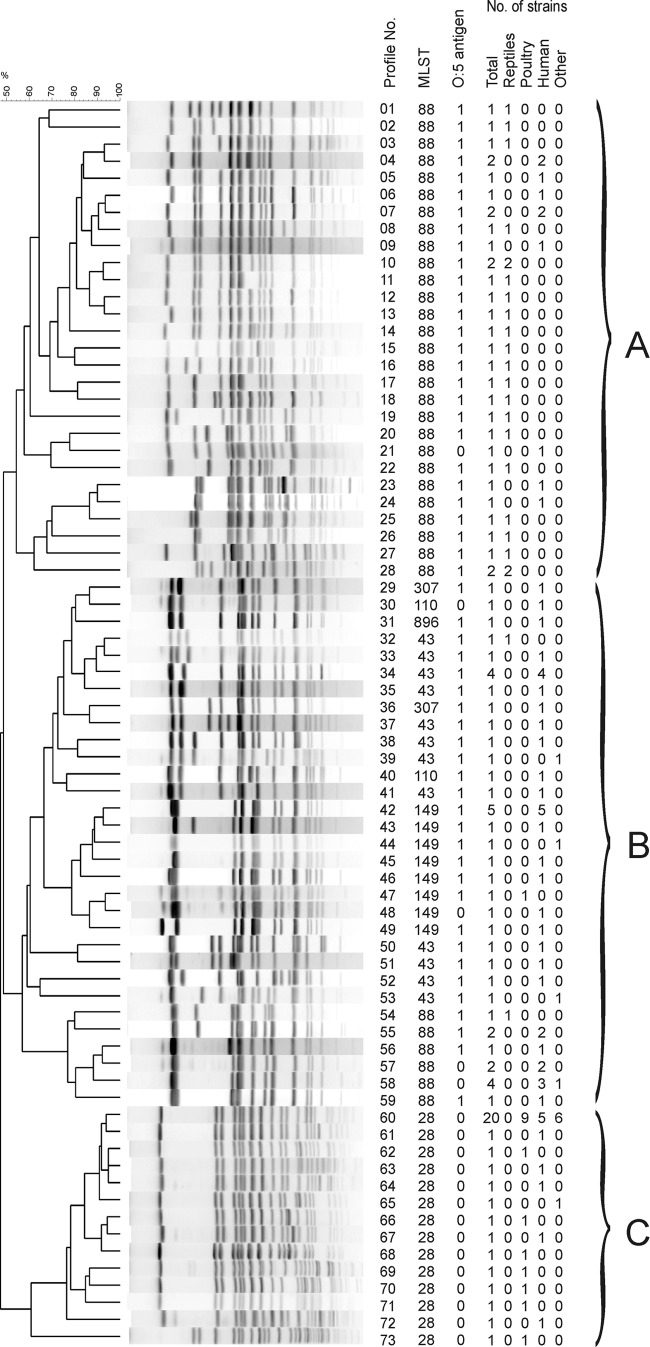
Seventy-three different XbaI profiles could be distinguished among the 108 strains analyzed (Fig. 1) (ID, 0.962 [95% CI, 0.937 to 0.988]). They were classified into three clusters (A, B, and C). Cluster A contained 32 strains of the 108 strains (30%), cluster B contained 43 strains (40%), and cluster C contained 33 strains (31%). XbaI profile number 60 harbored 20 strains of the 33 strains (61%) in cluster C and was previously described as an X8 profile (21, 28). These strains were isolated from poultry/poultry meat, other livestock, and humans but not from reptiles. All strains in cluster C lacked the oafA gene completely and therefore did not express the O:5 antigen. Strains isolated from reptiles clustered preferentially in cluster A. Human strains were distributed over all three clusters. The similarity coefficient (F value) ranged for cluster C from 0.6 to 0.95 and was slightly lower for clusters A (0.54 to 0.93) and B (0.56 to 0.93). The Simpson's index of discrimination (ID) was considerably higher for clusters A (ID, 0.992 [95% CI, 0.982 to 1.000]) and B (ID, 0.973 [95% CI, 0.952 to 0.995]) than for cluster C (ID, 0.640 [95% CI, 0.444 to 0.836]), indicating lower diversity within the poultry/poultry meat strains.
