Abstract
A new protocol for rapid, specific, and sensitive cell-based quantification of Vibrio cholerae/Vibrio mimicus in water samples was developed. The protocol is based on catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) in combination with solid-phase cytometry. For pure cultures, we were able to quantify down to 6 V. cholerae cells on one membrane with a relative precision of 39% and down to 12 cells with a relative precision of 17% after hybridization with the horseradish peroxidase (HRP)-labeled probe Vchomim1276 (specific for V. cholerae and V. mimicus) and signal amplification. The corresponding position of the probe on the 16S rRNA is highly accessible even when labeled with HRP. For the first time, we were also able to successfully quantify V. cholerae/V. mimicus via solid-phase cytometry in extremely turbid environmental water samples collected in Austria. Cell numbers ranged from 4.5 × 101 cells ml−1 in the large saline lake Neusiedler See to 5.6 × 104 cells ml−1 in an extremely turbid shallow soda lake situated nearby. We therefore suggest CARD-FISH in combination with solid-phase cytometry as a powerful tool to quantify V. cholerae/V. mimicus in ecological studies as well as for risk assessment and monitoring programs.
INTRODUCTION
Vibrio cholerae and Vibrio mimicus are important human pathogens and two very closely related species. Only recently, their classification as two different species has been confirmed (35). V. cholerae comprises more than 200 serotypes, but only serotypes O1 and O139 cause the severe diarrheal disease cholera (9, 32). Interestingly, several V. mimicus isolates carrying cholera toxin genes (ctxAB) have also been identified. It was suggested that horizontal transfers of virulence-related genes from an uncommon clone of V. cholerae have generated pathogenic V. mimicus strains carrying cholera toxin genes (35). All other V. cholerae serotypes, summarized as non-O1/non-O139, and V. mimicus strains are usually associated with less-severe gastrointestinal infections as well as blood, wound, and ear infections (7, 16, 28, 35). Besides their role as important human pathogens, both organisms are found worldwide in brackish water, coastal areas, and estuarine environments (8, 35) and are therefore considered to be natural components of the bacterial community in aquatic ecosystems.
Rapid and precise methods for detection and quantification of pathogenic bacteria are fundamental for monitoring and risk assessment in regard to the use of water as well as for studying their ecology in the environment. For quantification of the V. cholerae/V. mimicus clade, various methods have been developed during the last decades. Culture-based techniques for detecting these pathogens, followed by biochemical confirmation via API 20E or PCR, are still regularly used (5, 8). These techniques often include an enrichment step with alkaline peptone water and can then be used quantitatively only as laborious most-probable-number (MPN) techniques. In addition, they cannot be applied when Vibrio cells turn into the viable but nonculturable (VBNC) status. The VBNC state is an important life strategy of Gram-negative bacteria, and it has been shown that V. cholerae and V. mimicus enter this state in response to nutrient deprivation and low temperatures (31). Due to this phenomenon, whereby cells are still present in the environment but cannot be detected by culture, culture-independent methods are needed. Traditional PCR is a common molecularly based detection alternative to cultivation. Nevertheless, conventional PCR requires further product characterization by gel electrophoresis, and, even more important, it is a nonquantitative method (14). Real-time PCR is a strong improvement of traditional PCR and can be effectively used for quantification of microorganisms (26). Variable DNA extraction efficiencies as well as PCR inhibition, especially in environmental samples, are still a problem and need to be further improved (26).
Alternatively, fluorescence in situ hybridization (FISH) and the direct fluorescent antibody assay (DFA) are direct cell-based detection methods. DFA is a fluorescent monoclonal antibody-staining application and can be used for detection and enumeration of V. cholerae serotype O1 (15). Still, this method is not available for enumeration of V. cholerae serotypes other than O1. FISH with rRNA-targeted oligonucleotide probes, combined with epifluorescence microscopy (EFM), is generally accepted as a direct, cultivation-independent method to quantify microorganisms in the environment (2, 3, 29). Over the past 15 years, FISH has become a very strong tool in observing microbial communities in their environment and is therefore indispensable in the field of microbial ecology. Epifluorescence microscopy (EFM) and flow cytometry are widely used in microbiology, and EFM has become a standard technique for quantification of fluorescence-labeled microorganisms (23, 25, 34). Nevertheless, its use is very limited when it comes to detection of rare events. Low numbers of bacteria demand increasing sample volumes that need to be filtered in order to quantify them accurately with EFM (25). Filtrating high volumes of water on membranes has its clear limits. It is a very time-consuming procedure, and additionally, more volume most often means increased background and autofluorescence potentially interfering with the enumeration of the labeled microorganisms. So far, solid-phase cytometry (SPC) is the only technique that enables accurate quantification of rare events in combination with EFM for validation of the results (23). Since SPC can theoretically detect one labeled cell on a membrane (23), the sample volume can be kept rather low. This fact is an important advantage over flow cytometry and fluorescence microscopy, for which, in contrast to solid-phase cytometry, large volumes need to be concentrated for detection of rare events. Unfortunately, previous studies (24, 34) have shown that the fluorescence signals of standard FISH-labeled microorganisms were too weak to be detected with solid-phase cytometry. Catalyzed reporter deposition-FISH (CARD-FISH) is one methodical improvement to increase the signal intensity of microorganisms (30, 33, 37). The rRNA-targeted oligonucleotide probe is labeled with horseradish peroxidase (HRP). Signal amplification is achieved by dimerization of fluorescence-labeled multiple-tyramide molecules by one HRP molecule and subsequent binding of the highly reactive intermediates to electron-rich moieties of proteins at or near the site of the peroxidase binding site (1). It has been shown that this tyramide signal amplification results in an up-to-20-fold increase of the signal from that of normal FISH (22, 33). Solid-phase cytometry has already been successfully applied for the quantification of bacterial populations occurring at low concentrations in drinking water (23), for Legionella pneumophila (4), and for fecal indicator organisms like Escherichia coli (6).
