Abstract
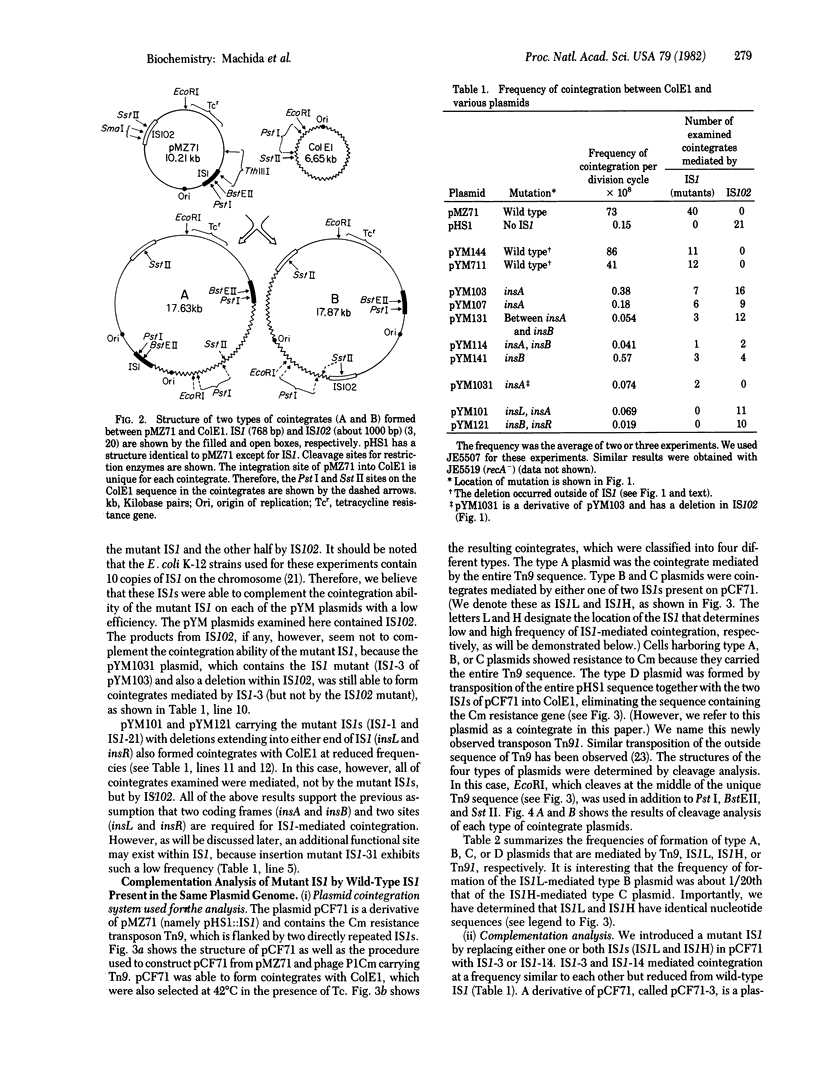
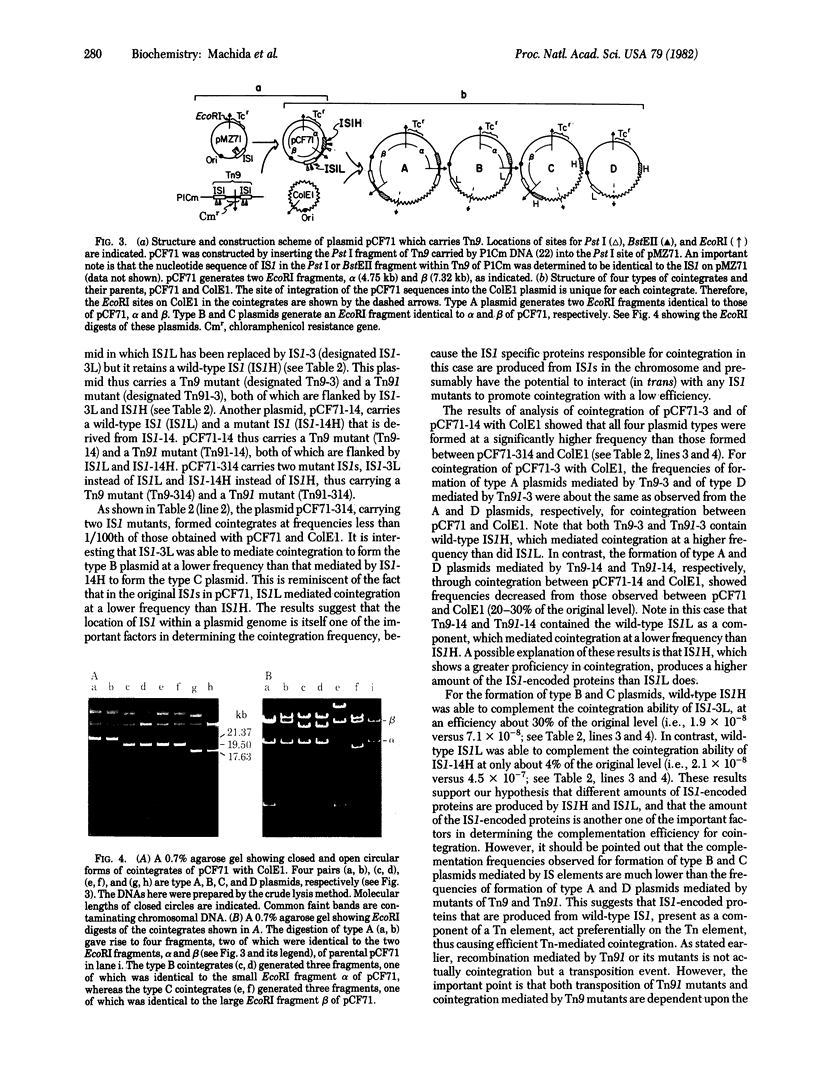
We demonstrate that mutants with deletions at either end of the insertion sequence IS1 lose the ability to mediate cointegration of two plasmids, whereas mutants with deletions or an insertion within IS1 can mediate cointegration at a reduced frequency. These results, together with the nucleotide sequence analysis of the IS1 mutants, indicate that the two ends of IS1 (insL and insR) and two genes (insA and insB) that are encoded by IS1 are required for cointegration. Using a plasmid carrying two copies of IS1, we found that the individual IS1s mediate cointegration at different characteristic frequencies, and that each of two parts of plasmid DNA segments flanked by the two IS1s is a transposon, mediating plasmid cointegration at a unique frequency. When one IS1 was replaced with a mutant IS1, the remaining wild-type IS1 complemented the cointegration ability of the mutant IS1 as well as a resulting mutant transposon that was then flanked by a wild-type IS1 and a mutant IS1. The efficiency of this complementation reflected the characteristic ability of an individual IS1 present on the plasmid to promote cointegration. The results suggest that the IS1-encoded proteins are produced in different amounts, depending on the location of IS1 in the plasmid, and that these amounts determine the efficiency of complementation of the cointegration ability of a mutant IS1 as well as a mutant transposon. However, the location of an individual IS1 itself can also determine the frequency of cointegration in the presence of a given amount of the IS1 proteins. On the basis of the observation that the cointegration ability of a mutant IS1 is less efficiently complemented than is the ability of a mutant transposon, we also suggest that the IS1-encoded proteins can function in trans, but act preferentially on the IS1 or transposon sequence from which they are produced in promoting cointegration.
Keywords: IS1 and Tn9 mutants, complementation analysis, amounts of IS1 proteins, position effect, preferential binding
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Fiandt M., Szybalski W., Malamy M. H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119(3):223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):125–133. doi: 10.1101/sqb.1981.045.01.021. [DOI] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch H. J., Starlinger P., Brachet P. Two kinds of insertions in bacterial genes. Mol Gen Genet. 1972;119(3):191–206. doi: 10.1007/BF00333858. [DOI] [PubMed] [Google Scholar]
- Johnsrud L., Calos M. P., Miller J. H. The transposon Tn9 generates a 9 bp repeated sequence during integration. Cell. 1978 Dec;15(4):1209–1219. doi: 10.1016/0092-8674(78)90047-8. [DOI] [PubMed] [Google Scholar]
