Abstract
Identification and design of new cellulolytic enzymes with higher catalytic efficiency are a key factor in reducing the production cost of lignocellulosic bioalcohol. We report here identification of a novel β-glucosidase (Gluc1C) from Paenibacillus sp. strain MTCC 5639 and construction of bifunctional chimeric proteins based on Gluc1C and Endo5A, a β-1,4-endoglucanase isolated from MTCC 5639 earlier. The 448-amino-acid-long Gluc1C contained a GH superfamily 1 domain and hydrolyzed cellodextrin up to a five-sugar chain length, with highest efficiency toward cellobiose. Addition of Gluc1C improved the ability of Endo5A to release the reducing sugars from carboxymethyl cellulose. We therefore constructed six bifunctional chimeric proteins based on Endo5A and Gluc1C varying in the positions and sizes of linkers. One of the constructs, EG5, consisting of Endo5A-(G4S)3-Gluc1C, demonstrated 3.2- and 2-fold higher molar specific activities for β-glucosidase and endoglucanase, respectively, than Gluc1C and Endo5A alone. EG5 also showed 2-fold higher catalytic efficiency than individual recombinant enzymes. The thermal denaturation monitored by circular dichroism (CD) spectroscopy demonstrated that the fusion of Gluc1C with Endo5A resulted in increased thermostability of both domains by 5°C and 9°C, respectively. Comparative hydrolysis experiments done on alkali-treated rice straw and CMC indicated 2-fold higher release of product by EG5 than that by the physical mixture of Endo5A and Gluc1C, providing a rationale for channeling of intermediates. Addition of EG5 to a commercial enzyme preparation significantly enhanced release of reducing sugars from pretreated biomass, indicating its commercial applicability.
INTRODUCTION
The energy captured by plants in the form of cellulosic biomass, which is the most abundant carbon source on earth, may provide a renewable and alternative option for transportation fuel (6, 13). The cellulosic biomass could be hydrolyzed enzymatically into the constituent d-glucose and then fermented into bioethanol (5, 10). However, hydrolysis of cellulosic biomass is a complex process and requires synergistic action of three classes of enzymes, namely, endoglucanases (EC 3.2.1.4), exoglucanases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21) (4, 7). Endoglucanases and exoglucanases produce cellodextrins or cellobiose, and the resulting products serve as the substrate for β-glucosidases to produce glucose (3, 32–34), which serves as a direct substrate for ethanolic fermentation. One major hurdle in commercial production of cellulosic ethanol in recent years has been the high cost of these cellulase enzymes (31).
The majority of the enzymes used for the hydrolysis of lignocellulosic biomass are derived from fungi due to their capability to produce copious amounts of cellulases and hemicellulases (8, 30). However, isolation and characterization of novel glycoside hydrolases from bacteria are now widely exploited because of their efficient heterologous production, high specific activity, and less stringent pH requirement (9, 26, 27). The metagenomic work has greatly facilitated identification of unique bacterial cellulases, especially from the guts of herbivores (17, 36). The possibility of cellulolytic bacterial species being engineered for consolidated bioprocessing has further raised interest in identification of robust bacterial cellulase producers (22, 25).
Considering that hydrolysis of cellulosic biomass will require a consortium of cellulases (35), minimizing the number of polypeptides in the consortium is likely to reduce the enzyme cost. Single polypeptides carrying more than one cellulolytic activity that have either been isolated from nature (12, 29) or constructed in the laboratory (2, 11, 18, 19, 23, 24) would certainly have advantages as candidate enzymes in the cellulase consortium.
We earlier cloned and expressed endoglucanase (Endo5A) and endoxylanase (Xyl11D) from a Paenibacillus strain isolated from the gut of cotton bollworm and made its chimeras (1). We now report characterization of β-glucosidase (Gluc1C) from the same species and construction of chimeras between Endo5A and Gluc1C. We further evaluated the thermostability and bifunctional nature of these chimeras and show that the close proximity of Endo5A and Gluc1C in the chimera helps in channeling of the intermediates and enhancing cellulolytic activity.
MATERIALS AND METHODS
Bacterial strains, plasmids and reagents.
Escherichia coli DH5α (Invitrogen) was used as the host strain for gene cloning and protein expression. Genes encoding β-glucosidase and endoglucanase were derived from Paenibacillus sp. strain MTCC 5639 (the same as ICGEB2008) (1). pQE30 (Qiagen) was used as the expression vector for cloning and expressing recombinant enzymes. Enzymes for molecular biology work were procured from New England BioLabs (NEB), and PCRs were performed using high-fidelity Phusion DNA polymerase (Finnzymes).
Cloning of β-glucosidase.
Phylogenetic analysis of the endoglucanase (Endo5A) (GenBank accession no. HQ657203) and xylanase (Xyl11D) (GenBank accession no. HQ657204) sequence of Paenibacillus sp. strain MTCC 5639 showed a high level of similarity to those of Paenibacillus polymyxa. We therefore designed primers Gluc1C-BamHI-F and Gluc1C-SalI-R (Table 1) based on the β-glucosidase gene sequence of Paenibacillus polymyxa (GenBank accession no. M60211) and amplified the β-glucosidase (Gluc1C) gene from the Paenibacillus sp. strain MTCC 5639 genome. The gene for Gluc1C was further cloned in pQE30 and sequenced commercially (Macrogen), and the nucleotide sequence was deposited in the GenBank database.
Table 1.
List of primers used for amplification of various constructs
| Primer name | Primer sequence |
|---|---|
| Gluc1C-BamHI-F | ACTGGATCCATGAGCGAGAATACCTTTA |
| Gluc1C-SalI-R | CACTGTCGACTTAAAACCCGTTCTTCGC |
| Endo-BamHI-EG1-F | AGGATCCGCCAGCGTGAAAGGATA |
| Endo-SacI-EG1-R | AGAGCTCTTCGGCGCTTGCTTTCG |
| Gluco-SacI-EG1-F | AGAGCTCATGAGCGAGAATACCTTTA |
| Gluco-SacI-EG2-R | AGAGCTCAAACCCGTTCTTCGCCAT |
| Endo-SacI-EG2-F | AGAGCTCGCCAGCGTGAAAGGATA |
| Endo-SalI-EG2-R | TACAGTCGACCTATTCGGCGCTTGCTTTCG |
| Endo-SacI-EG3-R | AGAGCTCCGACCCACCACCGCCCGAGCCACCGCCACCTTCGGCGCTTGCTTTCG |
| Gluco-SacI-EG4-R | AGAGCTCGACCCACCACCGCCCGAGCCACCGCCACCAAACCCGTTCTTCGCCAT |
| Endo-SacI-EG5-R | AGAGCTCCGAGCCACCGCCACCCGACCCACCACCGCCCGAGCCACCGCCACCTTCGGCGCTTGCTTTCG |
| Gluco-SacI-EG6-R | AGAGCTCCGAGCCACCGCCACCCGACCCACCACCGCCCGAGCCACCGCCACCAAACCCGTTCTTCGCCAT |
Design and construction of fusion proteins.