Fig 1.
UPGMA dendrogram of PFGE profiles identified in 108 S. enterica serovar Paratyphi B (dT+) strains after digestion with XbaI. Profiles were numbered serially from 1 to 73. The number of strains belonging to each source (total, reptile, poultry, human, and other), corresponding MLSTs, and present (1) or absent (0) O:5 antigen are shown on the right side. Assigned clusters A to C are indicated by curly brackets.
MLST.
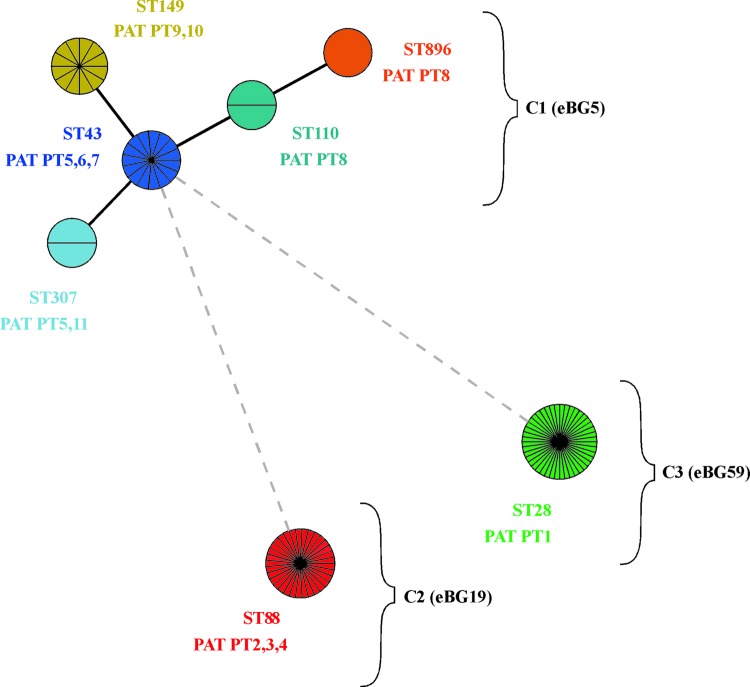
Seven different sequence types (STs) were identified, namely, ST28, -43, -88, -110, -149, -307, and -896 (ID, 0.722 [95% CI, 0.675 to 0.769]). Forty-three strains (39.8%) of the 108 strains investigated belonged to ST88, 33 strains (30.5%) belonged to ST28, 12 strains (11.1%) belonged to ST149, 15 strains (13.8%) belonged to ST43, and 2 strains (1.8%) belonged to ST307 and ST110 each. One single strain was assigned to ST896. STs were categorized by BioNumerics in three clonal complexes (Fig. 2). The founder of the largest complex (C1) was identified to be ST43.
Fig 2.
Minimal spanning tree of MLST data on 108 S. enterica serovar Paratyphi B (dT+) isolates. Each circle refers to one ST subdivided into one pie slice per strain. STs that share six identical gene alleles are linked by a black line, and STs sharing only one or no common gene allele are linked by a gray dotted line. Based on their similarity, STs were grouped in three complexes (C1, C2, and C3). According to the nomenclature of Achtman et al. (1), C1 is designated eBG5, C2 is eBG19, and C3 is eBG59. Pathogenicity array types (PATs) found in each ST are shown below ST designations.
ST43 differed from ST149 in one nucleotide in the purE allele and from ST307 in one nucleotide in the thrA allele. ST110 had 5 nucleotides different from ST43 in the dnaN allele. ST896 differed in two alleles from ST43 (dnaN, five nucleotides, and aroC, one nucleotide). The other two clonal complexes were represented by only one ST each, ST88 (complex C2) and ST28 (complex C3), with six and seven different alleles compared to ST43, respectively.
