Abstract
The biosynthesis of the major carotenoid spirilloxanthin by the purple nonsulfur bacterium Rhodospirillum rubrum is thought to occur via a linear pathway proceeding through phytoene and, later, lycopene as intermediates. This assumption is based solely on early chemical evidence (B. H. Davies, Biochem. J. 116:93–99, 1970). In most purple bacteria, the desaturation of phytoene, catalyzed by the enzyme phytoene desaturase (CrtI), leads to neurosporene, involving only three dehydrogenation steps and not four as in the case of lycopene. We show here that the chromosomal insertion of a kanamycin resistance cassette into the crtC-crtD region of the partial carotenoid gene cluster, whose gene products are responsible for the downstream processing of lycopene, leads to the accumulation of the latter as the major carotenoid. We provide spectroscopic and biochemical evidence that in vivo, lycopene is incorporated into the light-harvesting complex 1 as efficiently as the methoxylated carotenoids spirilloxanthin (in the wild type) and 3,4,3′,4′-tetrahydrospirilloxanthin (in a crtD mutant), both under semiaerobic, chemoheterotrophic, and photosynthetic, anaerobic conditions. Quantitative growth experiments conducted in dark, semiaerobic conditions, using a growth medium for high cell density and high intracellular membrane levels, which are suitable for the conventional industrial production in the absence of light, yielded lycopene at up to 2 mg/g (dry weight) of cells or up to 15 mg/liter of culture. These values are comparable to those of many previously described Escherichia coli strains engineered for lycopene production. This study provides the first genetic proof that the R. rubrum CrtI produces lycopene exclusively as an end product.
INTRODUCTION
The production of the carotenoid (crt) lycopene, which is used industrially as a natural colorant and also as a food additive, using microbial sources, is presently of great interest (29, 35, 46). Until now, most microbial production strategies have mostly used Escherichia coli as a host strain (1, 2, 10, 13, 24, 26, 30, 33–35, 43, 45, 48, 56–58, 64). However, a few studies using yeasts have also been reported (36, 49, 62). In early studies (46) with E. coli as a host strain, lycopene levels of up to about 0.5 mg/g (dry weight) of cells have been reported, whereas in Candida utilis, after extensive reengineering of the ergosterol pathway, up to 7.8 mg of lycopene/g (dry weight) has been achieved (49). More recently, using a combination of rational systems biological design and random screening approaches, lycopene levels of up to 18 mg/g (dry weight) have been successfully achieved in nonlinearly batch-fed E. coli cultures (2). In another approach, the Stephanopoulos group introduced up to 40 consecutive copies of heterologous carotenoid biosynthesis pathway genes into the E. coli chromosome, which allowed up to 14 mg of lycopene/g (dry weight) to be produced (56). In an alternative approach, Farmer and Liao (10) demonstrated that the carbon flow to lycopene in E. coli could be increased by the introduction of a synthetic regulatory circuit derived from the Ntr regulon, which senses an intracellular signaling molecule, acetyl phosphate. However, even these latter, very successful examples of lycopene production in E. coli have required extensive reengineering of the organism. In addition, these latter strategies have as yet only been reported for small shake cultures (about 50 to 250 ml) or small-scale bioreactors.
The implicit rationale for using E. coli as a production strain for a highly hydrophobic terpenoid such as lycopene is not completely clear. Although E. coli is the organism of choice due to the wealth of genetic detail that is available for this organism, together with its high growth rate and general robustness, the production of hydrophobic terpenoids in this strain poses several severe problems. First, lycopene biosynthesis requires three unique enzymes, geranylgeranyl pyrophosphate (GGPP) synthase (CrtE), phytoene synthase (CrtB), and phytoene desaturase (CrtI), which are not present in E. coli and must be therefore maintained either in trans or introduced chromosomally. These types of constructions often perform well at a small- to medium-scale but are frequently prone to stability problems when adapted to the industrial scale. Second, GGPP synthase uses the substrate farnesyl pyrophosphate (FPP) to produce GGPP, which is an essential intermediate in the quinone (both ubiquinone and menaquinone) pathways. Thus, a common observation is that increased flux to a recombinant phytoene synthase causes the GGPP pool to be depleted, thus leading to growth problems (37). This problem must be alleviated by complicated genetic strategies such as an extensive redesign of the pyruvate/melavonate pathways using rational and random screening techniques (2, 56) or by using a synthetic feedback loop using the metabolite acetyl phosphate as a sensor of energy metabolism (10). The latter approach has the potential difficulty that acetyl phosphate is also a global regulator of metabolism, where many of its roles, particularly in anaerobically operative pathways, have yet to be defined (see reference 61 for a review). However, the highest yields reported (2, 56) have usually required a fed-batch strategy, which is often difficult to apply during upscaling. Third, it is probable that the maximal attainable levels of terpenoid (in this case lycopene) are limited to the available volume of the cytoplasmic membrane. In fact, the localization of the carotenoid in E. coli has never been studied in detail. This factor is probably much more important than often expected, since a considerable perturbation of the natural membrane would be expected to affect electron transport and thus viability.
An interesting alternative strategy would be to utilize purple photosynthetic bacteria, which naturally overproduce large amounts of carotenoids in specific compartments (e.g., the photosynthetic membrane), for the production of industrially interesting carotenoids. However, only a single attempt with this strategy (see below) has been reported thus far (22). The reason for the low activity in this area is probably that a considerable degree of genetic engineering is still mandatory, since, as noted recently by Takaichi (55), the natural carotenoid products in photosynthetic purple bacteria are generally distinct from those of plants, and that processes requiring controlled light conditions are not easily accessible for industrial production systems.
In a recent review, Takaichi (55) classified the carotenoid biosynthesis pathways in purple bacteria into two types: (i) the spirilloxanthin (spx) pathway, which encompasses both “normal” (Fig. 1) and “unusual” pathways (see reference 55), which lead either to spx (e.g., found in Rhodospirillum rubrum) or spheroidene (e.g., in Rhodobacter [Rb.] capsulatus [3, 4, 63] and Rhodobacter sphaeroides [22]), and (ii) the okenone pathway (e.g., in Allochromatium oekenii [47]), which leads to okenone or other ketocarotenoids as final products. For both pathways, the first step unique to carotenoid biosynthesis is the oxidation of phytoene, catalyzed by the enzyme phytoene desaturase (CrtI; see Fig. 1). However, the final product of this reaction varies depending upon the source. Thus, the CrtIs of Rb. capsulatus and Rb. sphaeroides have been shown to produce exclusively neurosporene as a product (5, 28), whereas the CrtI of Rubrivivax (Rv.) gelatinosus oxidizes phytoene to 90% neurosporene and 10% lycopene (21), and the CrtI from the nonphotosynthetic Pantoea (formerly called Erwinia) yields exclusively lycopene (13).