Six fusion constructs consisting of Endo5A and Gluc1C, EG1 to EG6, were designed based on change in position and introduction of a linker in between. Two kinds of glycine-serine linkers were used in the construct: GGGGSGGGGS [named (G4S)2] and GGGGSGGGGSGGGGS [named (G4S)3]. The fusion constructs were amplified using the primers listed in Table 1 as follows. EG1 or Endo5A-Gluc1C, where Endo5A was at the N terminus and Gluc1C was at the C terminus, was constructed by amplifying Endo5A and Gluc1C using primer sets Endo-BamHI-EG1-F/Endo-SacI-EG1-R and Gluco-SacI-EG1-F/Gluc1C-SalI-R, respectively. EG2 or Gluc1C-Endo5A, where Gluc1C was at the N terminus and Endo5A was at the C terminus, was constructed by amplifying Gluc1C and Endo5A using primer sets Gluco1C-BamHI-F/Gluco-SacI-EG2-R and Endo-SacI-EG2-F/Endo-SalI-EG2-R, respectively. EG3 or Endo5A-(G4S)2-Gluc1C, where Endo5A was at the N terminus and Gluc1C was at the C terminus with (G4S)2 in between, was constructed by amplifying Endo5A and Gluc1C using primer sets Endo-BamHI-EG1-F/Endo-SacI-EG3-R and Gluco-SacI-EG1-F/Gluc1C-SalI-R, respectively. EG4 or Gluc1C-(G4S)2-Endo5A, where Gluc1C was at the N terminus and Endo5A was at the C terminus with (G4S)2 in between, was constructed by amplifying Gluc1C and Endo5A using primer sets Gluco1C-BamHI-F/Gluco-SacI-EG4-R and Endo-SacI-EG2-F/Endo-SalI-EG2-R, respectively. EG5 or Endo5A-(G4S)3-Gluc1C, where Endo5A was at the N terminus and Gluc1C was at the C terminus with (G4S)3 in between, was constructed by amplifying Endo5A and Gluc1C using primer sets Endo-BamHI-EG1-F/Endo-SacI-EG5-R and Gluco-SacI-EG1-F/Gluc1C-SalI-R, respectively. EG6 or Gluc1C-(G4S)3-Endo5A, where Gluc1C was at the N terminus and Endo5A was at the C terminus with (G4S)3 in between, was constructed by amplifying Gluc1C and Endo5A using primer sets Gluco1C-BamHI-F/Gluco-SacI-EG6-R and Endo-SacI-EG2-F/Endo-SalI-EG2-R, respectively. All of the amplified products were digested with restriction enzymes, as noted in the names of the primers, and cloned at the corresponding restriction sites of pQE30 to obtain plasmids pQE-EG1 to pQE-EG6.
Enzyme expression and purification.
The plasmid constructs pQE-Endo5A (1), pQE-Gluc1C, and pQE-EG1 to pQE-EG6 mentioned above were used to transform E. coli DH5α, and recombinant proteins were expressed at the 1-liter shake-flask level in Luria-Bertani (LB) medium containing ampicillin upon induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The cells were lysed via sonication, and recombinant proteins were purified on Ni-nitrilotriacetic acid (NTA) resin (Qiagen) as per the manufacturer's guidelines. The purified fractions were pooled, dialyzed against the assay buffer, and used for further characterization.
Enzyme assays.
β-Glucosidase activity was determined by incubating the enzyme with 5 mM para-nitrophenyl-d-glucopyranoside (pNPG) in 50 mM citrate buffer (pH 6.0) in 0.55 ml at 50°C for 15 min (14). The reaction was stopped by adding 1 ml of 1% sodium carbonate, and the absorbance was measured at 400 nm. One unit of β-glucosidase activity was defined as the amount of enzyme that produced 1 μmol of para-nitrophenol per minute. Endoglucanase activity of the recombinant proteins was measured by the 3,5-dinitrosalicylic acid (DNSA)-based Nelson and Somogyi method as described earlier (1). One unit of endoglucanase was defined as 1 μmol of reducing sugar released from carboxymethyl cellulose (CMC) per minute. To evaluate the impact of β-glucosidase on enhancement of Endo5A activity, increasing amounts of Gluc1C were added to 8 mU of Endo5A, and the amount of reducing sugar released was estimated by the DNSA method. To evaluate the endoglucanase activities of recombinant chimeras, 25 mM glucono-δ-lactose, an inhibitor for β-glucosidase activity, was added to the reaction mixture. The protein concentration was measured according to the bicinchoninic acid assay (BCA) method with bovine serum albumin (BSA) as the standard.
Measurement of pH and temperature optima and thermostability of the enzymes.
The activities of purified recombinant enzymes were measured under different pH conditions to determine the optimal pH. Four buffers of various pH ranges—50 mM citrate phosphate buffer (pH 3.0 to 6.0), 50 mM sodium phosphate buffer (pH 6.0 to 7.0), 50 mM Tris-HCl buffer (pH 7.0 to 9.0), and 50 mM carbonate buffer (pH 10.0)—were used in the assay, and enzyme activity was determined as described above. The optimal temperature for enzyme activity was determined by incubating reaction mixtures over a temperature range of 20 to 70°C and determining their activity. The thermal stability of the β-glucosidase domain was determined by incubating the enzymes at various temperatures for 30 min, cooling them down to ambient temperature, and then initiating the reactions by the addition of the substrate. The formation of p-nitrophenol was determined as described before. To analyze the in silico thermostability of β-glucosidase, the sequences of Gluc1C and EG5 were submitted to the I-TASSER web server, and the structural parameters obtained were analyzed using Discovery Studio 3.1 Visualizer software as detailed earlier (21).
Determination of kinetic parameters.
For determination of Km and Vmax, seven different substrate concentrations were used in the range of 0.15 to 10 mM pNPG for β-glucosidase and nine different substrate concentrations were used in the range of 0.1 to 0.9 mg/ml CMC for endoglucanase. The Km and Vmax were determined directly from the hyperbolic curve fitting of the Michaelis-Menten equation generated using Sigma Plot (Jandel Scientific). kcat was determined by the formula Vmax/Et, where Et is the total enzyme concentration in μmol ml−1.
Pretreated biomass hydrolysis.