It was obvious that strains isolated from poultry/poultry meat were, with one exception, always associated with ST28. Of the 16 strains isolated from humans between 2002 and 2010 and not expressing the O:5 antigen, 10 strains were assigned to ST28, 4 were assigned to ST88, and 1 was assigned to ST110 and ST149 each. All human O:5-antigen-negative strains distinct from ST28 harbored a nonfunctional oafA gene due to the 7-bp deletion within the ORF, whereas the oafA gene in ST28 strains was completely absent (Fig. 1). ST88 was identified with one exception in strains isolated from reptiles (53%) and humans (44%). The exceptional strain was isolated from fish. Strains assigned to the first clonal complex originated mainly from humans (84%), and single strains were from reptile, sludge, chicken, fertilizer, and spice (Table 2).
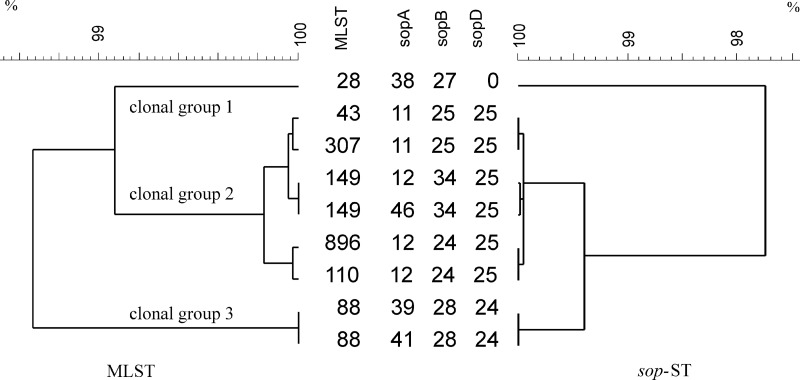
sop-ST.
For sequencing of the virulence genes sopA, sopB, and sopD, 35 strains were chosen representing the diversity of XbaI PFGE profiles. Among them, six different sopA alleles, five different sopB alleles, and two different sopD alleles resulting in seven different combinations of sopA, sopB, and sopD (sop-STs) were found (Fig. 3). Strains of the first clonal complex with the founder ST43 comprised four highly similar sop-STs. Strains with ST88 (second clonal complex) showed two different sop-STs (39-28-24 and 41-28-24) where the sopA allele 41 differed from allele 39 only in a 1-bp deletion at position 615 leading to a frameshift. All strains with sopA allele 41 clustered in one branch of the PFGE tree in cluster B. All ST28 strains (third clonal complex) were sopD negative and had identical sopA and sopB alleles (38-27-0). The sopA allele 11 had a major deletion of 47 base pairs, indicating a recombination event. However, the three major clonal complexes identified by MLST were in agreement with the clustering of sop-ST-based single nucleotide polymorphisms (SNPs) within all three genes (Fig. 3).
Fig 3.
UPGMA dendrograms of 35 S. enterica serovar Paratyphi B (dT+) strains based on the merged DNA sequences. On the left side, the MLST dendrogram with corresponding sequence types (STs) is shown, and on the right side the sop dendrogram with corresponding sop-ST pattern types (sopA-sopB-sopD) is shown. The scale indicates the similarity of merged DNA sequences.