- Johnsrud L. DNA sequence of the transposable element IS1. Mol Gen Genet. 1979 Jan 31;169(2):213–218. doi: 10.1007/BF00271673. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Foster T. J., Davis M. A., Hanley-Way S., Halling S. M., Lundblad V., Takeshita K. Genetic organization of Tn10 and analysis of Tn10-associated excision events. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):225–238. doi: 10.1101/sqb.1981.045.01.035. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick M., Wishart W., Ohtsubo H., Heffron F., Ohtsubo E. Plasmid cointegrates and their resolution mediated by transposon Tn3 mutants. Gene. 1981 Nov;15(2-3):103–118. doi: 10.1016/0378-1119(81)90120-7. [DOI] [PubMed] [Google Scholar]
- Nyman K., Nakamura K., Ohtsubo H., Ohtsubo E. Distribution of the insertion sequence IS1 in gram-negative bacteria. Nature. 1981 Feb 12;289(5798):609–612. doi: 10.1038/289609a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Rosenbloom M., Schrempf H., Goebel W., Rosen J. Site specific recombination involved in the generation of small plasmids. Mol Gen Genet. 1978 Feb 16;159(2):131–141. doi: 10.1007/BF00270886. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H., McCormick M., Machida C., Machida Y. Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):283–295. doi: 10.1101/sqb.1981.045.01.041. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H. Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci U S A. 1980 Feb;77(2):750–754. doi: 10.1073/pnas.77.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Nyman K., Doroszkiewicz W., Ohtsubo E. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature. 1981 Aug 13;292(5824):640–643. doi: 10.1038/292640a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Isolation of inverted repeat sequences, including IS1, IS2, and IS3, in Escherichia coli plasmids. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2316–2320. doi: 10.1073/pnas.73.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Zenilman M., Ohtsubo E. Insertion element IS102 resides in plasmid pSC101. J Bacteriol. 1980 Oct;144(1):131–140. doi: 10.1128/jb.144.1.131-140.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner J. L., Guyer M. S. Transposition of IS1-lambdaBIO-IS1 from a bacteriophage lambda derivative carrying the IS1-cat-IS1 transposon (Tn9). Mol Gen Genet. 1980 Apr;178(1):111–120. doi: 10.1007/BF00267219. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya T., Sato S. A spite specific endonuclease from thermus thermophilus 111, Tth111I. Nucleic Acids Res. 1980 Jan 11;8(1):43–56. doi: 10.1093/nar/8.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]