Cellulosic biomass used for the enzyme amenability assay was prepared by a process of alkaline fractionation (A. M. Lali et al., 2010, patent application PCT/IN2010/000355) from rice straw. Solid biomass (7% [wt/vol]) was reacted with 10% (wt/vol) aqueous NaOH at 130°C for 20 min. The recovered residue was repeatedly washed with water to remove the residual alkali and further neutralized to pH 7.0 for the reaction. For enzymatic hydrolysis, the pretreated rice straw (11.8 g/liter) was incubated with different amounts of Endo5A, Endo5A-Gluc1C mixture, and EG5 in 50 mM sodium phosphate buffer (pH 7.0) for 4 h at 40°C, since β-glucosidase activity is stable at this temperature for a longer duration. Sugar released upon addition of EG5 to commercial cellulase NS50013 (Novozyme) was determined by incubating pretreated rice straw with either NS50013 (1 filter paper unit and 6 CMC units per g biomass) alone or NS50013 along with EG5 (6 CMC units per g biomass). The glucose, cellobiose, and cellotriose released were analyzed by high-pressure liquid chromatography (HPLC) (Agilent) using an Aminex 85H column (Bio-Rad) with 5 mM H2SO4 as the eluent.
CD spectroscopy.
A spectropolarimeter (Jasco, Easton, MD) with a rectangular quartz cell with a 0.1-cm path length was used to perform circular dichroism (CD) spectroscopy. Spectra were acquired using an 8-s time response and a 100-nm/s scan speed. The spectra obtained were averaged for five acquisitions. Proteins were analyzed at 1.5 μM in 50 mM sodium phosphate buffer (pH 7.5). All spectra, with correction for the buffer background, were acquired from 200 to 250 nm. The temperature of the sample was varied during the experiments and measured by a sensor built into the cuvette holder. The experimental data obtained for the CD signal at 220 nm with respect to temperature were normalized (Origin software), and a sigmoidal curve (SigmaPlot) was plotted to calculate the melting temperature (Tm).
Nucleotide sequence accession number.
The nucleotide sequence of the gene coding for Gluc1C has been deposited in the GenBank database under accession no. JQ713769.
RESULTS
Characterization of a β-glucosidase from Paenibacillus sp. strain MTCC 5639.
Paenibacillus ICGEB2008 (MTCC 5639) (1), isolated from the gut of Helicoverpa armigera, showed significant pNPG activity (2.4 mU/108 cells) in the intracellular fraction. We, therefore, designed primers to amplify the β-glucosidase gene from the genome of Paenibacillus sp. strain MTCC 5639 based on the sequence available from the neighboring strain, cloned the amplified product in an E. coli vector, and sequenced it. An open reading frame (ORF) of 1,347 bp was detected in the amplified product that encoded a 448-amino-acid-long 51.7-kDa polypeptide with a theoretical pI of 5.06. The ORF sequence (GenBank accession no. JQ713769) exhibited 99% identity at the nucleotide and protein level with Bacillus polymyxa β-glucosidase (GenBank accession no. M60211.1) and contained the glycosyl hydrolase superfamily 1 domain without any signal sequence. We expressed full-length β-glucosidase (Gluc1C) of Paenibacillus sp. strain MTCC 5639 in E. coli along with the 6-histidine tag, purified it through metal affinity chromatography, and characterized its activity (Fig. 1 and Table 2). Gluc1C hydrolyzed oligosaccharides of up to five chain lengths, although the efficiencies of hydrolysis of cellobiose and cellotriose were significantly higher than those of cellotetraose and cellopentaose (see Fig. S1 and Table S1 in the supplemental material).
Fig 1.
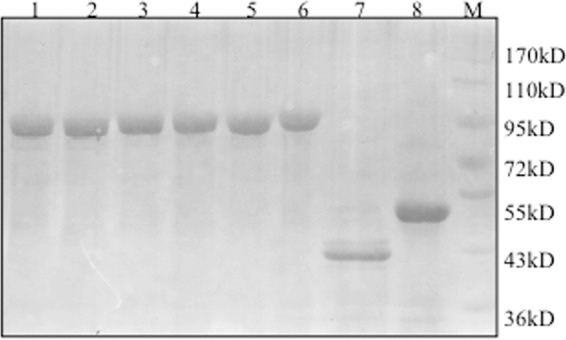
SDS-PAGE gel profile of recombinant enzymes after affinity purification. Lane 1, EG1; lane 2, EG2; lane 3, EG3; lane 4, EG4; lane 5, EG5; lane 6, EG6; lane 7, Endo5A; lane 8, Gluc1C; lane M, molecular mass marker.
Table 2.
Specific activities of recombinant enzymes
| Activity and recombinant protein | Sp act (U mg−1) | Molar sp act (U μmol−1) | Fold change in activity |
|---|---|---|---|
| β-Glucosidase | |||
| Gluc1C | 5.8 | 307 | 1.0 |
| EG1 (Endo5A-Gluc1C) | 6.4 | 608 | 1.9 |
| EG2 (Gluc1C-Endo5A) | 4.4 | 418 | 1.3 |
| EG3 [Endo5A-(G4S)2-Gluc1C] | 3.2 | 304 | 0.98 |
| EG4 [Gluc1C-(G4S)2-Endo5A] | 4.1 | 389 | 1.2 |
| EG5 [Endo5A-(G4S)3-Gluc1C] | 10.5 | 997 | 3.2 |
| EG6 [Gluc1C-(G4S)3-Endo5A] | 4.8 | 456 | 1.5 |
| Endoglucanase | |||
| Endo5A | 24.2 | 1,016 | 1.0 |
| EG1 (Endo5A-Gluc1C) | 19.7 | 1,871 | 1.8 |
| EG2 (Gluc1C-Endo5A) | 21.8 | 2,071 | 2.0 |
| EG3 [Endo5A-(G4S)2-Gluc1C] | 20.5 | 1,947 | 1.9 |
| EG4 [Gluc1C-(G4S)2-Endo5A] | 21.6 | 2,054 | 2.0 |
| EG5 [Endo5A-(G4S)3-Gluc1C] | 22.6 | 2,147 | 2.1 |
| EG6 [Gluc1C-(G4S)3-Endo5A] | 21.7 | 2,061 | 2.0 |
We had shown previously that endoglucanase (Endo5A) of Paenibacillus sp. strain MTCC 5639 yields cellobiose upon incubation with CMC as the substrate (1). It was likely that addition of Gluc1C in the same reaction mixture would increase the reducing sugar concentration since Gluc1C would convert this cellobiose into glucose. Indeed, there was a gradual increase in reducing sugar concentration upon increasing addition of Gluc1C (Fig. 2). We therefore envisaged that the construction of a bifunctional fusion protein with the catalytic domains from Endo5A and Gluc1C would be likely to have enhanced cellulolytic activity and reduced production cost.
Fig 2.