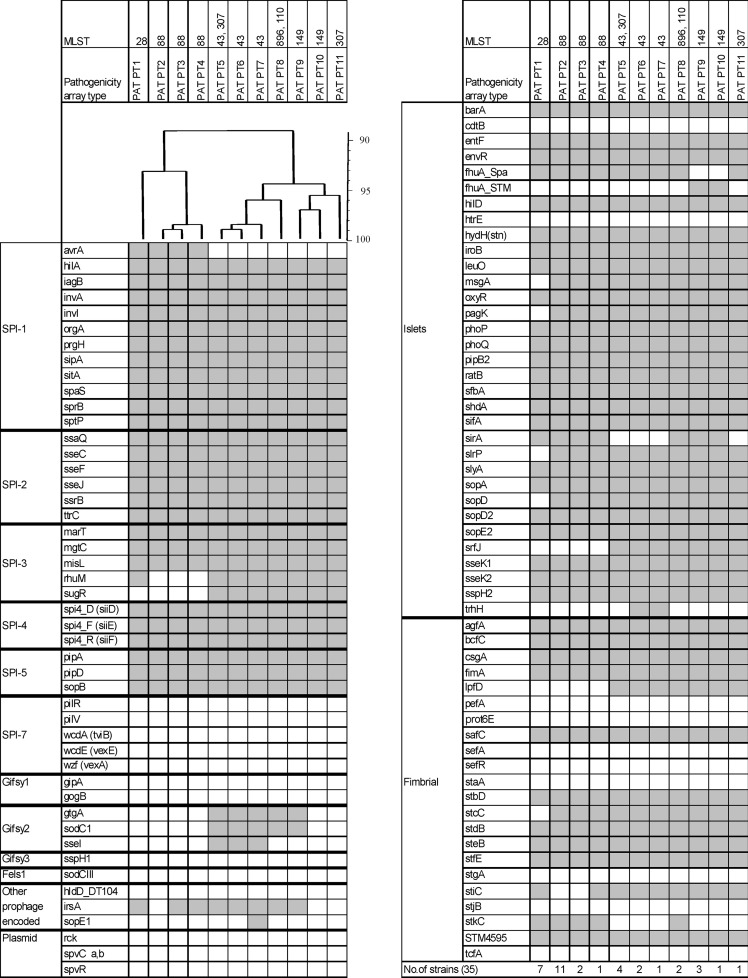
Determination of pathogenicity genes.
Eleven different pathogenicity array types (PATs) were identified among the same 35 strains tested as for sop-ST (Fig. 4). PATs differed in up to 21 targets from 102 markers tested. Almost all PATs corresponded to specific MLST types (Fig. 2). Only two PATs (PT5 and PT8) were found in two STs belonging to clonal complex 1. Markers targeting within the Salmonella pathogenicity islands (SPIs) 1 to 5 were partially absent in SPI-1 and SPI-3. PAT PT1 (exclusively associated with ST28 strains) lacked the sugR gene, and PAT PT2 to PT4 (ST88 strains) lacked the rhuM and sugR genes, both located in SPI-3. All strains belonging to clonal complex 1 (PAT PT5 to PT11) lacked the avrA gene present in SPI-1 (encoding a protein inhibiting the key proinflammatory immune response). In addition, markers for msgA, pagK, slrP, sopD, and stcC were specifically absent in PAT PT1. Three PATs (PT2 to PT4) were linked to ST88 strains which differed only in up to two targets (irsA and stiC). Strains tested belonging to clonal complex 1 showed seven different PATs (PT5 to PT9). These differed in up to nine pathogenicity gene markers. Strains with PAT PT6 and PAT PT7 probably contained a variant of the Salmonella genomic island 1 (SGI1) since their resistance genes, blaPSE1, floR, sul1, tet(G), aadA2-3-8, and qacEΔ were detected in combination with the presence of markers trhH and intSG1 (data not shown). Markers indicating the presence of a virulence plasmid (pSLT) were negative in all strains tested.
Fig 4.
Virulence determinants of 35 S. enterica serovar Paratyphi B (dT+) strains analyzed. On the left side the analyzed genes are indicated and grouped according to their particular genomic location (SPI-1 to SPI-7; prophages Gifsy-1, Gifsy-2, Gifsy-3 and Fels-1; plasmids; and islets) or function (fimbrial). At the top, assigned PATs and corresponding STs are indicated. Immediately below this, an UPGMA dendrogram shows the similarity of PATs in percentages. The hybridization result of each PAT is shown by column. A white box indicates the absence, and a gray box indicates the presence of the target sequence.
DISCUSSION
Population structure of S. enterica serovar Paratyphi B (dT+) in Germany.