The aim of this study was to develop a new CARD-FISH protocol for rapid and sensitive detection and enumeration of V. cholerae/V. mimicus cells in environmental samples with solid-phase cytometry. With this new method, we were able to quantify V. cholerae/V. mimicus at extremely low numbers (a few cells per filtration volume) for pure cultures and starved cells spiked into environmental samples. Moreover, we were also successful in enumerating V. cholerae/V. mimicus cells directly in extremely turbid lake water samples, a process which is highly useful for ecological studies as well as for risk assessment and monitoring programs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For protocol development, a variety of V. cholerae non-O1/non-O139 strains isolated from the lake Neusiedler See and epidemic O1 and O139 strains were grown in 20 ml Luria Bertani (LB) broth and put on a shaker at 37°C until an optical density at 600 nm (OD600) of 0.6 to 0.8 to obtain a concentration of ∼6 × 108 V. cholerae cells ml−1 (approximately 2 h). Additional Vibrio species used for testing the specificity and hybridization efficiency of the CARD-FISH probe were also grown in LB broth until an OD600 of 0.6 to 0.8 (exponential growth phase). A concentration of approximately 106 Vibrio cells ml−1 was used for these experiments. For spiking experiments, V. cholerae cells were grown until late stationary growth phase to produce cells with low rRNA content, which is expected for environmental bacteria. A single colony from an LB agar plate was grown overnight in 5 ml LB broth and put on a shaker at 37°C. From this preculture, 50 μl was inoculated into 50 ml fresh LB broth and cultured at the same temperature on a shaker for 20 to 24 h.
Quantification of pure cultures.
Liquid cultures of V. cholerae non-O1/non-O139 strains and an O1 classical strain (strain O395) were stepwise diluted 1:10 in sterile 1× phosphate-buffered saline (PBS) to achieve concentrations of 6 × 106 down to 6 cells ml−1. For epifluorescence microscopy (EFM), 10 ml of the 6 × 103-cells ml−1 suspension and 1 ml of the 6 × 104-, 6 × 105-, and 6 × 106-cells ml−1 suspensions were used. For solid-phase cytometry (SPC), 1 ml of the 6 × 100-, 6 × 101-, 6 × 102-, and 6 × 103-cells ml−1 suspensions were taken. All investigations were done in triplicate.
Spiking experiment.
Cells with low rRNA content (preparation was done as described above) were spiked into either sterile 1× PBS or freshly collected lake water to achieve concentrations of ∼104, 103, and 102 cells ml−1. Spiked and unspiked lake water samples were prepared for CARD-FISH.
Environmental samples.
For testing the newly developed CARD-FISH protocol for quantification of V. cholerae/V. mimicus in environmental samples by solid-phase cytometry, water samples were taken from the Austrian lake Neusiedler See and two shallow soda lakes situated along the eastern shore of the lake. Kirschner et al. (21) had recently demonstrated the permanent autochthonous existence of Vibrio cholerae non-O1/non-O139 strains in the lake Neusiedler See. Up to now, no V. mimicus strains have been isolated from the lake. All these lakes are known for their high turbidity (total suspended solids up to 3 g liter−1), leading to high concentrations of background particles on filtered water samples. Detailed information on the lakes can be found in the works by Eiler et al. (10) and Kirschner et al. (21). Water samples were collected in clean, sterile, 500-ml glass flasks from two different sampling points of the Neusiedler See as well as from two shallow soda lakes (Zicklacke and Oberer Stinker) during summer 2011. For CARD-FISH, 3 to 8 replicate subsamples of appropriate volume (between 10 and 100 μl, depending on the turbidity of the water) were taken and filled up with 1× PBS to reach a final volume of 1 ml. Subsamples were fixed with 4% paraformaldehyde (PFA) to reach a final concentration of 1% PFA. PFA fixation was carried out overnight (12 to 16 h) at 4°C. After fixation, subsamples (in total, 1.5 ml each) were filtered on appropriate membranes. In addition, cultivation-based quantification of V. cholerae was done by membrane filtration to allow a comparison between the results obtained with the newly developed method and the results of the traditional culture technique. In brief, samples were filtered through 0.45-μm-pore-size cellulose nitrate filters (Ø, 47 mm; Sartorius, Germany). For the Neusiedler See, 100 μl, 1 ml, and 10 ml of each water sample were filtered and the filter was directly placed on thiosulfate citrate bile sucrose (TCBS; Merck, Darmstadt, Germany) agar plates and incubated for 18 h at 37°C (5). According to the WHO guidelines for drinking water quality, TCBS is a highly selective differential medium most commonly used for the isolation of V. cholerae (36). For the shallow soda lakes, 10, 100, and 1,000 μl were filtered. All water samples were filled up to 10 ml with sterile 1× PBS for an even distribution of the sample on the filter. Yellow, flat, 1- to 3-mm diameter colonies were counted and streaked onto plates with nutrient agar without NaCl (3% beef extract, 5% peptone, 15% agar). After incubation overnight at 37°C, colonies growing on agar without NaCl were considered presumptive V. cholerae (21). Representative presumptive isolates were confirmed by species-specific ompW-based PCR (5). In addition, green colonies on TCBS were picked and screened for V. mimicus via API 20E.
Probe.
The 16S rRNA probe Vchomim1276 (5′-ACT TTG TGA GAT TCG CTC CAC CTC G-3′) that is specific for V. cholerae and V. mimicus and that was used in this study has been described previously (13, 17, 20). For FISH, the probe was monolabeled with 6-carboxyfluorescein (FAM), and for CARD-FISH, the probe was HRP labeled and amplification was based on tyramide signal amplification. In this study, tyramide was labeled with Alexa 488. The specificity of the FAM-monolabeled Vchomim1276 probe has been previously tested (20).