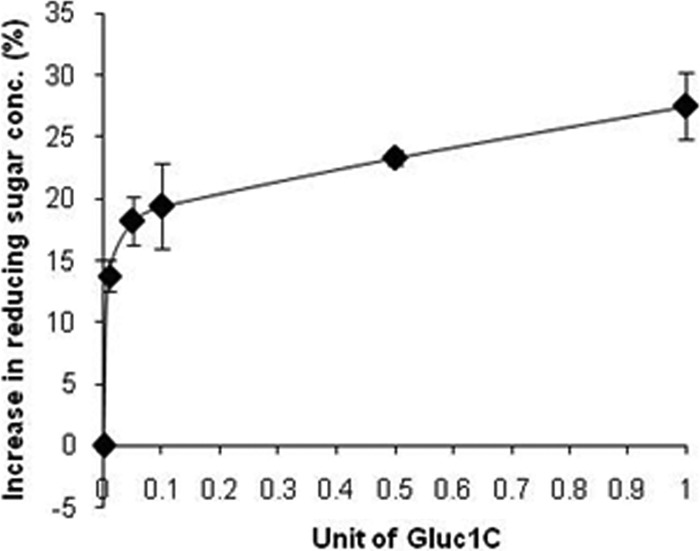
Impact of Gluc1C addition to Endo5A on reducing sugar concentration. Recombinant Endo5A and Gluc1C purified through metal affinity chromatography were used for the assay. Endo5A (0.008 U) was incubated with CMC as the substrate in the presence of various amounts of Gluc1C. The graph has been plotted as the percentage of increase in reducing sugar concentration upon addition of Gluc1C compared to that with Endo5A alone.
Design, synthesis, and characterization of bifunctional fusion proteins.
It has often been observed previously that the fusion of two or more catalytic domains leads to reduction in catalytic activity of one or both of the domains (2, 18, 19). We, therefore, decided to construct six fusion proteins, each containing a catalytic domain of Endo5A and Gluc1C, as mentioned in Materials and Methods. We selected a commonly used glycine-serine linker of two chain lengths to join the two enzymes, and we changed the positions of the enzymes as well. The resultant constructs EG1 to EG6 were expressed in E. coli with a 6-histidine tag at the N terminus and purified through metal affinity chromatography (Fig. 1). Differences in specific activities were observed among all of the constructs (Table 2). There was no direct relationship of chain length of linkers with either endoglucanase or β-glucosidase activity. However, the longest-chain-length linker, (G4S)3, yielded the highest activity. The EG5 construct with the Endo5A-(G4S)3-Gluc1C fusion protein resulted in a 3.2-fold higher molar specific activity for β-glucosidase and a 2-fold higher molar specific activity for endoglucanase than its individual counterpart.
Evaluation of kinetic parameters of bifunctional fusion proteins.
We further studied the reaction processes and catalytic events for the fusion proteins by determining Km, a measure of substrate affinity, and kcat/Km, a measure of catalytic efficiency. With respect to β-glucosidase activity, the Km and kcat/Km of the fusion constructs ranged from 2.7 to 8.4 mM and 0.62 to 6.40 mM−1 s−1, respectively (Table 3). These results indicated that fusion of two catalytic domains had either a favorable or adverse impact on the substrate affinity and catalytic efficiency of enzymes, depending upon the domain positioning and the kind of linkers used. Similar results were obtained for endoglucanase activity, where the Km and kcat/Km of the fusion constructs ranged from 0.76 to 3.00 mg ml−1 and 1.0 ×105 to 2.0 ×105 ml mg−1 s−1, respectively (Table 3). The β-glucosidase activity was sensitive to its position as it showed decline in affinity and catalytic efficiency when Gluc1C was placed at the N terminus. Neither the positioning nor the linker size showed any trend for endoglucanase activity in the fusion. EG5 stood out among all of the fusion constructs for having 1.5-fold higher substrate affinity and 2-fold higher catalytic efficiency for the β-glucosidase domain than Gluc1C alone. In addition, EG5 constructs showed similar Km values and 2-fold higher catalytic efficiency for the endoglucanase domain than the Endo5A alone. The favorable results of EG5 prompted us to characterize it further.
Table 3.
Kinetic parameters of recombinant enzymes
| Activity and recombinant protein | Km (mM or mg ml−1)a | kcat/Km (mM−1 s−1 or ml mg−1 s−1 × 105)b |
|---|---|---|
| β-Glucosidase | ||
| Gluc1C | 4.2 | 3.23 |
| EG1 (Endo5A-Gluc1C) | 3.8 | 1.90 |
| EG2 (Gluc1C-Endo5A) | 8.1 | 0.93 |
| EG3 [Endo5A-(G4S)2-Gluc1C] | 6.7 | 0.70 |
| EG4 [Gluc1C-(G4S)2-Endo5A] | 8.4 | 0.62 |
| EG5 [Endo5A-(G4S)3-Gluc1C] | 2.7 | 6.40 |
| EG6 [Gluc1C-(G4S)3-Endo5A] | 4.3 | 0.89 |
| Endoglucanase | ||
| Endo5A | 0.97 | 1.0 |
| EG1 (Endo5A-Gluc1C) | 1.14 | 1.9 |
| EG2 (Gluc1C-Endo5A) | 2.01 | 1.5 |
| EG3 [Endo5A-(G4S)2-Gluc1C] | 3.00 | 1.7 |
| EG4 [Gluc1C-(G4S)2-Endo5A] | 2.25 | 1.7 |
| EG5 [Endo5A-(G4S)3-Gluc1C] | 0.96 | 2.0 |
| EG6 [Gluc1C-(G4S)3-Endo5A] | 0.76 | 1.8 |
Km was calculated in mM for β-glucosidase and mg ml−1 for endoglucanase activity.
kcat/Km was calculated in mM−1 s−1 for β-glucosidase and ml mg−1 s−1 for endoglucanase activity.
Biochemical and biophysical characterization of EG5.