The application of genotypic methods to 108 S. enterica serovar Paratyphi B (dT+) strains isolated from diverse sources between 2002 and 2010 in Germany revealed that the serovar is polyphyletic. Polyphyletic serovars originate from more than one common ancestor, and they consist of several distantly related STs within one serovar. Phylogenetic trees based on data from PFGE, MLST, sop-ST, and the pathogenicity gene repertoire resulted, independently of the method, in three divergent lineages (referred to also as clonal complexes). The first clonal complex is composed of five closely related STs (ST43, ST110, ST149, ST307, and ST896) which correlated to PFGE cluster B (Fig. 1) and four closely related sop-STs. The other two clonal complexes were each represented by one single ST (ST88 and ST28) associated with PFGE clusters A and C. An exceptional set of 10 ST88 strains were grouped in cluster B instead of cluster A. Three of these strains had a unique sop-ST41-28-24 instead of sop-ST39-28-24. sopA allele 41 differed from sopA allele 39 by a deletion of one nucleotide, leading to a frameshift mutation. This shows that PFGE is not a reliable mirroring of the phylogeny of S. enterica serovar Paratyphi B (dT+) but is a highly discriminative method for short-term evolutionary studies since the Simpson index for discrimination was highest for PFGE in the strain set tested.
The three clonal complexes identified and characterized here were also recently detected within an international set of S. enterica serovar Paratyphi B strains based on MLST analysis (1, 31). Achtman et al. (1) have grouped STs into clonal complexes on the basis of an eBurst analysis. According to this nomenclature, clonal complex 1 was designated eBG5, clonal complex 2 was named eBG19, and clonal complex 3 was named eBG59. Specific information on the sources of strains and their origins or the differences in O:5 antigen expression for S. enterica serovar Paratyphi B (dT+) were not considered by Achtman et al. (1). However, the study contributed to resolve the evolutionary relationship between the d-tartrate fermenting (dT+) and nonfermenting (dT−) variants of S. enterica serovar Paratyphi B. The dT− variant is regarded as a cause of typhoid-like illness in humans, whereas the dT+ variant is associated with gastroenteritis in animals and humans (2, 22). Interestingly, dT− strains belonged to one of the same complex, eBG5, as the dT+ variant, but they are represented by other sequence types.
Recent studies indicated that genetic diversity within S. enterica is derived also from homologous recombination in addition to mutational changes (9, 11, 14). However, in this study it was obvious that the tree structures (genetic relatedness) resulting from independent genotypic typing methods (MLST, sop-ST, and DNA microarray) were highly congruent (Fig. 3 and 4). The conclusion is that the three complexes evolved independently within specific niches or hosts with a small opportunity for recombination between them. We speculate that humans, as the only common host for all three clonal complexes, might have formerly played a role in recombination leading to the same serovar expression (O:4,[5],12:b:1,2) shared by all three complexes.
Association of lineages with origin of strains.
The three divergent lineages could clearly be associated with the origin of the strains. Strains isolated from humans were found in all three lineages, with preference in clonal complex 1 (ST43 founder). Strains which were isolated from reptiles belonged to clonal complex 2 (ST88), and strains which were isolated mainly from poultry and occasionally from other livestock (pig and cattle) belonged to clonal complex 3 (ST28). However, the selection of the 108 epidemiologically unrelated strains did not reflect the frequency of isolation within a specific source. Despite the high number of S. enterica serovar Paratyphi B (dT+) strains received at the NRL-BfR isolated from poultry/poultry meat and the potentially high exposure of poultry meat to humans, NRZ-RKI obtained only 10 human strains which could be associated with the typical poultry-adapted ST28 (O:5-antigen-negative) complex within the last 9 years (Table 2). This indicates that apparently ST28 strains are not as pathogenic for humans as the other clonal complexes (represented by ST88 and ST43 as founders) which were frequently isolated from humans. A reason might be that ST28 strains lacked a number of unique virulence genes (msgA, pagK, slrP, sopD, and stcC) compared to the other STs (Fig. 4). In contrast, the other clonal complexes (C2 and C3) possess virulence determinants that possibly enable them to interact with a broad range of hosts and the environment.