Fig 1.
Carotenoid biosynthesis pathway in R. rubrum (end product, spirilloxanthin [spx]). The known pathway to spheroidene/spheroidenone in Rb. capsulatus and Rb. sphaeroides, which branches at the third CrtI-mediated oxidation step, is also shown (dashed box) for comparison. For the latter pathway, several intermediate steps have been omitted for convenience. The gene products which catalyze the various steps are also indicated (small solid-line boxes).
In the single study using Rb. sphaeroides as a host organism (22), the gene encoding CrtI from Erwinia (Ew.) herbicola was introduced into Rb. sphaeroides on a conjugable plasmid in trans and the transconjugant was shown to produce lycopene in the intracytoplasmic membrane (ICM), which contains the photosynthetic apparatus. However, the levels of lycopene reported for semiaerobic cultures appeared to be extremely low (ca. 20% of those obtainable with the wild-type strain).
In R. rubrum, the product of the CrtI reaction is assumed to be lycopene (55), based upon chemical analysis of carotenoid intermediates performed in early studies (8). Thus far, however, no genetic or functional proof has been provided to confirm this suggestion. In this exploratory study, we show that a crtC crtD deletion mutant of R. rubrum indeed produces lycopene exclusively as a final product, thus confirming the early proposal of Davies (8). We also show that the lycopene levels attainable in R. rubrum are equivalent to those reported for several highly engineered E. coli strains reported as “starting points” for “superproducing” E. coli strains (2, 10, 56). We also provide biochemical evidence, using purified light-harvesting 1 (LH1) complexes, which are localized exclusively in the ICM in vivo, that the lycopene produced is specifically bound to this protein.
MATERIALS AND METHODS
Growth conditions.
Bacterial strains and plasmids are listed in Table 1. E. coli cultures were grown in Luria-Bertani medium at 37°C (44). Antibiotics were added as required at the following concentrations: ampicillin (sodium salt) at 100 μg/ml and kanamycin sulfate at 50 μg/ml. Initially, R. rubrum strains were inoculated into Sistrom medium A (here designated M medium) (51) containing 20 mM potassium succinate as a carbon source and cultivated phototrophically in closed bottles (Pyrex) at 30°C. For semiaerobic growth, R. rubrum strains were cultivated in modified M medium: either M2S medium (containing 40 mM NH4+-succinate as carbon source), M2SF medium (containing 40 mM NH4+-succinate, and 16.7 mM [0.3%] fructose as carbon source) or M2SF+ medium (containing 60 mM NH4+-succinate and 111.3 mM [2%] fructose as a carbon source) (18). Small-scale growth experiments were performed in 100-ml medium, whereas medium-scale experiments were performed using 3-liter cultures.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1 MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrn173 endA1 supE44 thi1 recA1 gyrA96 relA1 lac) | Stratagene |
| R. rubrum | ||
| S1 | Wild type | 7 |
| ST4 | S1-derived crtD mutant; Kanr | 27 |
| SLYC18 | S1-derived crtC crtD site-directed mutant; Kanr | This study |
| Plasmids | ||
| pBsKSII+ | High-copy cloning vector, ColE1; Ampr | Stratagene |
| pBsLGKan | pBsKSII+ derivative, carrying the kanamycin resistance cassette; Kanr | 32 |
| pRK404 | Derivative of pRK290; mob+ | 9 |
| pRK2013 | Mobilizing helper plasmid, tra+; Kanr | 12 |
| pBsSGE5 | pBsKSII+ derivative, containing the 6.37-kb EcoRV fragment from the pSC4 cosmid | This study |
| pBsSGE5K1 | pBsSGE5 derivative, containing the kanamycin resistance cassette in the opposite direction to the crtD gene | This study |
| pBsSGE5K2 | pBsSGE5 derivative, containing the kanamycin resistance cassette in the same direction as the crtD gene | This study |
| pRKSGE5 | pRK404 derivative, containing the 6.42-kb HindIII/XbaI insert from pBsSGE5 | This study |
| pSUP202 | Suicide vector, ColE1, mob+; Ampr Cmr Tetr | 50 |
| pSUPSGE5K1 | pSUP202 derivative, containing the HindIII/SalI insert of pBsSGE5K1, Ampr Kanr Cmr Tetr | This study |
| pSUPSGE5K2 | pSUP202 derivative, containing the HindIII/SalI insert of pBsSGE5K2, Ampr Kanr Cmr Tetr | This study |
Cmr, chloramphenicol resistance; Tetr, tetracycline resistance; Ampr, ampicillin resistance; Kanr, kanamycin resistance.
For growth curves using either M2SF or M2SF+ media, R. rubrum strains were grown chemoheterotrophically in the dark in 250-ml baffled Erlenmeyer flasks containing 100-ml medium with a 4-ml inoculum (obtained from anaerobically grown late-exponential-phase cultures) at 30°C, with shaking at 150 rpm (2-cm throw). The optical densities (ODs), A660 and A882, were measured using a single-beam spectrophotometer with a 4-mm path-length cuvette. In a control experiment, we ascertained that the ODs measured in a 4-mm cuvette correlate linearly with dry weights up to a value of 1.4. Therefore, cell cultures showing a high OD (>1.4) were diluted prior to the measurement, so as not to exceed this value.
Construction of the lycopene-producing mutant, SLYC18.
A 6.37-kb EcoRV fragment, containing crtC, crtD, and crtE, was obtained from the cosmid pSC4 (Fig. 2), which was isolated from a cosmid gene bank containing chromosomal DNA from the R. rubrum wild-type S1, and which encodes about half of the photosynthetic gene cluster. The EcoRV fragment was subcloned into the SmaI site of pBluescript (pBs) KSII+ to yield the plasmid pBsSGE5 (Fig. 2). Subsequently, a 1.2-kb BstEII fragment was replaced with a 1.5-kb HindIII/SalI blunt-ended fragment, derived from Tn5, which contains the npt gene encoding neomycin phosphotransferase (32), yielding plasmids pBsSGE5K1 and pBsSGE5K2, with the npt gene in the opposite and same orientation as the crtD promoter, respectively. The BstEII fragment deletion simultaneously eliminates large regions of both the crtC and crtD genes, thus leading to their functional arrest. Finally, the inserts of pBsSGE5K1 and pBsSGE5K2 were subcloned into the conjugable suicide vector pSUP202 (50) to yield the plasmids pSUPSGE5K1 and pSUPSGE5K2, respectively, which were subsequently triparentally conjugated with R. rubrum S1 (12, 32). Selection of doubly recombinant npt interposon mutants was determined by their Kanr Tets phenotypes. The chromosomal localization of the npt gene was confirmed by using Southern hybridization (44).