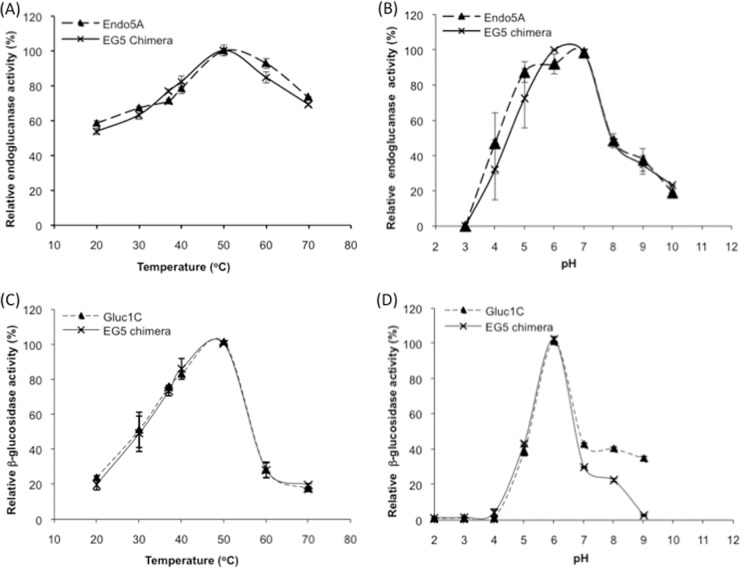
The bifunctional EG5 construct that contained the Endo5A domain at the N terminus and the Gluc1C domain at the C terminus with a triplicate glycine-serine repeat in between [Endo5A-(G4S)3-Gluc1C] demonstrated higher specific activity and catalytic efficiency for both of its substrates than either Endo5A or Gluc1C alone. Interestingly, the expression level of EG5 in E. coli was also found to be higher than that of Endo5A or Gluc1C alone (see Table S2 in the supplemental material). The inhibition kinetics for β-glucosidase activity using glucono-δ-lactose as an inhibitor indicated a similar Ki (∼1.2 mM) for both Gluc1C and EG5 (see Fig. S2 in the supplemental material). The EG5 construct also showed similar pH and temperature optima for its activities, as shown by Endo5A or Gluc1C alone: i.e., 50°C and pH 6 to 7 for endoglucanase activity (Fig. 3A and B) and 50°C and pH 6 for β-glucosidase activity (Fig. 3C and D). We noticed that while the endoglucanase activity of recombinant enzymes showed >60% of optimal activity between 30°C and 70°C (Fig. 3A), the β-glucosidase activity of recombinant enzymes declined sharply at higher temperatures (Fig. 3C). We therefore evaluated the thermal stability of the β-glucosidase activity of Gluc1C and EG5 at various temperatures. The β-glucosidase activity of the EG5 construct showed higher thermostability at all temperatures tested compared to Gluc1C (Fig. 4A and B). The EG5 construct demonstrated a 4-fold higher half-life (7.5 min) at 55°C than the Gluc1C half-life (1.9 min), indicating that the Gluc1C domain of EG5 attained some sort of structural stability upon fusion. We further studied the stability of secondary structures of EG5 at various temperatures using CD spectroscopy. A melting curve was generated by gradually increasing the temperature and monitoring the signal at 222 nm (Fig. 4C). The melting temperature (Tm) of Gluc1C was found to be 45.2°C, while Endo5A exhibited a Tm of 64.3°C. The EG5 chimeric protein exhibited an expected biphasic melting curve corresponding to its β-glucosidase and endoglucanase domains (Fig. 4C). The Tms for β-glucosidase and the endoglucanase domain of EG5 corresponded to 50.5°C and 73.5°C, respectively. The 5°C and 9°C upward shifts in Tm for Gluc1C and the Endo5A domain of EG5, respectively, clearly indicated higher thermostability of the EG5 chimeric protein and partly explained the rationale for the 4-fold higher half-life of β-glucosidase activity at 55°C. We further performed in silico analysis of EG5 structure by predicting its three-dimensional structure using the I-TASSER web server and comparing the structural parameters that affect thermostability of proteins (21) using Discovery Studio 3.1 Visualizer software. The β-glucosidase domain of EG5 showed a significant increase in values of parameters that had been shown earlier (21) to contribute to the thermostability of proteins (Table 4).
Fig 3.
Temperature and pH optima of recombinant enzymes. The metal affinity-purified recombinant enzymes were tested for endoglucanase activity (A and B) and β-glucosidase activity (C and D) at various temperatures (A and C) and pHs (B and D), and the optimal temperature and pH for their maximum activities were determined.
Fig 4.
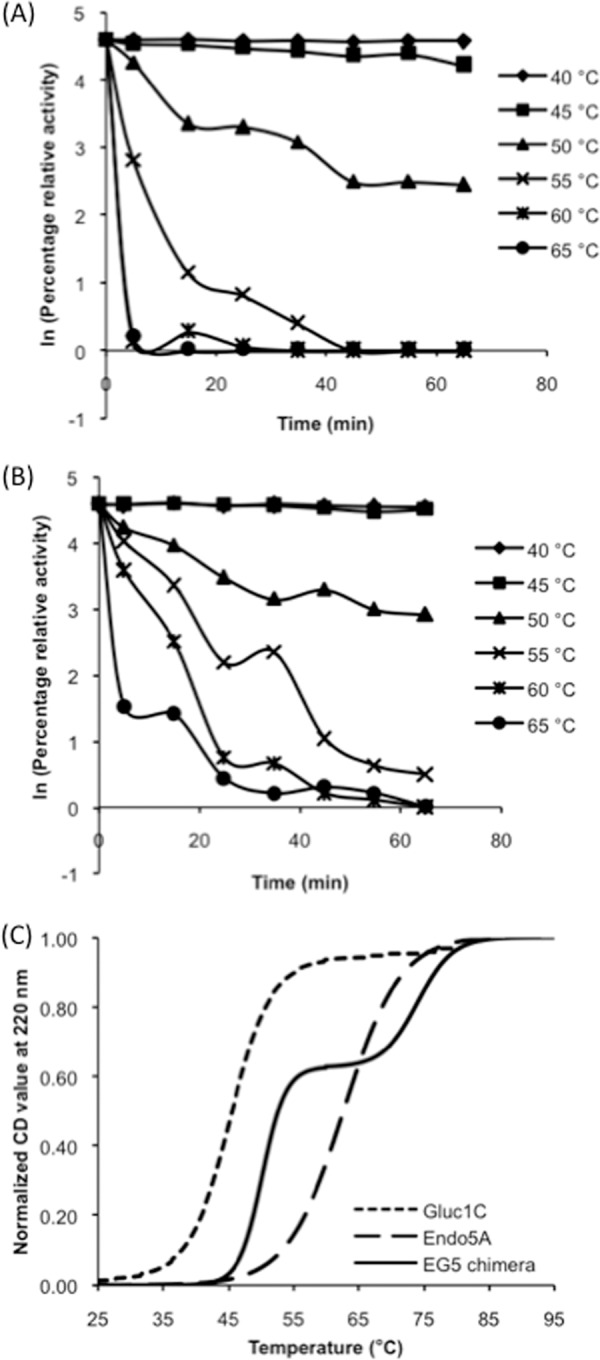
Thermostability of activities and structures of recombinant enzymes. The β-glucosidase activities of recombinant Gluc1C (A) and EG5 (B) were tested at various temperatures with respect to time. (C) The stability of secondary structures of Gluc1C, Endo5A, and the EG5 chimera was determined by denaturing the recombinant enzymes with increasing temperature and measuring the CD signals at 220 nm.
Table 4.
In silico analysis of parameters affecting the structure of β-glucosidase in EG5 chimera
| Parametera | Result for enzyme: |
% increase in EG5 | |
|---|---|---|---|
| Gluc1C | EG5 | ||
| Frac-nonpol buried area | 0.26 | 0.28 | 7.8 |
| Frac-exp-pol area | 0.28 | 0.31 | 10.7 |
| Helical content (%) | 38.7 | 40.5 | 4.6 |
| Hydrophobicity | 0.62 | 0.67 | 8.1 |
“Frac-nonpol buried area” denotes the contribution of nonpolar residues to the buried surface area, and “Frac-exp-pol area” denotes the contribution of polar residues to the exposed surface area. “Helical content” refers to the percentage of residues that have an α-helical conformation in the protein. Hydrophobicity was calculated as the fraction of the buried nonpolar area out of the total nonpolar area.