FISH.
Hybridization was performed according to the protocol described by Kirschner et al. (20). In brief, samples were fixed with 4% PFA, filtered on white polycarbonate membrane filters (Ø, 25 mm; pore size, 0.2 μm; Whatman), and air dried. One section of the filter was hybridized with the probe Vchomim1276, monolabeled with FAM, for 1.5 h to 2 h at 46°C (the hybridization solution was composed of 0.9 M NaCl, 20 mM Tris-HCl, 35% formamide, and 0.01% SDS). Filter sections were transferred into a prewarmed washing buffer (80 mM NaCl, 20 mM Tris-HCl, 5 mM EDTA, and 0.01% SDS) and incubated for 10 min at 48°C. The sections were rinsed with ethanol (94%) and air dried on paper in the dark. For counterstaining, a DAPI (4′,6-diamidino-2-phenylindole) mix (5.5 parts Citifluor, 1 part Vectashield, and 0.5 parts 1× PBS, with DAPI at a final concentration of 1 μg ml−1) was used. Microscope slides can be stored at −20°C for at least 6 months.
CARD-FISH.
The newly developed protocol is based on the CARD-FISH protocol described by Wilhartitz et al. (37), with modifications. Samples were fixed with 4% PFA and filtered on CB04 filters (black; Ø, 25 mm; pore size, 0.4 μm; AES Chemunex). Air-dried filters can either be used immediately or stored at −20°C in sterile 1.5-ml reaction tubes until use. CB04 filters were placed on labeling pads (AES Chemunex), soaked in 650 μl of 50%, 80%, and 94% ethanol solution in petri dishes, and incubated at room temperature for 4 min each.
For permeabilization, filters were placed on 100 μl of lysozyme solution (10 mg ml−1 lysozyme, 0.1 M Tris-HCl, and 0.05 M EDTA) in petri dishes and incubated for 45 min at 37°C. After permeabilization, filters were washed in sterile deionized reverse-osmosis water. To inhibit potentially present intracellular peroxidases, filters were put on labeling pads soaked in 650 μl of 0.01 M HCl for 20 min at room temperature. After another washing step in sterile deionized reverse-osmosis water, filters were shortly dipped in ethanol (94%) and air dried. At this point, the process can be stopped and filters can be stored at −20°C in sterile 1.5-ml reaction tubes.
For hybridization, filters were put on glass slides. Thirty microliters of hybridization solution (0.9 M NaCl, 20 mM Tris-HCl, 10% dextran sulfate, 0.02% SDS, 1% blocking reagent [Boehringer Mannheim, Germany], and 55% formamide) containing the Vchomim probe labeled with HRP instead of FAM (final concentration, 5 ng μl−1) was pipetted onto the filters. For limiting evaporation during hybridization, cover slides were mounted. The glass slide containing the filter and cover slide was put into a slide holder. Hybridization was carried out for 2 h at 46°C in the dark. Filters were washed for 15 min at 37°C on labeling pads soaked in 650 μl prewarmed washing buffer (13 mM NaCl, 20 mM Tris-HCl, 5 mM EDTA, and 0.01% SDS). Thereafter, filters were transferred to labeling pads soaked in 650 μl PBS-Triton buffer (0.05% Triton X-100) and incubated for 15 min at room temperature.
For amplification, the filters were placed on 100 μl of substrate mix and incubated in the dark at 37°C for 15 min. The substrate mix consists of an amplification buffer (10% dextran sulfate, 2 M NaCl, and 0.1% blocking reagent), 0.0015% freshly prepared H2O2, and tyramide-Alexa 488 (1 mg ml−1; Invitrogen, Germany). Filters were then transferred to labeling pads soaked in 650 μl PBS-Triton buffer and incubated for 10 min at room temperature. After another washing step with sterile deionized reverse-osmosis water, filters were shortly dipped in ethanol (94%) and air dried. Filters were either analyzed immediately with the solid-phase cytometer or stored at −20°C in sterile 1.5-ml reaction tubes in the dark until use.
For environmental samples and spiking experiments with starved cells, the lysozyme treatment was prolonged to 60 min and amplification was carried out for 45 min to increase signal intensity. In order to reduce background interference, filters were pretreated for 10 min in the dark with a 0.01% concentration of Evans Blue (Sigma-Aldrich, Vienna, Austria) before the scan.
Cell enumeration with EFM and SPC.
For epifluorescence microscopy, filters were examined under a Nikon Eclipse 8000 microscope at ×1,250 magnification; at least 20 microscopic fields were counted. For solid-phase cytometry, a ChemScan RDI (AES-Chemunex, Ivry-sur-Seine, France) was used. Details on the applied system have been described elsewhere (23, 27, 34). For cell enumeration, a support pad saturated with 100 μl washing buffer was placed on a ChemScan RDI membrane holder. The filter with the CARD-FISH-labeled bacteria was positioned on top of that support pad. In brief, the CB04 filters are scanned with an argon laser (488-nm emission wavelength) within 3 min. The fluorescent light is collected by three photomultiplier tubes (PMT) at different wavelengths (500 to 530 nm, 540 to 570 nm, and 575 to 625 nm). For Alexa 488, the fluorescence is collected in the green channel (500 to 530 nm). All fluorescent events undergo a screening by computer software, for which several discriminant parameters can be adjusted by the operator to differentiate between labeled microorganisms and other fluorescent particles. The most important discriminant parameters are the number of “lines” and the number of “samples” for the shape and the peak intensity of the signal. The ratio of the fluorescent light detected in the secondary channel to the fluorescent light detected in the primary channel (fluorescence from the probe Alexa 488) is another important discriminant parameter and is referred to as the S/P ratio. When the discriminant parameters were tested and adjusted, 100 fluorescence events from the raw data were validated, and labeled V. cholerae/V. mimicus cells and fluorescent particles were manually discriminated with the microscope equipped with a motorized stage.