Fig 2.
Construction of the plasmids pBsSGE5K1 and pBsSGE5K2. The gene organization of the cosmid initially isolated, pSC4, which complemented the crtD mutant ST4, is indicated. The partial crt cluster (crtCDEF) is indicated (dark gray), as is the pSC4-derived EcoRV fragment used to construct the plasmid pBsSGE5. Site-directed deletion of the crtC and crtD genes was performed by replacing a BstEII fragment overlapping both genes with the npt cassette. Both npt orientations relative to the crt cluster were obtained (indicated).
Absorption spectroscopy.
The absorption spectra of intact cells and also extracted carotenoids were determined by using 2-mm path-length cuvettes with a Jasco V-560 UV/VIS spectrophotometer equipped with a photodiode detector for turbid samples. Intact cells were measured after suspension in M medium containing 80% glycerol. For comparisons of relative amounts of carotenoids in cells, the suspensions to be measured were adjusted to the same OD (usually A660 [2-mm path-length cuvette] = 0.5) at 660 nm.
Biochemical methods. (i) Dry weight determination.
Dry weights were determined from 1- to 2-ml aliquots taken from growing cultures by first filtering them through preweighed filters (Versapor-200 [Pall, USA]), which were then dried in an oven at 80°C for 24 h to achieve a constant weight.
(ii) Protein determination.
The total protein of cell culture aliquots was determined by a modified Lowry procedure (40) using bovine serum albumin as a standard. Generally, 100- or 50-μl aliquots of cell cultures were used for protein determination.
(iii) Purification of the LH1 complex.
LH1 complexes were isolated by established procedures (41). Briefly, the ICM fraction, isolated from a crude cell extract by differential centrifugation, was extracted three times using 0.3% (wt/vol) lauryldimethylamine-N-oxide (LDAO). The first two extractions primarily solubilize the reaction center (RC), and the third extract is highly enriched in the LH1 complex. The LDAO fraction was diluted to 0.1% LDAO and then dialyzed extensively, followed by DEAE-cellulose chromatography using 50 mM Tris-HCl, pH 8.0, as the initial equilibration buffer. Fractions eluted with the same buffer in the presence of 0.2 and 0.3% (wt/vol) n-octyl-β-d-glucoside (βOG), respectively, in the presence of 250 mM NaCl contained highly purified LH1 complexes. The purified LH1 complexes were dialyzed extensively to remove detergent, pelleted by ultracentrifugation, and then stored at −85°C. For the determination of the total pigments, the purified LH1 complexes (1 volume) were extracted with 3 volumes of methylene chloride-methanol (MeOH; 1:2 [vol/vol]) for 20 min, and then phase separation was induced by the further addition of H2O-methanol (1.8:1.8 [vol/vol]). Phase separation was completed after a short centrifugal step (3,000 × g). The total organic solvent-soluble pigments were obtained from the lower fraction and then dried under N2. The procedure was repeated three times, and all fractions were pooled. The dried extract was then dissolved in diethyl ether prior to spectral measurement.
(iv) Carotenoid extraction.
Portions (2 ml) of a stationary-phase cell culture were centrifuged briefly, and the cell pellets were extracted twice with 1 ml of MeOH to remove the bacteriochlorophyll a (BChla). Three sequential hexane extractions were then performed to obtain the carotenoids (see Fig. S1 in the supplemental material for comparative spectra in hexane). The entire extraction process was performed under low-light conditions under N2 to prevent photobleaching and degradation. The MeOH and hexane extracts were initially evaporated under vacuum. In some cases, crt extinction coefficients not reported in the literature were determined in the present study (see Table S1 in the supplemental material). The extinction coefficients used were ε1 cm488 nm = 140 mM−1 cm−1 (spx in petroleum ether), ε1 cm502 nm = 184 mM−1 cm−1 (lycopene in diethyl ether), and ε1 cm502 nm = 172 mM−1 cm−1 (lycopene in hexane) (see Table S1 in the supplemental material). For BChla, an extinction coefficient of 90 mM−1 cm−1 (in diethyl ether [52]) was used.
(v) Thin-layer chromatography (TLC).
Samples (100 μl) were dried under N2 and then redissolved in 100 μl of pentane-ethanol (100:2 [vol/vol]) and applied to silica gel plates (DC-Fertigplatten SIL G-25 [Merck]), which were subsequently developed using pentane-ethanol (100:2 [vol/vol]) as a running solvent.
(vi) High performance liquid chromatography-mass spectroscopy (HPLC-MS).
Extraction of carotenoid pigments was performed essentially as described above, except that the final carotenoid fraction, following the two initial MeOH extractions, was obtained by a further two sequential extractions with methylene chloride. The determination and identification of carotenoid pigments in cell extracts were conducted using an Agilent 1100 quaternary HPLC system with two detectors—a diode array detector (DAD) and a single-quadrupole MSD SL mass spectrometer (Agilent, Palo Alto, CA)—coupled in series. For data acquisition and analysis, the Agilent LC/MSD ChemStation software (version Rev.B02.01-SR2) was applied. HPLC separation was achieved on a Grom-Sil 300 ODS column (150 by 4 mm; inner diameter, 5 μm [Alltech Grom GmbH, Rottenburg-Hailfingen, Germany]) using the solvents A (acetonitrile), B (MeOH–2-propanol (4:1 [vol/vol]), ammonium acetate [10 mM]), C (H2O), and D (tetrahydrofuran). The applied elution profile was as follows: first 30% solvent A, followed by 20% solvent B, 50% solvent C, a linear gradient to 30% solvent A, 50% solvent B, and then 20% solvent D was run within 20 min at a flow rate of 1 ml/min. This condition was maintained for 20 min for eluting all carotenoid pigments. The column temperature was 20°C, controlled by a column thermostat. After passing the DAD flow cell, the HPLC mobile phase was directly introduced into the mass spectrometer via an atmospheric pressure chemical ionization ion source. A nitrogen gas generator was used for supplying nitrogen from pressurized air as a drying gas. The nebulizer pressure was 60 lb/in2. The drying gas flow was 5 liter/min, and the gas temperature was 300°C (vaporizer temperature, 325°C). The capillary voltage was 4,000 V in both positive and negative modes. A corona current of positive 10 μA and negative 15 μA was applied. Ions were monitored in the scan mode, covering a mass range from 200 to 1,200 mass units with negative polarity. The fragmentor voltage was 100 V. The identity of lycopene was confirmed with a reference standard obtained from Sigma-Aldrich (St. Louis, MO).