Channeling effect of EG5.
When the EG5 chimeric protein containing 8 mU of endoglucanase and 4 mU of β-glucosidase activity was incubated with CMC in the absence of glucono-δ-lactose (inhibitor for β-glucosidase), it released 19% higher reducing sugar compared to the EG5 chimera in the presence of glucono-δ-lactose (Fig. 5A), indicating a favorable role of Gluc1C in the EG5 chimera in releasing reducing sugars. More importantly, EG5 released 10% higher reducing sugar than the physical mixture of Endo5A and Gluc1C containing similar amounts of endoglucanase and β-glucosidase. This observation suggested that the close proximity of Gluc1C with Endo5A in the EG5 chimera helped channeling of hydrolyzed products between the two catalytic domains and resulted in improved hydrolysis.
Fig 5.
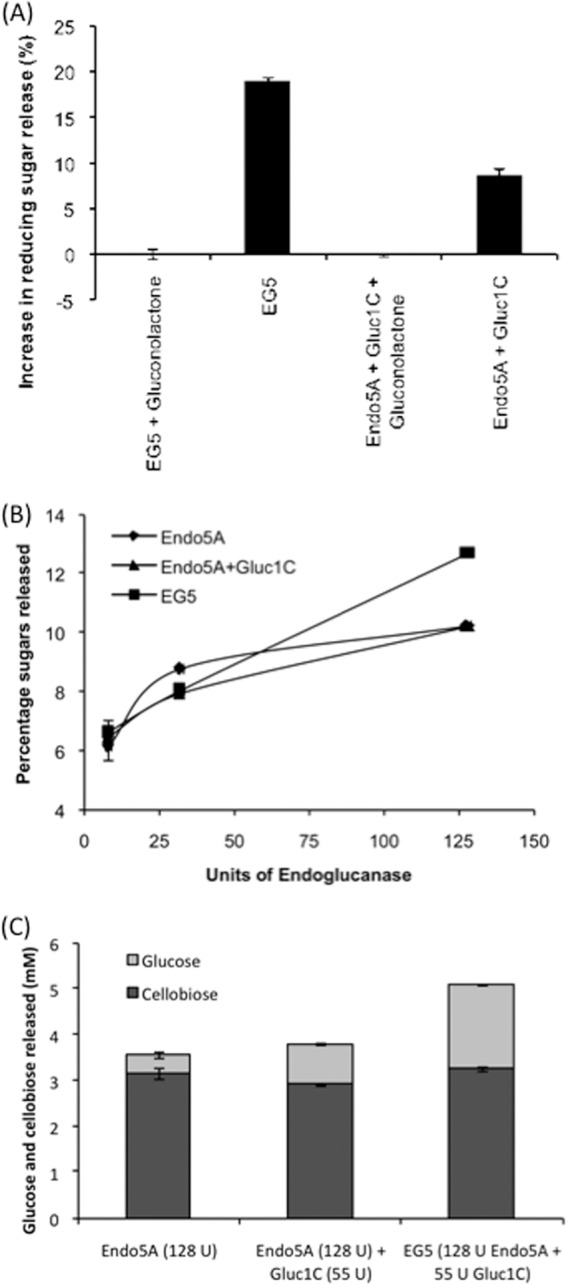
Channeling effect of EG5 on cellulosic hydrolysates. (A) Reducing sugar release from CMC as the carbon source. The EG5 fusion protein, containing 8 mU of endoglucanase and 4 mU of β-glucosidase, was incubated with CMC in the presence and absence of gluconolactone, and the relative increase in reducing sugar release was plotted. Data from the physical mixture of Endo5A and Gluc1C containing 8 mU of endoglucanase and 4 mU of β-glucosidase are included in the graph for comparison. The EG5 chimera released 10% more reducing sugar than the physical mixture of the two enzymes. (B) Comparison of sugars (glucose and cellobiose) released from pretreated biomass by Endo5A, the Endo5A-Gluc1C mixture, and EG5. Enzymes corresponding to 8, 32, and 128 U of endoglucanase, which contained 3.5, 14, and 55 U of β-glucosidase, respectively, in the case of the Endo5A-Gluc1C mixture and EG5, were incubated with alkali-treated rice straw, and the sugars (glucose and cellobiose) released were monitored by HPLC. (C) Amounts of glucose and cellobiose released from pretreated biomass by recombinant enzymes at 128 U of endoglucanase and 55 U of β-glucosidase.
We further evaluated hydrolysis of lignocellulosic biomass using Endo5A, Gluc1C, and EG5 chimeric enzyme. We used alkali-treated rice straw for this purpose and monitored cellulose hydrolysates using HPLC. We could only detect cellobiose and glucose in the hydrolysate through HPLC. There was a successive increase in sugar release upon increasing the concentration of Endo5A, which showed a sign of saturation toward higher amounts of Endo5A (Fig. 5B). Endo5A mixed with Gluc1C showed a similar pattern of sugar release. EG5, on the other hand, showed a linear increase in sugar release with increasing enzyme amounts, and the amount of sugar released at 128 U of endoglucanase was 24% higher than that with Endo5A alone or the Endo5A-Gluc1C mixture (Fig. 5B). Upon analysis of the amount of glucose present in the sugar mixture, EG5 yielded the highest level of glucose, 5-fold higher than Endo5A and 2-fold higher than the Endo5A-Gluc1C mixture (Fig. 5C). Here also, the larger amount of glucose produced by the same amount of EG5 is an indication of intermediate channeling for cellulose hydrolysate.
Evaluation of EG5 for commercial application.
We compared the specific CMCase activity of EG5 with that of the commercial enzyme NS50013 at pH 4.8 and 7.0. The commercial preparation exhibited a specific activity of 3.1 U/mg at pH 4.8 and a negligible activity at pH 7.0 (Fig. 6A). EG5 yielded 6- and 70-fold higher specific activities at pH 4.8 and 7.0, respectively, than the NS50013 preparation. We further evaluated the impact of addition of EG5 in commercial preparation on the hydrolysis of pretreated biomass. Addition of EG5 (6 CMC units/g biomass) to NS50013 (1 FPU plus 6 CMC units/g biomass) resulted in a nearly 50% increase in total reducing sugar release and a 20% increase in glucose (C1) to cellotriose (C3) chain-length sugar release (Fig. 6B).
Fig 6.