The discriminant settings were set as follows: number of “lines,” 1.5 to 3.5; number of “samples,” 4 to 10; peak intensity, 200 to 1,500; and S/P ratio, 0.35 to 0.95. After a full scan, the fluorescent events are rendered on raw data and net result maps. Each spot can be visualized and confirmed by epifluorescence microscopy (Nikon Eclipse 50i). For this purpose, the sample holder with the scanned filter is moved to a motorized stage mounted on the microscope. For microscopic validation, 300 representative spots or all spots on the net result map were looked at and confirmed as either positive results or particles. After this step, the total number of labeled bacteria on the membrane was calculated. For determination of the precision of enumerating fluorescent particles with the ChemScan RDI, green fluorescent latex beads (1-μm diameter; standard G; AES Chemunex) were used. The beads were filtered on CB04 filters and immediately analyzed. Thirty subsamples of each concentration (∼103, ∼102, and ∼101 beads ml−1) were scanned and validated.
RESULTS
Sensitivity and specificity of FISH and CARD-FISH (qualitative).
Sensitivity testing revealed positive FISH and CARD-FISH signals for V. mimicus and all investigated V. cholerae strains (Table 1). Furthermore, the hybridization efficiency was found to be ≥95%, as almost all cells visible with DAPI also showed bright fluorescence signals with the FISH and CARD-FISH probes (see Fig. S1 in the supplemental material). Specificity testing confirmed that the probe Vchomim1276 was specific for the V. cholerae/V. mimicus clade, not detecting other Vibrio strains (Table 1). FISH signals showed too weak fluorescence intensity (signal intensity ≤ 150 arbitrary ChemScan RDI fluorescence units) for the analysis with SPC, and FISH was therefore not considered to be suitable for quantification of V. cholerae/V. mimicus cells. In contrast, sufficient fluorescence intensity for SPC analysis was achieved by applying the CARD-FISH protocol (signal intensity ≥ 400 arbitrary ChemScan RDI fluorescence units).
Table 1.
V. cholerae strains and other Vibrio species used for specificity and sensitivity testing (qualitative) of the probe Vchomim1276, labeled with FAM (FISH) or HRP (CARD-FISH)
| Strain | Origin | Signala with: |
|
|---|---|---|---|
| FISH | CARD-FISH | ||
| V. cholerae non-O1/non-O139 | NCCB 36033 | + | + |
| V. cholerae non-O1/non-O139 | Lake Neusiedler See | + | + |
| V. cholerae non-O1/non-O139 | Lake Neusiedler See | + | + |
| V. cholerae non-O1/non-O139 | Lake Neusiedler See | + | + |
| V. cholerae non-O1/non-O139 | Lake Neusiedler See | + | + |
| V. cholerae non-O1/non-O139 | Lake Neusiedler See | + | + |
| V. cholerae O1 El Tor | ATCC 39315b | + | + |
| V. cholerae O1 Classic | O395b | + | + |
| V. cholerae O139 | MO10b | + | + |
| V. cholerae O1 | MJ-1750c | + | + |
| V. cholerae O1 | MK-1948c | + | + |
| V. cholerae O1 | MK-2268c | + | + |
| V. cholerae O1 | ML-1148c | + | + |
| V. cholerae O1 | ML-2835c | + | + |
| V. cholerae O139 | MJ-5791c | + | + |
| V. cholerae O139 | MJ-5872c | + | + |
| V. cholerae O139 | MK-4706c | + | + |
| V. mimicus | ATCC 33653b | + | + |
| Vibrio fluvialis | CIP103355b | − | − |
| Vibrio alginolyticus | ATCC 17749b | − | − |
| Vibrio parahaemolyticus | CIP75.2b | − | − |
| Vibrio vulnificus | ATCC 27562b | − | − |
+, bright EFM signal; −, no EFM signal.
Gift from Christopher Grim.
Gift from Munirul Alam.
Quantification of V. cholerae cells from pure cultures.
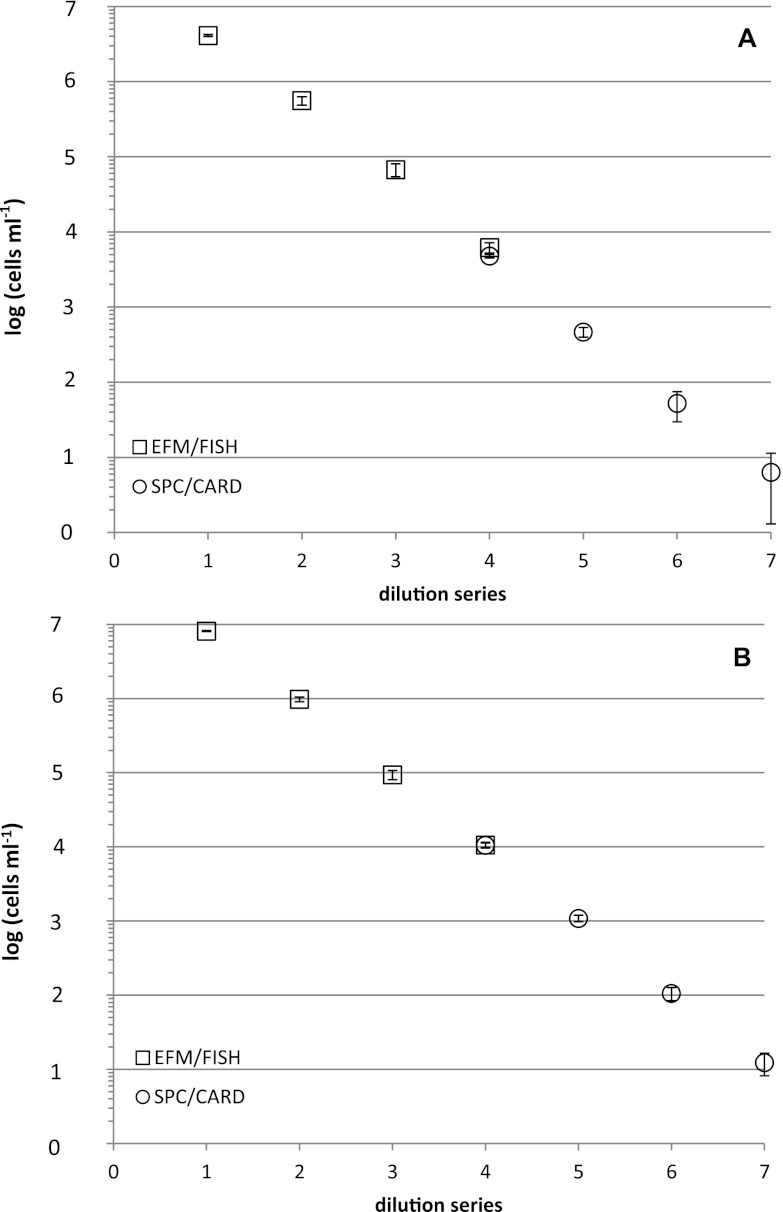