RESULTS
Isolation of the lycopene-producing strain SLYC18.
Transconjugants were only obtained using pSUPSGE5K2, where the npt gene is in the same orientation as crtD. Approximately 400 transconjugants were isolated using an initial kanamycin selection on Sistrom (M) agar under dark, aerobic conditions. Both purple and brown colonies were observed. An initial spectral analysis (Fig. 3; see below for a detailed analysis) showed the purple colonies to contain spx, whereas the brown colonies exhibited absorption spectra resembling those previously reported for LH1-bound 3,4,3′,4′-tetrahydrospirilloxanthin (thspx [14, 27]) in the carotenoid region. All of the purple colonies examined had the Kanr Tetr phenotype and were therefore probably single recombinants. Double recombinant brown colonies were identified by their Kanr Tets phenotype, and one of these, designated SLYC18, was chosen for further analysis.
Fig 3.
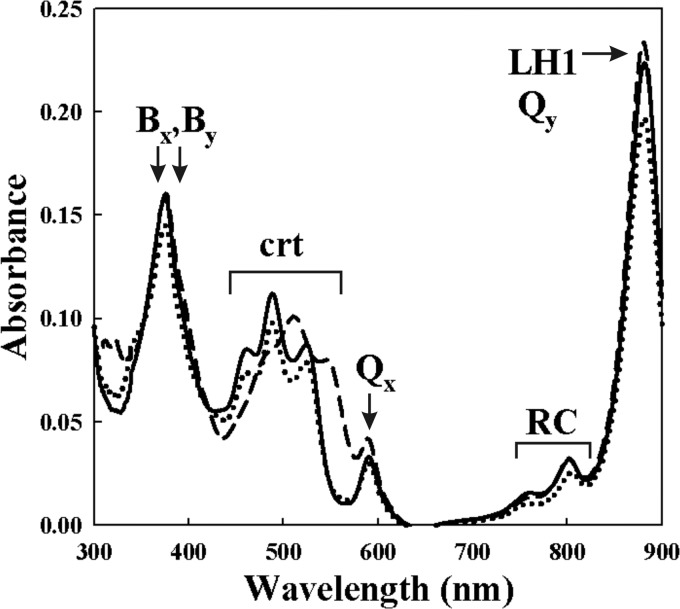
Whole-cell spectra of strains SLYC18 (solid line), ST4 (dotted line), and the wild-type S1 (dashed line), containing different carotenoids. The cell spectra were normalized to the same A660 for comparison. Indicated are the absorption peaks corresponding to crt, as well as the BChla near-IR maxima (Qx and Qy) and Soret bands (Bx and By) of the LH1 and RC.
HPLC analysis of the carotenoids extracted from late-exponential-phase or stationary-phase cells of SLYC18 with organic solvents showed a dominant peak, which was confirmed by both spectral and MS analysis to be lycopene (Fig. 4A and B). By comparison, carotenoid extracts obtained from cells of several purple strains were shown to be dominated by spx, showing HPLC-MS profiles corresponding to the wild-type S1 (Fig. 4C and D).
Fig 4.
HPLC-MS analysis of extracted carotenoids from SLYC18 (A and B) and S1 (C and D). The insets show the absorption spectra of the dominant peaks (peak 4 [in panel A] and peak 2 [in panel C], respectively). The masses (M− mode) shown in panel B correspond to lycopene (m/z 536.4), 3,4-didehydrorhodopin (m/z 551.2), and an unassigned species (m/z 442.4). (D) shows the mass spectrum (M+H+ mode) of the dominant peak 2 (spx, m/z 597.2) in panel C. The remaining peaks in panel C were also assigned from their mass spectra (data not shown): peak 1, OH-spx (m/z 568.8); peak 3, anhydrorhodovibrin (m/z 567.2); peak 4, lycopene; and peak 5, 3,4-didehydrorhodopin (m/z 553.4).
Finally, we confirmed by Southern hybridization (data not shown) that only a single chromosomal insertion of the npt gene was present in the chromosome. The mutant could also be complemented (see Fig. S2A in the supplemental material) by the plasmid pRKSGE5, which contains the 6.37-kb EcoRV fragment from the cosmid pSC4, containing crtC, crtD, and crtE. The complemented mutant was purple in color and grew almost normally (i.e., as the wild-type S1) under anaerobic, photoheterotrophic conditions, as well as under dark, chemoheterotrophic semiaerobic conditions (see Fig. S2A in the supplemental material). The carotenoid spectrum of both total cells and also the extracted pigments of the complemented strain was identical to that observed for the wild type (see Fig. S2B [whole-cell spectrum] and Fig. S3A [extracted pigments] in the supplemental material). Furthermore, TLC analysis of the extracted pigments of the complemented strain showed only the pink band due to spx, with no trace of the orange lycopene band observed for the extracted pigments of SLYC18 (see Fig. S3B in the supplemental material). We conclude that the complemented cells contain spx levels identical to those for the wild-type strain S1.
We note, also, in passing, that there appears to be no polar effect of the interposon lesion on the downstream R. rubrum cobZ gene variant (cobZRR). Lesions in this gene lead to a very distinct phenotype (R. Ghosh, manuscript in preparation); cobZRR mutants are photosynthetically incompetent and excrete a brown BChla precursor in large quantities. In contrast, SLYC18 grows well photosynthetically and produces no brown precursor.
Detailed spectral analysis of the SLYC18 mutant.