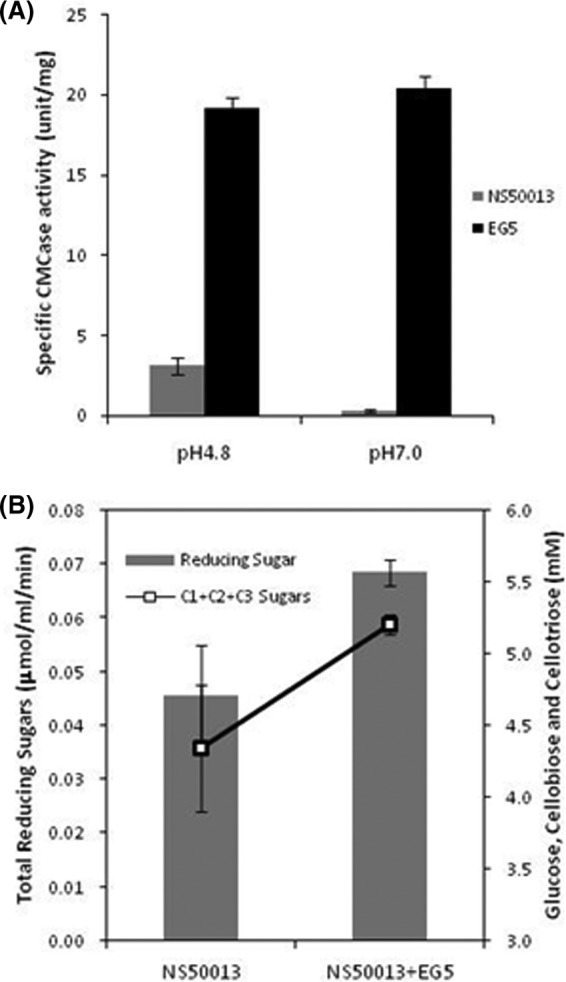
(A) Comparison of the specific activity of EG5 with that of commercial cellulase NS50013 at pH 4.8 and 7. (B) Increase in release in sugars from pretreated biomass upon addition of EG5 to NS50013. The total amounts of reducing sugars released were determined through the DNSA method, while release of glucose (C1), cellobiose (C2), and cellotriose (C3) was determined through HPLC.
DISCUSSION
Production of cellulolytic enzymes in a cost-effective manner is crucial for the economic viability of lignocellulosic ethanol. Since a consortium of enzymes is required to perform the hydrolysis, construction of bifunctional enzymes is likely to reduce the production cost significantly. We constructed a series of chimeras consisting of an endoglucanase (Endo5A) domain and a β-glucosidase (Gluc1C) domain and analyzed their enzymatic potential.
Gluc1C for the chimeric proteins was cloned from Paenibacillus sp. strain MTCC 5639 (1) and expressed in E. coli. Gluc1C showed 99% homology with BglB (GenBank accession no. AAA22264) of Paenibacillus polymyxa (15), and its biochemical properties, such as optimal pH and temperature for enzyme activity, Km, and kcat, were found to be similar to those of type I β-glucosidase, as described previously (14–16). It had been mentioned earlier that bacterial β-glucosidase could hydrolyze cellodextrin (16, 20). Gluc1C was found to hydrolyze cello-oligosaccharides with a chain length of up to five sugars, with highest efficiency toward cellobiose (see Fig. S1 and Table S1 in the supplemental material).
During hydrolysis of cellulosic biomass, the substrates for Gluc1C are generated in the form of cellobiose or cellodextrin via the action of endo- or exoglucanase, which are then hydrolyzed into monomers. This synergism is important for reaching the end product (28). Along a similar line, we found that addition of Gluc1C in the Endo5A-catalyzed reaction resulted in a higher reducing sugar concentration (Fig. 2). This prompted us to construct a bifunctional chimera based on Endo5A and Gluc1C. Two such kinds of chimeras have been reported in the literature (18, 23). In one case, a fusion of endoglucanase and β-glucosidase from Thermotoga maritima was made and reported to have both activities, albeit they were 70% less than those of the individual enzymes (18). In the second case, two types of endoglucanase and an exoglucanase of Clostridium thermocellum were fused with a β-glucosidase of Clostridium cellulovorans (23). Here, one of the combinations, in which cellulosomal endoglucanase was fused with β-glucosidase (CtCD-CcBG), resulted in improved thermal stability of the β-glucosidase domain. In our study, we made six constructs containing Endo5A and Gluc1C with different linkers and positions and assessed their bifunctional characteristics. Among the six constructs we made, the constructs that have the Gluc1C domain at the C terminus resulted in higher ligand affinity for pNPG compared to its N terminus positioning (Table 3). This positioning was also found to be favorable in the previous report for β-glucosidase activity (18). The remarkable enhancement in specific activity, substrate affinity, and catalytic efficiency was demonstrated by one of the constructs, EG5 [Endo5A-(G4S)3-Gluc1C]. The extent of improvement in kinetic parameters for both endoglucanase and β-glucosidase activities (Table 2 and Table 3) was significantly higher than those of the other fusion constructs reported before (18, 23). EG5 also demonstrated a greater half-life for β-glucosidase activity at higher temperatures (Fig. 4A and B), possibly due to higher thermostability of the protein, as observed by analyzing the melting curve through CD spectroscopy. The in silico analysis of the structural parameters of EG5 further confirmed the higher thermostability for the β-glucosidase domain (Table 4).
An important aspect of any cellulolytic enzyme preparation is the enzymes' ability to hydrolyze the potential substrate for lignocellulosic alcohol. We tested the hydrolysis of alkali-treated rice straw using the same units of Endo5A, the Endo5A-Gluc1C mixture, and the EG5 chimera. Here, EG5 not only showed 24% higher sugar release compared to the other enzymes (Fig. 5B), but it also yielded 5-fold and 2-fold higher levels of glucose than Endo5A and the Endo5A-Gluc1C mixture, respectively (Fig. 5C). This result clearly indicated the channeling effect of intermediates in EG5, which led to a higher glucose concentration. Furthermore, addition of EG5 to a commercial cellulase preparation significantly enhanced its ability to release reducing sugars, indicating the commercial applicability of EG5.
In conclusion, we have shown here characterization of a β-glucosidase isolated from a Paenibacillus species and construction of a series of chimeras consisting of endoglucanase and β-glucosidase domains. One of the chimeras, EG5, showed outstanding activity against its substrates and significantly higher thermostability. This chimera also released more glucose from the cellulosic biomass than the enzyme mixture and therefore is a potential candidate for an enzyme cocktail in the manufacturing of lignocellulosic alcohol.
Supplementary Material
ACKNOWLEDGMENTS
We thank Akash Saini for assisting with the CD spectroscopy.
We acknowledge financial support for this work from the Department of Biotechnology, Government of India.