Dilution series were carried out with cells from pure cultures to test the quantitative sensitivity of the new CARD-FISH protocol combined with SPC. Due to practical reasons (the ChemScan RDI system is able to quantify maximally 5 × 104 particles per membrane), the four highest concentrations were determined with EFM and FISH while the four lowest concentrations were determined with SPC and CARD-FISH, with one overlapping concentration. Figure 1A shows the dilution series of the V. cholerae non-O1/non-O139 test strain. Cell numbers per ml started at a concentration of 4.10 × 106 cells ml−1 (first dilution step) and went down to a final value of 6 cells ml−1 (last dilution step). Coefficients of variation (CV %) ranged between 2.9% for the highest concentration, determined via SPC, and 39.7% for the lowest concentration (6 cells). Figure 1B shows the same experiment carried out with a V. cholerae serotype O1 strain. Cell numbers per ml ranged from 8.10 × 106 after the first dilution step to 1.2 × 101 cells ml−1 after the last dilution step. Coefficients of variation ranged between 3.3% for the highest concentration, determined via SPC, and 16.9% for the lowest concentration (12 cells). In both cases, regression curves with excellent predictive abilities were obtained. Coefficients of determination (r2) lay between 0.9938 and 0.9976 for the V. cholerae non-O1/non-O139 experiment and between 0.9986 and 0.9996 for the V. cholerae O1 experiment. The number of observed CARD-FISH cells matched the expected numbers according to the dilution series. No cell losses due to a potential burst of cells during the application of the CARD-FISH procedure were observed. This can be demonstrated by comparing cell numbers from EFM/FISH detection and SPC/CARD-FISH detection.
Fig 1.
Enumeration of V. cholerae non-O1/non-O139 (A) and V. cholerae O1 (B) with EFM combined with FISH and SPC combined with CARD-FISH in 10-fold dilution series (n = 3). Error bars indicate double standard deviations.
To further evaluate the levels of precision, as gained by the dilution experiments with cells from pure cultures, precision levels of the ChemScan RDI were also determined for three concentrations with fluorescent latex beads. Results from latex beads were in agreement with results from bacterial cells. Table 2 demonstrates that the coefficient of variation is low (less than 6.5%) for 102 and 103 beads per filter. Even with a concentration of ∼101 beads per filter, the coefficient of variation was below 30%.
Table 2.
Determination of variation coefficients, carried out with fluorescent beadsa
| Concn of beads, scale | Concn of beads, detected |
CV % | ||
|---|---|---|---|---|
| Mean value | Minimum value | Maximum value | ||
| ∼103 | 1,238 | 1,136 | 1,336 | 3.6 |
| ∼102 | 122 | 112 | 133 | 6.0 |
| ∼101 | 13 | 8 | 19 | 26.5 |
For each concentration (in beads ml−1), 30 subsamples were analyzed.
Spiking experiments under natural sample matrix conditions.
Spiking experiments were carried out to test the performance of the newly developed SPC/CARD-FISH protocol with cells having a low rRNA content and to examine the recovery rate of cells spiked into a natural environmental water sample. As shown in Table 3 the recovery rate was between 93% for the lowest spiked cell concentration and 98% for the highest cell concentration. V. cholerae/V. mimicus background cell numbers in unspiked lake water were below 10 cells ml−1 and were considered in the calculation of the recovery rate.
Table 3.
Concentrations of V. cholerae cells grown until late stationary growth phase (low rRNA content) and spiked into 1× PBS or into freshly collected lake water
| Concn, scale (cells ml−1) | Concn of cells (cells ml−1), spiked intoa: |
Recovery rate (%) | |||
|---|---|---|---|---|---|
| 1× PBS |
Lake water |
||||
| Mean | SD | Mean | SD | ||
| ∼104 | 15,721 | 200 | 15,408 | 243 | 98.0 |
| ∼103 | 1,680 | 80 | 1,619 | 91 | 96.1 |
| ∼102 | 179 | 18 | 171 | 18 | 92.7 |
SD, standard deviation.
Quantification of V. cholerae/V. mimicus cells in environmental samples.