The absorption spectra of whole cells (Fig. 3) showed, as reported previously for the thspx-expressing mutant, ST4 (27), that the near-infrared (near-IR) Qy absorption maximum of the LH1 complex is present at 882 nm, which is characteristic for crt-containing LH1 complexes (11, 60). The LH1 peak maximum at 882 nm is easily distinguishable from that of LH1 complexes lacking carotenoids, which occurs at 874 nm (11, 17, 32, 60). The absorption maxima of the observable RC at 750 and 802 nm, corresponding to the RC-bound bacteriopheophytin (BPh) and accessory BChla, respectively, are unchanged in both position and relative intensity for all three strains. Although the small absorption peak (at 870 nm) corresponding to the special pair of the RC is not observable in the cell spectrum (since the LH1 peak is dominant), we assume the RC to be intact and functional, since SLYC18 grows normally (i.e., with respect to S1) under photoheterotrophic conditions. Also, the relative intensities of the three crt peaks compared to those (Qx and Qy) of BChla are very similar in both the wild-type and the two crt mutants. It is now well established (6, 17, 41) that spx, which is present in the wild-type strain, is always protein bound, with ca. 94% (16 mol of spx/mol of LH1 complex) bound to the LH1 complex and 6% (1 mol of spx/mol of RC) bound to the RC. For practical purposes, therefore, the observed spectra in Fig. 3 correspond primarily to that of the LH1 complex. In the case of ST4, we have previously shown (14) that thspx is also bound to the LH1 complex and RC and that this generates the characteristic peak maximum of BChla at 882 nm and a relative intensity of this peak maximum to those of thspx as described above. The very strong similarity between the ST4 and SLYC18 cell spectra suggests strongly that, in SLYC18, lycopene is also bound to pigment-protein complexes. Fractionation of SLYC18 cells after breakage with a French press-type apparatus, followed by differential centrifugation (17, 32), showed the lycopene to be located exclusively in the membrane fraction, of which the ICM, containing the LH1 and RC, comprises ca. 80%.
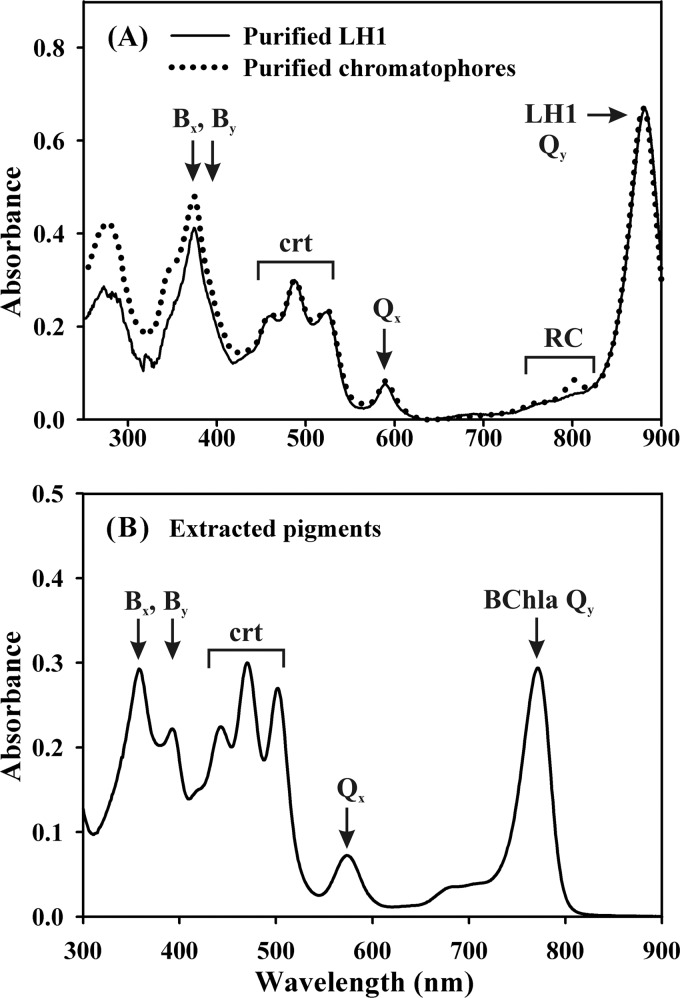
Finally, we purified the LH1 complex from SLYC18 and analyzed its spectroscopic properties and pigment content. As shown in Fig. 5A, the purified LH1 complex shows an absorption spectrum which is essentially identical to that of total ICM (which in isolated form are called chromatophores), including the position of the LH1-Qy maximum at 882 nm, which is characteristic of native aggregated LH1 rings (53), and the relative intensities of the peaks of LH1-bound lycopene to that of the LH1 peak maximum at 882 nm. For all of the LH1 complexes obtained from spx-, thspx-, and lycopene-containing strains, this latter ratio is essentially constant. The absorption maxima of the LH1-bound lycopene correspond perfectly to those of intact cells (see Fig. 3). Finally, the pigment stoichiometry of the LH1 complex, determined from the characteristic absorption maxima of BChla (771 nm) and lycopene (502 nm) of the total extracted pigments redissolved in diethyl ether (Fig. 5B), was calculated to be 2.27 ± 0.07, corresponding well to the expected value of 2 mol of BChla/mol of crt (41). Comparison of the biochemical and spectroscopic data therefore indicates clearly that, in vivo, lycopene is almost exclusively bound to LH1 complexes in the ICM.
Fig 5.
(A) Comparison of the absorption spectrum of isolated SLYC18 chromatophores (the ICM fraction [dotted line]) to that of the purified LH1 complex (solid line) obtained from the ICM fraction. Note that the peak at 802 nm, which is observed for the ICM and is due to the absorption of the accessory BChla of the RC, is not present in the LH1 preparation. (B) Absorption spectrum of the pigment fraction after methylene chloride-MeOH extraction (see Materials and Methods) of the purified LH1 complexes. The spectrum was obtained using diethyl ether as a solvent.
Growth physiology of the crt mutants.
In many studies using purple photosynthetic bacteria, it is natural to focus on their growth characteristics obtained under light, anaerobic conditions. Although SLYC18 indeed grows well photosynthetically, in the present study we focused on the dark, semiaerobic growth condition for lycopene production since we believe that the latter is most readily adapted to a large-scale industrial environment. These conditions are facilitated by the availability of M2SF and M2SF+ medium (18), which allows exceptionally high levels of pigment production under semiaerobic growth conditions, which has not been achieved by any other purple bacterium thus far. However, the present study is the first to use this medium for the production of a “non-natural” pigment product, so we felt it useful to study the growth characteristics in detail in this case.