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adlakha N, Rajagopal R, Kumar S, Reddy VS, Yazdani SS. 2011. Synthesis and characterization of chimeric proteins based on cellulase and xylanase from an insect gut bacterium. Appl. Environ. Microbiol. 77:4859–4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An JM, et al. 2005. Evaluation of a novel bifunctional xylanase-cellulase constructed by gene fusion. Enzyme Microb. Technol. 36:989–995 [Google Scholar]
- 3. Beguin P, Aubert JP. 1994. The biological degradation of cellulose. FEMS Microbiol. Rev. 13:25–58 [DOI] [PubMed] [Google Scholar]
- 4. Bhat MK, Bhat S. 1997. Cellulose degrading enzymes and their potential industrial applications. Biotechnol. Adv. 15:583–620 [DOI] [PubMed] [Google Scholar]
- 5. Clarke AJ. 1997. Biodegradation of cellulose: enzymology and biotechnology. Technomic Publishing Co., Inc., Lancaster, PA [Google Scholar]
- 6. Coughlan MP. 1990. Cellulose degradation by fungi. Microbial enzymes and biotechnology, 2nd ed Elsevier, New York, NY [Google Scholar]
- 7. Dashtban M, Maki M, Leung KT, Mao C, Qin W. 2010. Cellulase activities in biomass conversion: measurement methods and comparison. Crit. Rev. Biotechnol. 30:302–309 [DOI] [PubMed] [Google Scholar]
- 8. Dashtban M, Schraft H, Qin W. 2009. Fungal bioconversion of lignocellulosic residues: opportunities and perspectives. Int. J. Biol. Sci. 5:578–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Demain AL, Newcomb M, Wu JH. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dowe N. 2009. Assessing cellulase performance on pretreated lignocellulosic biomass using saccharification and fermentation-based protocols. Methods Mol. Biol. 581:233–245 [DOI] [PubMed] [Google Scholar]
- 11. Fan Z, Werkman JR, Yuan L. 2009. Engineering of a multifunctional hemicellulase. Biotechnol. Lett. 31:751–757 [DOI] [PubMed] [Google Scholar]
- 12. Flint HJ, Martin J, McPherson CA, Daniel AS, Zhang JX. 1993. A bifunctional enzyme, with separate xylanase and beta(1,3-1,4)-glucanase domains, encoded by the xynD gene of Ruminococcus flavefaciens. J. Bacteriol. 175:2943–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geddes CC, Nieves IU, Ingram LO. 2011. Advances in ethanol production. Curr. Opin. Biotechnol. 22:312–319 [DOI] [PubMed] [Google Scholar]
- 14. Gonzalez-Candelas L, Aristoy MC, Polaina J, Flors A. 1989. Cloning and characterization of two genes from Bacillus polymyxa expressing beta-glucosidase activity in Escherichia coli. Appl. Environ. Microbiol. 55:3173–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gonzalez-Candelas L, Ramon D, Polaina J. 1990. Sequences and homology analysis of two genes encoding beta-glucosidases from Bacillus polymyxa. Gene 95:31–38 [DOI] [PubMed] [Google Scholar]
- 16. Hashimoto W, et al. 1998. Molecular cloning of two genes for beta-d-glucosidase in Bacillus sp. GL1 and identification of one as a gellan-degrading enzyme. Arch. Biochem. Biophys. 360:1–9 [DOI] [PubMed] [Google Scholar]
- 17. Hess M, et al. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467 [DOI] [PubMed] [Google Scholar]
- 18. Hong SY, et al. 2007. Construction of the bifunctional enzyme cellulase-beta-glucosidase from the hyperthermophilic bacterium Thermotoga maritima. Biotechnol. Lett. 29:931–936 [DOI] [PubMed] [Google Scholar]
- 19. Hong SY, et al. 2006. Assembling a novel bifunctional cellulase-xylanase from Thermotoga maritima by end-to-end fusion. Biotechnol. Lett. 28:1857–1862 [DOI] [PubMed] [Google Scholar]
- 20. Isorna P, et al. 2007. Crystal structures of Paenibacillus polymyxa beta-glucosidase B complexes reveal the molecular basis of substrate specificity and give new insights into the catalytic machinery of family I glycosidases. J. Mol. Biol. 371:1204–1218 [DOI] [PubMed] [Google Scholar]
- 21. Kumar S, Tsai CJ, Nussinov R. 2000. Factors enhancing protein thermostability. Protein Eng. 13:179–191 [DOI] [PubMed] [Google Scholar]
- 22. la Grange DC, den Haan R, van Zyl WH. 2010. Engineering cellulolytic ability into bioprocessing organisms. Appl. Microbiol. Biotechnol. 87:1195–1208 [DOI] [PubMed] [Google Scholar]
- 23. Lee HL, Chang CK, Teng KH, Liang PH. 2011. Construction and characterization of different fusion proteins between cellulases and beta-glucosidase to improve glucose production and thermostability. Bioresour. Technol. 102:3973–3976 [DOI] [PubMed] [Google Scholar]
- 24. Lu P, Feng MG. 2008. Bifunctional enhancement of a beta-glucanase-xylanase fusion enzyme by optimization of peptide linkers. Appl. Microbiol. Biotechnol. 79:579–587 [DOI] [PubMed] [Google Scholar]
- 25. Lynd LR, van Zyl WH, McBride JE, Laser M. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 16:577–583 [DOI] [PubMed] [Google Scholar]
- 26. Maki M, Leung KT, Qin W. 2009. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 5:500–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maki ML, Broere M, Leung KT, Qin W. 2011. Characterization of some efficient cellulase producing bacteria isolated from paper mill sludges and organic fertilizers. Int. J. Biochem. Mol. Biol. 2:146–154 [PMC free article] [PubMed] [Google Scholar]
- 28. Ng IS, Tsai SW, Ju YM, Yu SM, Ho TH. 2011. Dynamic synergistic effect on Trichoderma reesei cellulases by novel beta-glucosidases from Taiwanese fungi. Bioresour. Technol. 102:6073–6081 [DOI] [PubMed] [Google Scholar]
- 29. Perez-Avalos O, Sanchez-Herrera LM, Salgado LM, Ponce-Noyola T. 2008. A bifunctional endoglucanase/endoxylanase from Cellulomonas flavigena with potential use in industrial processes at different pH. Curr. Microbiol. 57:39–44 [DOI] [PubMed] [Google Scholar]
- 30. Sanchez C. 2009. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol. Adv. 27:185–194 [DOI] [PubMed] [Google Scholar]
- 31. Stephanopoulos G. 2007. Challenges in engineering microbes for biofuels production. Science 315:801–804 [DOI] [PubMed] [Google Scholar]
- 32. Teeri TT. 1997. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 15:160–167 [Google Scholar]
- 33. Wood TM, BK 1988. Methods for measuring cellulase activities. Methods Enzymol. 160:87–112 [Google Scholar]
- 34. Wood TM. 1989. Enzyme systems for lignocellulose degradation. Mechanisms of cellulose degradation by enzymes from aerobic and anaerobic fungi. Elsevier, New York, NY [Google Scholar]
- 35. Zhou J, et al. 2009. Optimization of cellulase mixture for efficient hydrolysis of steam-exploded corn stover by statistically designed experiments. Bioresour. Technol. 100:819–825 [DOI] [PubMed] [Google Scholar]
- 36. Zhu L, Wu Q, Dai J, Zhang S, Wei F. 2011. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. U. S. A. 108:17714–17719 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.