Reptiles could play a major role as a potential source for human salmonellosis caused by S. enterica serovar Paratyphi B because ST88 isolates were frequently found in both sources. This sequence type was continuously and specifically isolated in Germany from reptiles within the last 9 years. Only one of the 24 reptile strains analyzed was assigned to another type, namely, ST43. Unfortunately, information provided with strains for typing at the NRL-BfR was incomplete and did not allow investigation of whether they originated from reptiles living in a zoological garden or in a private terrarium. However, there are many studies worldwide reporting reptile-associated salmonellosis and sepsis (4, 7, 29). Because reptiles are known to shed Salmonella frequently and because they have become more and more popular in households and petting zoos, they are recognized as an emerging public health problem (4).
A potential source for human infection caused by the first clonal complex (ST43 as founder) could not be identified. It was only sporadically isolated from various sources within the last 9 years. Possibly, human-to-human transmission might play a role for dissemination or other unidentified sources such as vegetables (32), fish aquariums (25), or goat's milk cheese (10).
Despite the strong association of the clonal complexes with the origin, there was no exclusive host adaptation observed. Host-adapted variants typically causes systemic disease in a limited number of related species, as thought for the d-tartrate-nonfermenting variant of S. enterica serovar Paratyphi B. In contrast, host-associated or -restricted variants are primarily associated with one or two closely related host species and are able to persist in the population but may also infrequently cause illness in other hosts (34). Host adaptation can be triggered by the specific organization of the immune system in birds, mammals, or cold-blooded vertebrates, leading to an adapted pathogenicity gene repertoire of the serovar or variant (3, 24).
Antimicrobial resistance in S. enterica serovar Paratyphi B (dT+).
A multidrug-resistant subgroup of S. enterica serovar Paratyphi B (dT+) has become relatively widely distributed throughout the world. Strains encode the Salmonella genomic island 1 (SGI1) originally described in S. enterica serovar Typhimurium DT104 (5). They are usually resistant to ampicillin, chloramphenicol, streptomycin, spectinomycin, sulfonamides, and tetracyclines and have not yet been found in poultry but in tropical fish aquaria (25) and cattle (13). In France, the SGI1-containing clone was frequently found in clinical isolates of S. enterica serovar Paratyphi B (dT+), but no potential sources of infection could be identified (35). In this study, strains with SGI1 were rarely identified. Only three strains isolated from humans harbored typical SGI1-associated resistance and other genes. All of them belonged to ST43. Unfortunately, a comparison of the antimicrobial resistance patterns gave no hints on any infection source from food or animals.
Another multidrug-resistant clone has been genotypically described in this and previous studies (21, 27). Strains belonged invariably to ST28 (clonal complex 3) and are mostly isolated from poultry/poultry meat. They possess a chromosomally located Tn7-like class 2 integron carrying a dfrA1-sat1-aadA1 gene cassette. Since the manifestation of this clone in poultry in the mid-1990s in several European countries, strains have apparently in addition accumulated quinolone resistance and extended-spectrum β-lactam resistance (35) (B. Guerra, personal communication). Three strains investigated (one chicken and two human strains) harbored blaCMY-2-like, blaCTX-M2-like, and blaTEM1-like genes, indicating that variety of genes lead to β-lactam resistance within this complex.
In conclusion, S. enterica serovar Paratyphi B (dT+) is an example of a serovar that can consist of groups with divergent evolutionary paths. Here, data indicated that three clonal complexes evolved independently from each other in different niches. By recombination events in recent time, we speculate that complex 2 and 3, represented by only one ST, acquired structural surface antigens (e.g., O, H1, and H2 antigens) leading to S. enterica serovar Paratyphi B (dT+) expression and, consequently, to disruption of the phylogeny within this serovar. Serotyping is therefore for polyphyletic serovars of limited value for the study of population structure and the description of risk characters. Furthermore, we hypothesize that clonal complex 1 is much older than complex 2 or 3 because of its observed genetic diversity. Sequencing of the core genome could clarify this hypothesis. This study showed also that the different complexes were associated with unique sources. Each complex had a specific pathogenicity gene repertoire which might contribute to the frequency of isolation of the complexes from humans and animals. Preferentially by contact with reptiles and to a less extent by exposure to poultry or poultry meat, S. enterica serovar Paratyphi B (dT+) poses a health risk for humans. However, other nonidentified sources might play a role for human salmonellosis caused by S. enterica serovar Paratyphi B (dT+).