To compare the physiology of the wild-type and crt mutants, we grew them in two different media: M2SF and M2SF+. M2SF has been described previously (18) to yield high cell densities with high levels of ICM that are normally only observed under anaerobic, photoheterotrophic conditions with conventional media. M2SF+ is a modification of M2SF, using even higher concentrations of both fructose (111.3 mM) and succinate (60 mM), which leads to 4-fold-higher cell densities than with M2SF, while maintaining the phototrophic levels of ICM/cell. A very useful monitor of semiaerobic growth is the ratio A882/A660, which indicates the relative level of ICM (proportional to the amount of LH1 complex/cell). The A882/A660 ratio reflects the internal redox physiology (i.e., both the intracellular pO2 and the ratio between reduced and oxidized ubiquinone in the ICM) of the cell, which influences the signal transduction events that regulate ICM expression (20). Thus, in a typical semiaerobic growth curve, performed using shake flasks and a high ICM-containing inoculum, the A882/A660 ratio initially decreases (corresponding to the repression of ICM synthesis at high pO2, while maintaining cell growth) until the cell density is high enough to reduce the local pO2 to <0.5% (18). At this point, ICM synthesis resumes, concomitant with a rise in the A882/A660 ratio. During this growth phase, both the “aerobic”, oxidative mode and the “anaerobic”, reductive mode of metabolism are utilized (18, 20). The A882/A660 turning point is designated here as a “trough.”
Typical semiaerobic growth curves (100-ml cultures) in both M2SF and M2SF+ media are shown for the wild type and mutants in Fig. 6. The M2SF growth profiles indicate the major physiological differences between strains. In all cases, a lag phase is observed, although for the wild type this phase is short (∼14.7 h) and within the normal range observed for cultures where the inoculum has been stored for about 12 to 24 h at 4°C before inoculation. However, lag phases exhibited by the mutants are significantly longer (ST4 [22.8 h] and SLYC18 [52.5 h]) than is ever observed for wild-type cells. Interestingly, the position of the A882/A660 trough does not correlate perfectly with the cell density. Whereas both S1 and SLYC18 exhibit the trough at an A660 of ∼0.28, the ST4 trough occurs at about twice this value (Fig. 6A and B). The long lag phase observed for SLYC18 may indicate that the rate of O2 consumption is lower in this strain in comparison to those of S1 and ST4 or that low-level oxidation of carotenoids may be toxic when the cell density is low. However, after the appropriate time, all of the cultures reach the same final cell densities, and the ICM content in the late exponential and stationary phase is comparable. We note, in passing, that the carotenoid-dependent lag phase does not correspond to the number of reducing equivalents removed from the carotenoids during the transition from phytoene to the final product (S1, 12 [H]; SLYC18, 8 [H]; and ST4, 8 [H]).
Fig 6.
Growth curves of wild-type (S1 [●]) and crt mutants (SLYC18 [○]) and ST4 [▼]) in M2SF medium (A) and M2SF+ medium (C). For each strain, in each growth medium, three independent growth curves were performed, but in the figure, only the average curve with error bars is shown. For each growth curve, the development of ICM (indicated by A882/A660) is also shown in panels B and D. The dashed lines have been included to provide visual aids showing the correlation between the onset of the microaerophilic state experienced by the cells, corresponding to the increase in ICM/cell (A882/A660) and the growth phase of the cells in the respective media. For comparative purposes, we have also included the commonly used semilogarithmic plot as an inset.
Since we believe the length of the lag phase to have a distinct physiological meaning, related to the type of crt being produced, it would be useful to precisely determine its length using an objective procedure. We found the usually applied semilogarithmic plots to be both subjective and inaccurate, so for this reason we turned to a precise fitting procedure, which also yields reliable growth rate estimates. First, we noticed that the growth behavior following the lag phase fits the well-known logistic equation (see reference 38):
| (1) |
In equation 1, n is the number of cells at measurement time t, n0 is the initial cell number, K is the “carrying capacity” of the medium (an empirical constant that indicates the maximal cell density when the medium is exhausted), and r is the growth rate. The difficulty here is to objectively define the end of the lag phase. To do this, in a series of trials, each curve was fitted to equation 1, with a successively longer lag time as the theoretical “time zero” value. The success of the fit was judged by observing the variance (S2) of the residual, calculated as follows:
| (2) |
where nfit and nexptl correspond to the fitted and experimental A660 values at each time point, and N is the number of time points used for the fit to equation 1 above (see Fig. S4 in the supplemental material for an example). The fitted lag phase endpoint was defined as the time point corresponding to the smallest variance. This treatment also allowed us to determine the exponential growth rates (r) precisely for each strain. In fact, all strains show very similar growth rates (∼0.1 A660 h−1 [M2SF] and ∼0.06 A660 h−1 [M2SF+]) upon entering the exponential phase (see Table S2 in the supplemental material).
In M2SF+ medium, crt mutants exhibited a similar long lag phase (ca. 39.5 h for SLYC18 and 46.6 h for ST4, respectively) which was about twice that of the wild-type (21.8 h) (Fig. 6C, see Table S2 in the supplemental material), although active growth still followed equation 1 and all strains attained comparable cell densities (∼4-fold of those reached in M2SF medium). Also, the A882/A660 ratio in the late exponential phase was very similar to that observed in M2SF medium. The position of the A882/A660 trough with respect to the cell density appeared to be nearly the same in all strains (Fig. 6D). In comparison to the M2SF data, the M2SF+ data are relatively “noisy.” This is probably due to the fact that at very high cell densities, even during the short time required for manual sampling (generally about 2 to 3 min), the redox physiology can change significantly. We have noticed this effect repeatedly in other experiments utilizing very high cell densities.
Quantitation of carotenoid levels.
Carotenoid levels were determined using stationary-phase M2SF+-grown cells (Fig. 6C) by MeOH-hexane extraction followed by spectral analysis. The wild-type S1 yielded values of 1.41 mg of spx/g (dry weight) (Fig. 7A) and 2.17 mg of spx/g of protein (Fig. 7C), respectively. The latter value is very close to the value 2.45 mg of spx/g of protein, determined by Jensen et al. (23) for photoheterotrophic cultures of S1 and corresponds to the ratio of 65% protein/g (dry weight), which we have consistently observed for S1 cells growing in either M2SF or M2SF+. The lycopene and thspx levels per g of protein appear to be significantly higher (36 and 16%, respectively, Fig. 7C), but in fact these values only reflect a corresponding lowering of the percentage of total protein/g (dry weight) in SLYC18 and ST4 (53 and 46%, respectively). However, notwithstanding these considerations, the final levels of both lycopene and thspx per g (dry weight) still appear to be ca. 22% higher than those of spx in S1 (Fig. 7A), and the amount of lycopene and thspx obtainable per liter M2SF+ culture appears to be ca. 40% higher for the mutants than for the wild type (Fig. 7B). These large differences are inconsistent with the spectral analyses of whole cells, which when normalized to the same cell density show identical crt levels (and also LH1 levels) in all strains. This inconsistency can be partly rationalized by the fact that, for S1, the growth sampling was aborted prior to attainment of the stationary phase. In the last part of the growth curve, even relatively small differences in cell turbidity can reflect large differences in cell mass. Our decision to abort the S1 growth curves prematurely was dictated by the fact that the turbidity can also fall significantly in the stationary phase. This behavior is due partly to the loss of cell viability but is also due to the optical effects caused by cell elongation. However, other physiological differences may also play a significant role (see Discussion).