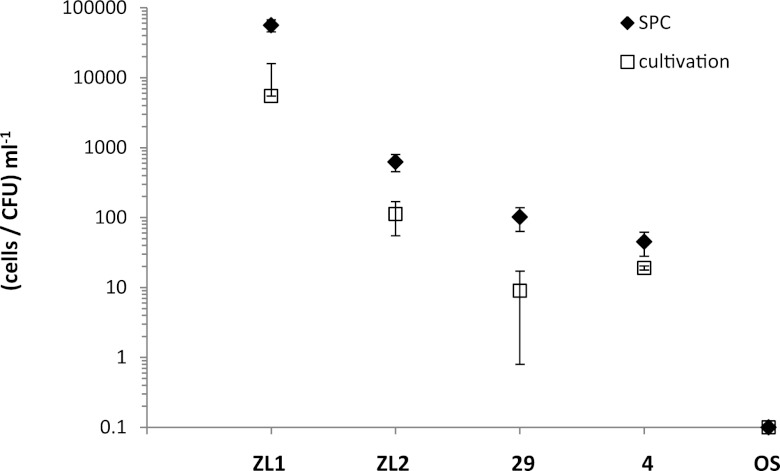
To show that the newly developed protocol is applicable to environmental samples, water samples from the lake Neusiedler See as well as from two shallow soda lakes were investigated. A comparison of the results determined by SPC and those obtained by cultivation are shown in Fig. 2. The concentrations of V. cholerae cells enumerated by cultivation ranged from 5.5 × 103 to 1.1 × 102 CFU ml−1 in the Zicklacke (samples were taken in August for ZL1 and in June for ZL2) and from 9 to 19 CFU ml−1 for sampling points 4 and 29 in the lake Neusiedler See. No V. mimicus-positive colonies were identified via API 20E. Cell numbers of V. cholerae/V. mimicus quantified by CARD-FISH combined with SPC were always higher than results from cultivation. Cell numbers ranged from 5.6 × 104 to 6.2 × 102 cells ml−1 for sampling points ZL1 and ZL2. For the sampling points 29 and 4 in the Neusiedler See, cell numbers were between 1.0 × 102 and 4.5 × 101 cells ml−1. Neither with cultivation nor with SPC was it possible to detect and enumerate V. cholerae/V. mimicus from the soda lake Oberer Stinker. The ratios of SPC counts to cultivation-based counts varied between 2.4 (sampling point 4) and 11.2 (sampling point 29).
Fig 2.
Quantification of V. cholerae by SPC and a cultivation-based method from water samples taken from the lake Neusiedler See (sampling points 29 and 4) and the shallow soda lakes Zicklacke (ZL1 and ZL2) and Oberer Stinker (OS). At the sampling point in Oberer Stinker, no V. cholerae cells were detected. For logarithmic scaling, the cell number here has been set as 0.1 (n = 3 to 8).
DISCUSSION
Our newly developed protocol for the quantification of V. cholerae/V. mimicus cells in water samples by using CARD-FISH combined with solid-phase cytometry revealed excellent applicability in terms of rapidity, specificity, and sensitivity.
Rapidity.
The development of rapid methods for the quantification of microorganisms, especially those occurring at low cell concentrations, is still a challenging topic. Cultivation-based methods with additional biochemical confirmation are very time-consuming and require, in the case of V. cholerae and V. mimicus, up to 3 days before results are available. Direct cell-based detection methods like CARD-FISH provide significantly faster results. We demonstrated that even though the CARD-FISH protocol is more time-consuming than the FISH protocol, results can still be obtained within 1 day.
Specificity and sensitivity (qualitative).
The specificity of the probe Vchomim1276, labeled with FAM for V. cholerae and V. mimicus, has been demonstrated before. This probe was already used for quantification of V. cholerae/V. mimicus via FISH on environmental zooplankton (13) and in laboratory experiments (20) as well as for RNA colony blot hybridization for enumeration of culturable V. cholerae/V. mimicus bacteria (13). As for the FISH probe, Vibrio species other than V. cholerae and V. mimicus did not hybridize with the CARD-FISH probe Vchomim1276, labeled with HRP. We also attempted to design new probes specific for either V. cholerae or V. mimicus. However, since the 16S rRNA sequences of these two species differ by only 6 out of 1,456 nucleotides (13), no suitable probes without cross-reaction between these two species were able to be designed (results not shown). For monitoring and risk assessment, a separation of these two closely related species may not be relevant, since they can induce similar diseases.
We demonstrated that the specific position on the 16S rRNA was easily accessible for the probe Vchomim1276, labeled with HRP, in all V. cholerae/V. mimicus strains tested. It can thus be assumed to be a highly sensitive probe for the detection of a broad variety of environmental as well as clinical and pandemic strains. Despite the fact that the HRP-labeled probe is a significantly larger molecule than fluorescent markers, no helper probes were needed to unfold the tertiary structure of the RNA (11).
Our results are consistent with previous studies which demonstrated that FISH gives a too weak signal for SPC (24, 34) and that CARD-FISH is a methodical improvement to increase the fluorescence signal (30). The choice of an appropriate amplification time during the CARD-FISH protocol is especially important for increasing the signal. This is a very sensitive step, since cells can burst when amplification is too long, and at the same time, the signal might not be as bright as possible if the amplification time is too short. We showed that by increasing the amplification time from 10 min (for cells grown until the exponential growth phase) to 45 min (for cells grown until late stationary growth phase), signal intensity can be kept high for these so-called “starved” cells (see Table S1 in the supplemental material). In addition, we demonstrated that there was no significant difference in signal intensity between starved cells spiked into lake water and environmental V. cholerae cells.
Limit of detection and quantification.
The enumeration of fluorescence-labeled microorganisms with SPC is highly sensitive, since this technique can theoretically detect one labeled cell on a membrane (method detection limit [MDL]) (23, 25). Due to the Poisson distribution of randomly distributed cells, the sample limit of detection (SLOD) based on a 95% detection probability would be 3 cells/particles per filtration volume (18). Operational variability (i.e., analytical losses) was assumed to be negligible (M. E. Stevenson, A. P. Blaschke, S. Schauer, M. Zessner, R. Sommer, A. H. Farnleitner, and A. K. T. Kirschner, submitted for publication). In our experiments, we demonstrated that during CARD-FISH, no cell loss occurred compared to that of the regular FISH protocol. This is of high importance, since for the CARD-FISH protocol, more working steps are involved, and especially during permeabilization, cell loss may occur through the bursting of cells. In addition, we found a high recovery rate (>92%) of starved V. cholerae cells spiked into natural lake water and a moderate variability (coefficient of variation < 30%) when enumerating low numbers (∼10) of bacterial targets and fluorescent latex beads. These results indicate that cell loss due to operational factors can be considered negligible in comparison to variability introduced by the Poisson distribution.