ACKNOWLEDGMENTS
This work was funded by the Bundesministerium für Bildung und Forschung (BMBF), FBI-Zoo 2 (01KI1012I and 01KI1012F).
We thank Gabriele Berendonk, Manuela Jaber, Marta Brohm, and Katharina Thomas for technical assistance. We thank Andreas Schroeter and Beatriz Guerra for scientific discussion about antimicrobial resistance.
Footnotes
Published ahead of print 10 August 2012
REFERENCES
- 1. Achtman M, et al. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8:e1002776 doi:10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barker RM, et al. 1988. Types of Salmonella paratyphi B and their phylogenetic significance. J. Med. Microbiol. 26:285–293 [DOI] [PubMed] [Google Scholar]
- 3. Bäumler AJ, Tsolis RM, Ficht TA, Adams LG. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertrand S, et al. 2008. Salmonella infections associated with reptiles: the current situation in Europe. Euro Surveill. 13:18902 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=18902 [PubMed] [Google Scholar]
- 5. Boyd D, et al. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breitenfeld V, Aleraj D. 1967. Clinical and bacteriological characteristics of salmonellosis caused by Salmonella java. Zentralbl. Bakteriol. Orig. 204:89–99 (In German.) [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention 2008. Multistate outbreak of human Salmonella infections associated with exposure to turtles—United States, 2007–2008. MMWR Morb. Mortal. Wkly. Rep. 57:69–72 [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7th ed CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Commission of the European Communities 2005. Commission regulation (EC) no. 1003/2005 of 30 June 2005 implementing regulation (EC) no. 2160/2003 as regards a Community target for the reduction of the prevalence of certain salmonella serotypes in breeding flocks of Gallus gallus and amending regulation (EC) no. 2160/2003. Official Journal of the European Union, L 170. Commission of the European Communities, Brussels, Belgium: http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2005:170:0012:0017:EN:PDF [Google Scholar]
- 10. Desenclos JC, et al. 1996. Large outbreak of Salmonella enterica serotype Paratyphi B infection caused by a goats' milk cheese, France, 1993: a case finding and epidemiological study. BMJ 312:91–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Didelot X, et al. 2011. Recombination and population structure in Salmonella enterica. PLoS Genet. 7:e1002191 doi:10.1371/journal.pgen.1002191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. European Food Safety Authority 2007. Report of the Task Force on Zoonoses data collection on the analysis of the baseline survey on the prevalence of Salmonella in broiler flocks of Gallus gallus, in the EU, 2005–2006. EFSA J. 98:1–85 doi:10.2903/j.efsa.2007.98r [Google Scholar]
- 13. Evans SJ, et al. 2005. Multiple antimicrobial resistant Salmonella enterica serovar Paratyphi B variant Java in cattle: a case report. Vet. Rec. 156:343–346 [DOI] [PubMed] [Google Scholar]
- 14. Falush D, et al. 2006. Mismatch induced speciation in Salmonella: model and data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2045–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaulin C, Vincent C, Alain L, Ismail J. 2002. Outbreak of Salmonella paratyphi B linked to aquariums in the province of Quebec, 2000. Can. Commun. Dis. Rep. 28:89–93, 96 [PubMed] [Google Scholar]
- 16. Grimont PAD, Weill F-X. 2007. Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella, Paris, France [Google Scholar]
- 17. Harris JR, et al. 2009. Multistate outbreak of infections associated with small turtle exposure, 2007–2008. Pediatrics 124:1388–1394 [DOI] [PubMed] [Google Scholar]
- 18. Hauser E, et al. 2011. Diversity of Salmonella enterica serovar Derby isolated from pig, pork, and humans in Germany. Int. J. Food Microbiol. 151:141–149 [DOI] [PubMed] [Google Scholar]