Fig 7.
Quantitative analysis of carotenoids from stationary phase cells grown in M2SF+ medium. All determinations were performed in triplicate for each of the individual cultures shown in Fig. 6C. Thus, each of the bar chart values and their corresponding error bars were obtained from a total of nine determinations— mg of crt/g (dry weight) of cells (A), mg of crt/liter of culture (B), and mg of crt/g of total protein (C)—as determined by a modified Lowry-Peterson procedure (40; see also Materials and Methods).
DISCUSSION
In this study, we have shown unambiguously that the final product of the R. rubrum phytoene desaturase reaction is indeed lycopene, as indicated by early chemical studies (8). Using LC-MS techniques, we could find no trace of neurosporene, which is the final physiological product of the phytoene desaturase present in both Rb. capsulatus (5, 19) and Rb. sphaeroides (28). Another type of CrtI has been found in Rv. gelatinosus (39), where the final products appear to be a mixture of lycopene (10%) and neurosporene (90%) (21), a finding consistent with the presence of both spx and spheroidene as the end carotenoids in this organism (42). Thus, the R. rubrum phytoene desaturase corresponds to a third type of CrtI, with the same enzymatic function as in the nonphotosynthetic Pantoea ananatis (31, 34) and Pantoea stewartii (15, 22). Thus far, only one other purple bacterial CrtI (from Rhodopseudomonas [Rp.] palustris) has been reported (55), which yields exclusively lycopene as the final product.
The reasons for the different product specificities of the various CrtIs are not yet clear. Although Wang and Liao (59) were able to isolate a random mutant of the Rb. sphaeroides CrtI that could produce ca. 90% lycopene and 10% neurosporene, the amino acid changes they reported do not correlate with the corresponding amino acids in either the Rv. gelatinosus or R. rubrum enzymes (see Fig. S5 in the supplemental material, homology set 1). Recently, Stickforth and Sandmann (54) showed that the Rv. gelatinosus enzyme could also be mutated to increase the lycopene production to 80%. However, here also the amino acid changes reported bear no relation to those present at the corresponding sequence positions of the R. rubrum CrtI (see Fig. S5 in the supplemental material). A sequence comparison of crt enzymes with known function shows that although the N- and C-terminal domains are reasonably conserved, there is considerable sequence variation within the central domains among the various enzymes (see Fig. S5 in the supplemental material). Also, the lycopene-producing bacterial CrtIs from R. rubrum and Rp. palustris are more similar to other purple bacterial enzymes than to those from the nonphotosynthetic Pantoea species. Finally, the Rv. gelatinosus CrtI appears to show more extensive sequence homologies to the CrtI enzymes from R. rubrum and Rp. palustris than to those from Rb. sphaeroides and Rb. capsulatus, despite the fact that its product specificity is much closer to those of the latter enzymes (see Fig. S5 in the supplemental material, homology set 2). Thus, the sequence differences between the various enzymes which lead to different substrate and product specificities must be subtle, since no characteristic sequence motifs can be readily detected.
The spectral analysis of whole cells and also isolated ICM indicates strongly that lycopene is incorporated quantitatively into the LH1 complex. First, the relative ratios of the carotenoid maxima to that of the near-IR absorption maximum of the LH1 complex are essentially identical to those of the wild type, as well as to those of the thspx mutant ST4. We have previously reported that the LH1 complex from ST4 is functionally intact and contains tightly bound thspx (16, 27). In fact, the thspx-LH1 complex shows a somewhat higher photochemical stability than that from S1 (data not shown). Second, the position of the near-IR peak maximum of the SLYC18 LH1 at 882 nm, which is a well-established diagnostic feature of LH1 containing bound carotenoid (11, 17, 32, 60), is identical to those of S1 and ST4. The prediction that the lycopene is indeed bound specifically to LH1 complexes was confirmed unambiguously by the analysis of purified LH1 complexes obtained from SLYC18. Here, the observed ratio of ∼2 mol of BChla/mol of lycopene is completely consistent with the structural (6) and biochemical (41) data available for the LH1 complex thus far. Finally, the spectrum of the isolated ICM from SLYC18, which is known to contain the LH1 and RC, is almost identical to that of the isolated LH1 complex from the same strain, as is the case for both spx-containing and thspx-containing strains. These observations contradict the conclusions of Fiedor et al. (11) and Kakitani et al. (25), based on in vitro reconstitution studies, that nonhydroxylated carotenoids such as lycopene cannot be incorporated efficiently into R. rubrum LH1 complexes.
The crt levels reported here are comparable to early attempts to produce carotenoids in engineered strains of E. coli (58) or yeasts (36, 49, 62), by the introduction of plasmid-borne crt genes from Pantoea species. More recent studies, using more sophisticated metabolic engineering techniques, have reported levels ranging from 14 to 22 mg of crt/g (dry weight) of E. coli cells (2, 56, 64).
The growth physiology of the R. rubrum crt mutants well illustrates the potential and relative simplicity of future crt production systems using phototrophic bacteria. Our choice of R. rubrum as a production strain was dictated partly by its natural high-level crt production capacity and partly by the now well-established fact that a massive redirection of carbon flux to pigments, as well as to phospholipid biosynthesis, is caused by the lowering of the pO2 to below 0.5%. The latter largely eliminates the necessity to redesign the early metabolic steps (as in E. coli) to achieve high pigment levels and simultaneously allows the enlargement of the membrane compartment. Finally, the availability of special media (M2SF and M2SF+) which allow ICM (and thus crt) to be expressed semiaerobically at levels normally observed only under anaerobic, photoheterotrophic growth conditions (18, 20), lends itself well to upscaling for an industrial process. In the present study, we have routinely used 3-liter semiaerobic shake cultures, which yield identical crt levels to those of 100-ml shake flask cultures grown in parallel. For all strains we have also obtained 10- to 50-liter batch cultures, which based on a comparison of their absorption spectra, appear to have identical crt levels (for all strains here) to those reported here.