According to ISO 8199:2005 (19), the lowest number of cells/particles that can be quantified is 4 cells/particles per volume, based on an acceptable relative precision of 50% (coefficient of variation = 0.50), which seems to be reasonable in microbiology. If one requires a higher relative precision of 30% (coefficient of variation = 0.30), then the limit of quantification would be 11 cells/particles per volume (Stevenson et al., submitted). Based on our data set (Table 2), we demonstrated that we can reliably quantify down to 13 fluorescent beads with a coefficient of variation of 26.5%. For pure cultures, we were able to quantify as few as 6 V. cholerae cells with a relative precision of 39% (coefficient of variation = 0.39) and quantify as few as 12 V. cholerae cells with a relative precision of 17% (coefficient of variation = 0.17). Thus, solid-phase cytometry exhibits a sensitivity similar to that of cultivation (23), with the significant advantage of detecting microorganisms with the VBNC status. The presence of these VBNC bacteria in the environment needs to be further investigated, since several authors suggest that pathogenic VBNC bacteria might be able to maintain their pathogenicity or regain it after resuscitation (12).
Application of the SPC/CARD-FISH protocol for environmental samples.
For the first time, solid-phase cytometry was successfully applied for the detection of cells belonging to the V. cholerae/V. mimicus clade in extremely turbid samples. Environmental samples with a high load of particles are enormously difficult to analyze via epifluorescence microscopy because of the large amount of background, fluorescent particles, and autofluorescent bacteria and/or algae. In this respect, SPC shows its clear advantages when it comes to quantification of rare events. Compared to EFM, a much smaller amount of sample volume can be analyzed, which automatically means a smaller amount of background, particles, and autofluorescence. Still, fluorescent particles and background should be kept as low as possible, since they may be classified by the SPC software as positive signals. One possibility to reduce the background is to pretreat the filter with Evans Blue (an azo dye). Even though all fluorescence events undergo a screening by computer software, not only can the targeted labeled cells be found in the net result map but also false-positive signals which fit into the chosen application. With the provided computer software and the connected microscope equipped with an automated stage, however, it is easily and quickly possible to manually validate the net results and discriminate visually between target cells and false-positive signals. Nevertheless, false positives may not be completely excluded, because our probe, which appears to be specific for the V. cholerae/V. mimicus clade, may potentially bind to the same sequence present in unknown targets. But this is a general issue when molecular biological methods which are based on the use of available sequence information are applied. It also needs to be pointed out that there is a possibility that target cells are overseen by the software and/or application used (false negatives). This may happen when the fluorescent signal is below the chosen threshold. It is important to set the discriminant parameters as narrow as possible. This is crucial, on the one hand, to detect all labeled microorganisms and, on the other hand, to exclude as many false-negative signals as possible from the net result data map.
In our samples, we observed a certain difference between cell numbers achieved by CARD-FISH combined with SPC and numbers of CFU determined by cultivation: concentrations from cultivation were mostly lower. This may result from several reasons. One reason may be that a CFU does not necessarily arise from one single cell. One CFU can also originate from two or more cells, whereas by SPC, every single cell can be detected. Another important point is the fact that selective agar like TCBS might not be an appropriate medium for stressed cells, and therefore, cell propagation might not be observed. The lower results can also be explained by the presence of a certain percentage of VBNC cells, which can be detected by SPC but not via cultivation.
Conclusion.
CARD-FISH in combination with solid-phase cytometry is a very powerful tool for the rapid and sensitive quantification of V. cholerae/V. mimicus cells, especially when V. cholerae/V. mimicus occur at low cell numbers, which is usually the case in environmental samples. SPC is the only cultivation-independent, cell-based method able to accurately quantify these rare events. Moreover, because the CARD-FISH protocol requires the presence of ribosomes, surrounding proteins, and an intact cell wall, the proposed protocol allows quantification of V. cholerae/V. mimicus cells, including cells in the VBNC state, in environmental samples.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christopher Grim (University of Maryland) and Munirul Alam (International Centre for Diarrheal Disease Research, Bangladesh) for providing a variety of Vibrio strains. Additionally, we thank Julia Baudart and Philippe Lebaron (Banyuls Oceanology Observatory) for helpful suggestions concerning the solid-phase cytometer ChemScan RDI.
The study was financed by the Austrian Science Fund (FWF, project number P21625-B20).
This study is a joint publication of the Interuniversity Cooperation Centre (ICC) Water and Health.