- 19. Hauser E, Junker E, Helmuth R, Malorny B. 2011. Different mutations in the oafA gene lead to loss of O5-antigen expression in Salmonella enterica serovar Typhimurium. J. Appl. Microbiol. 110:248–253 [DOI] [PubMed] [Google Scholar]
- 20. Huehn S, Bunge C, Junker E, Helmuth R, Malorny B. 2009. Poultry associated Salmonella enterica subsp. enterica serovar 4,12:d:− reveals high clonality and a distinct pathogenicity gene repertoire. Appl. Environ. Microbiol. 75:1011–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huehn S, Helmuth R, Bunge C, van Pelt W, Malorny B. 2009. Characterization of pathogenic and resistant genome repertoire reveals two clonal lines in Salmonella enterica subsp. enterica serovar Paratyphi B (+)-tartrate positive. Foodborne Pathog. Dis. 6:431–443 [DOI] [PubMed] [Google Scholar]
- 22. Kauffmann F. 1955. Zur Differentialdiagnose und Pathogenität von Salmonella java und Salmonella paratyphi B. Z. Hyg. Infektionskr. 141:546–550 [PubMed] [Google Scholar]
- 23. Kidgell C, et al. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39–45 [DOI] [PubMed] [Google Scholar]
- 24. Kingsley RA, Bäumler AJ. 2002. Pathogenicity islands and host adaptation of Salmonella serovars. Curr. Top. Microbiol. Immunol. 264:67–87 [PubMed] [Google Scholar]
- 25. Levings RS, Lightfoot D, Hall RM, Djordjevic SP. 2006. Aquariums as reservoirs for multidrug-resistant Salmonella Paratyphi B. Emerg. Infect. Dis. 12:507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malorny B, Bunge C, Helmuth R. 2003. Discrimination of d-tartrate-fermenting and -nonfermenting Salmonella enterica subsp. enterica isolates by genotypic and phenotypic methods. J. Clin. Microbiol. 41:4292–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miko A, Guerra B, Schroeter A, Dorn C, Helmuth R. 2002. Molecular charcaterization of multiresistent D-tartrate-positive Salmonella enterica serovar Paratyphi B isolates. J. Clin. Microbiol. 40:3184–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miko A, Pries K, Schroeter A, Helmuth R. 2003. Multiple-drug resistance in d-tartrate-positive Salmonella enterica serovar Paratyphi B isolates from poultry is mediated by class 2 integrons inserted into the bacterial chromosome. Antimicrob. Agents Chemother. 47:3640–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagano N, Oana S, Nagano Y, Arakawa Y. 2006. A severe Salmonella enterica serotype Paratyphi B infection in a child related to a pet turtle, Trachemys scripta elegans. Jpn. J. Infect. Dis. 59:132–134 [PubMed] [Google Scholar]
- 30. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 31. Sangal V, et al. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J. Bacteriol. 192:6465–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stratton J, et al. 2001. Outbreak of Salmonella paratyphi B var. java due to contaminated alfalfa sprouts in Alberta, British Columbia and Saskatchewan. Can. Commun. Dis. Rep. 27:133–137 [PubMed] [Google Scholar]
- 33. van Pelt W, et al. 2003. Explosive increase of Salmonella Java in poultry in the Netherlands: consequences for public health. Euro Surveill. 8:31–35 [DOI] [PubMed] [Google Scholar]
- 34. Wallis TS, Barrow PA. 25 July 2005, posting date Salmonella epidemiology and pathogenesis in food-producing animals. Chapter 8.6.2.1 In Böck A, et al. (ed), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: doi:10.1128/ecosal.8.6.2.1. http://www.ecosal.org [DOI] [PubMed] [Google Scholar]
- 35. Weill FX, Fabre L, Grandry B, Grimont PA, Casin I. 2005. Multiple-antibiotic resistance in Salmonella enterica serotype Paratyphi B isolates collected in France between 2000 and 2003 is due mainly to strains harboring Salmonella genomic islands 1, 1-B, and 1-C. Antimicrob. Agents Chemother. 49:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wollin R. 2007. A study of invasiveness of different Salmonella serovars based on analysis of the Enter-net database. Euro Surveill. 12:3275 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3275 [DOI] [PubMed] [Google Scholar]