Footnotes
Published ahead of print 10 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Amann R, Fuchs BM. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6:339–348 [DOI] [PubMed] [Google Scholar]
- 2. Amann R, Fuchs BM, Behrens S. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231–236 [DOI] [PubMed] [Google Scholar]
- 3. Amann RI, Ludwig W, Schleifer K-H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aurell H, et al. 2004. Rapid detection and enumeration of Legionella pneumophila in hot water systems by solid-phase cytometry. Appl. Environ. Microbiol. 70:1651–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baron S, Chevalier S, Lesne J. 2007. Vibrio cholerae in the environment: a simple method for reliable identification of the species. J. Health Popul. Nutr. 25:312–318 [PMC free article] [PubMed] [Google Scholar]
- 6. Baudart J, Lebaron P. 2010. Rapid detection of Escherichia coli in waters using fluorescent in situ hybridization, direct viable counting and solid phase cytometry. J. Appl. Microbiol. 109:1253–1264 [DOI] [PubMed] [Google Scholar]
- 7. Cheasty T, Said B, Threlfall EJ. 1999. V. cholerae non-O1: implications for man? Lancet 354:89–90 [DOI] [PubMed] [Google Scholar]
- 8. Chowdhury MAR, Yamanaka H, Miyoshi S, Aziz KMS, Shinoda S. 1989. Ecology of Vibrio mimicus in aquatic environments. Appl. Environ. Microbiol. 55:2073–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cottingham KL, Chiavelli DA, Taylor RK. 2003. Environmental microbe and human pathogen: the ecology and microbiology of Vibrio cholerae. Front. Ecol. Environ. 1:80–86 [Google Scholar]
- 10. Eiler A, et al. 2003. Factors controlling extremely productive heterotrophic bacterial communities in shallow soda pools. Microb. Ecol. 46:43–54 [DOI] [PubMed] [Google Scholar]
- 11. Fuchs BM, Glockner FO, Wulf J, Amann R. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garcia-Armisen T, Servais P. 2004. Enumeration of viable E. coli in rivers and wastewaters by fluorescent in situ hybridization. J. Microbiol. Methods 58:269–279 [DOI] [PubMed] [Google Scholar]
- 13. Grim CJ, et al. 2009. RNA colony blot hybridization method for enumeration of culturable Vibrio cholerae and Vibrio mimicus bacteria. Appl. Environ. Microbiol. 75:5439–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gubala AJ. 2006. Multiplex real-time PCR detection of Vibrio cholerae. J. Microbiol. Methods 65:278–293 [DOI] [PubMed] [Google Scholar]
- 15. Hasan JAK, et al. 1994. Cholera DFA—an improved direct fluorescent monoclonal antibody staining kit for rapid detection and enumeration of Vibrio cholerae O1. FEMS Microbiol. Lett. 120:143–148 [DOI] [PubMed] [Google Scholar]
- 16. Hasan NA, et al. 2010. Comparative genomics of clinical and environmental Vibrio mimicus. Proc. Natl. Acad. Sci. U. S. A. 107:21134–21139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heidelberg JF, Heidelberg KB, Colwell RR. 2002. Bacteria of the γ-subclass Proteobacteria associated with zooplankton in Chesapeake Bay. Appl. Environ. Microbiol. 68:5498–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. International Organization of Standardization 2000. Water quality—guidance on validation of microbiological methods, p 47 ISO/TR 13843:2000 International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 19. International Organization of Standardization 2005. Water quality—general guidance on the enumeration of micro-organisms by culture, p 38 ISO 8199:2005 International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 20. Kirschner AKT, et al. 2011. Interaction of Vibrio cholerae non-O1/non-O139 with copepods, cladocerans and competing bacteria in the large alkaline lake Neusiedler See, Austria. Microb. Ecol. 61:496–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirschner AKT, et al. 2008. Rapid growth of planktonic Vibrio cholerae non-O1/non-O139 strains in a large alkaline lake in Austria: dependence on temperature and dissolved organic carbon quality. Appl. Environ. Microbiol. 74:2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lebaron P, et al. 1997. A new sensitive, whole-cell hybridization technique for detection of bacteria involving a biotinylated oligonucleotide probe targeting rRNA and tyramide signal amplification. Appl. Environ. Microbiol. 63:3274–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lemarchand K, Parthuisot N, Catala P, Lebaron P. 2001. Comparative assessment of epifluorescence microscopy, flow cytometry and solid-phase cytometry used in the enumeration of specific bacteria in water. Aquat. Microb. Ecol. 25:301–309 [Google Scholar]
- 24. Lepeuple A-S, Delabre K, Gilouppe S, Intertaglia L, Roubin M-R. 2003. Laser scanning detection of FISH-labelled Escherichia coli from water samples. Water Sci. Technol. 47:123–129 [PubMed] [Google Scholar]
- 25. Lisle JT, Hamilton MA, Willse AR, McFeters GA. 2004. Comparison of fluorescence microscopy and solid-phase cytometry methods for counting bacteria in water. Appl. Environ. Microbiol. 70:5343–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lyon WJ. 2001. TaqMan PCR for detection of Vibrio cholerae O1, O139, non-O1, and non-O139 in pure cultures, raw oysters, and synthetic seawater. Appl. Environ. Microbiol. 67:4685–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mignon-Godefroy K, Guillet JG, Butor C. 1997. Solid phase cytometry for detection of rare events. Cytometry 27:336–344 [PubMed] [Google Scholar]
- 28. Morris JG., Jr 1990. Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol. Rev. 12:179–191 [DOI] [PubMed] [Google Scholar]
- 29. Moter A, Gobel UB. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41:85–112 [DOI] [PubMed] [Google Scholar]
- 30. Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125–139 [DOI] [PubMed] [Google Scholar]
- 32. Sack DA, Sack RB, Nair GB, Siddique AK. 2004. Cholera. Lancet 363:223–233 [DOI] [PubMed] [Google Scholar]
- 33. Schönhuber W, Fuchs B, Juretschko S, Amann R. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tobe K. 2006. Automated detection and enumeration for toxic algae by solid-phase cytometry and the introduction of a new probe for Prymnesium parvum (Haptophyta: Prymnesiophyceae). J. Plankton Res. 28:643–657 [Google Scholar]
- 35. Wang D, et al. 2011. Genome sequencing reveals unique mutations in characteristic metabolic pathways and the transfer of virulence genes between V. mimicus and V. cholerae. PLoS One 6:e21299 doi:10.1371/journal.pone.0021299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. WHO 2002. Vibrio cholerae, p 119–142 In Guidelines for drinking water quality, 2nd ed World Health Organization, Geneva, Switzerland [Google Scholar]
- 37. Wilhartitz I, et al. 2007. Prokaryotic community analysis with CARD-FISH in comparison with FISH in ultra-oligotrophic ground- and drinking water. J. Appl. Microbiol. 103:871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.