The significantly different levels of crt/g (dry weight) in the three strains is partly attributable to the time point of harvesting in the stationary phase. Although R. rubrum is often portrayed as a large elongated spiral bacterium with many “bends,” this form arises only when the cells are in the late exponential or stationary phase. Rapidly growing R. rubrum cultures show only a single “bend” and are quite short (∼2 μm). The physiological changes (i.e., protein and lipid content) in the elongation phase have not yet been documented but may be significant. Possibly, this factor is responsible for the very different protein levels present in this phase (Fig. 7C).
An understanding of the growth physiology of phototrophic crt-producing strains will be necessary to proceed to produce higher crt levels, as well as other, possibly oxygenated carotenoids in R. rubrum. Thus, a relevant question is: if the growth physiologies of all three strains examined are so similar, what determines the very different lag phases observed? At present, we favor the hypothesis that the lag phase reflects the relative sensitivity of the strains to pO2, mediated at the crt level, in the initial growth phase. First, we do not observe significant lag phase differences when the three strains are grown photoheterotrophically, indicating that the incorporation of non-natural carotenoids (for R. rubrum, we consider also lycopene to be “non-natural” since only trace amounts are observed in growing cultures of wild-type strains) into the LH1 complex and RC is not fundamentally toxic for the cells. Second, in bioreactor experiments, performed in parallel to the present study, the early reduction of the O2 level essentially abolishes the lag phase difference (R. Feuer, J. Bona-Lovacz, O. Sawodny, and R. Ghosh, unpublished data). Possibly, the incorporation of non-natural carotenoids into the LH1 complex induces a conformational strain upon the structure, thereby making the BChla, which is normally quite well protected in the protein environment, more accessible to oxidative degradation. It has been shown (17) that even low oxidative damage of the LH1 complex leads to a significant destruction of the ICM organization, which is associated with aerobic toxicity. After a certain time, a small portion of the population is able to tolerate the toxicity so to commence cell growth, thereby lowering the pO2 and alleviating growth inhibition. Presumably, the hydroxylated/methoxylated carotenoids (e.g., spx) lead to less structural strain than the nonhydroxylated/nonmethoxylated ones (e.g., lycopene), which explains the relative efficiencies of the in vitro reconstitution experiments (11, 25). We have also observed long lag phases in other nonrelated R. rubrum mutants where the lesion causes the LH1 complex to be abnormally sensitive to oxidative stress (C. Autenrieth and R. Ghosh, unpublished data). Future strategies might therefore focus on making the LH1 complex less sensitive to the incorportion of non-natural carotenoids.
Although we have shown that it is feasible to produce respectable quantities of lycopene in R. rubrum, the levels are still much lower than those observed for recently reported, highly optimized E. coli strains (2, 10). However, there is good reason to believe that the lycopene levels observed with R. rubrum correspond to minimal levels. In particular, the very similar levels of crt in all three strains examined, together with the spectral and biochemical evidence that only sufficient levels to saturate the LH1 complex are produced, suggest that some kind of feedback regulation at the posttranslational level is occurring. This is supported by the observation that almost all mutants with lesions in BChla but not crt biosynthesis do not produce carotenoids (R. Ghosh, unpublished data). It is quite possible that if one could release this putative “brake” then significantly higher levels of crt may be attainable. This area is the focus of our ongoing research.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gerasimoula Gerasimidou for expert technical assistance and Caroline Autenrieth for critical reading of the manuscript.
The BMBF-FORSYS-Partner program (grant 0315282) is acknowledged for generous financial support.
Footnotes
Published ahead of print 3 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Albermann C. 2011. High- versus low-level expression of the lycopene biosynthesis genes from Pantoea ananatis in Escherichia coli. Biotechnol. Lett. 33:313–319 [DOI] [PubMed] [Google Scholar]
- 2. Alper H, Miyaoku K, Stephanopoulos G. 2005. Construction of lycopene-overproducing Escherichia coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23:612–616 [DOI] [PubMed] [Google Scholar]
- 3. Armstrong GA, Alberti M, Leach F, Hearst JE. 1989. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol. Gen. Genet. 216:254–268 [DOI] [PubMed] [Google Scholar]
- 4. Armstrong GA. 1995. Genetic analysis and regulation of carotenoid biosynthesis, p 1135–1157 In Blankenship RE, Madigan MT, Bauer CE. (ed), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 5. Bartley GE, Scolnik PA. 1989. Carotenoid biosynthesis in photosynthetic bacteria. J. Biol. Chem. 264:13109–13113 [PubMed] [Google Scholar]
- 6. Brunisholz RA, et al. 1986. The membrane location of the B890-complex from Rhodospirillum rubrum and the effect of carotenoid on the conformation of its two apoproteins exposed at the cytoplasmic surface. Biochim. Biophys. Acta 849:295–303 [Google Scholar]
- 7. Cohen-Bazire G, Sistrom WR, Stanier RY. 1956. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Comp. Physiol. 49:25–68 [DOI] [PubMed] [Google Scholar]
- 8. Davies BH. 1970. A novel sequence for phytoene dehydrogenation in Rhodospirillum rubrum. Biochem. J. 116:93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ditta G, et al. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149–153 [DOI] [PubMed] [Google Scholar]
- 10. Farmer WR, Liao JC. 2000. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 18:533–537 [DOI] [PubMed] [Google Scholar]
- 11. Fiedor L, Akahane J, Koyama Y. 2004. Carotenoid-induced cooperative formation of bacterial photosynthetic LH1 complex. Biochemistry 43:16487–16496 [DOI] [PubMed] [Google Scholar]
- 12. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fraser PD, et al. 1992. Expression in Escherichia coli, purification, and reactivation of the recombinant Erwinia uredovora phytoene desaturase. J. Biol. Chem. 267:19891–19895 [PubMed] [Google Scholar]
- 14. Gaertner P, Port H, Branschaedel M, Ghosh R. 2004. FS-study on energy relaxation in light-harvesting (LH1) complexes from Rhodospirillum rubrum with carotenoids of different conjugation length. J. Luminescence 108:111–116 [Google Scholar]
- 15. Garcia-Asua G, Cogdell RJ, Hunter CN. 2002. Functional assembly of the foreign carotenoid lycopene into the photosynthetic apparatus of Rhodobacter sphaeroides achieved by replacement of the native 3-step phytoene desaturase with its 4-step counterpart from Erwinia herbicola. Mol. Microbiol. 44:233–244 [DOI] [PubMed] [Google Scholar]
- 16. Gerken U, et al. 2003. Membrane environment reduces the accessible conformational space available to an integral membrane protein. J. Phys. Chem. 107:338–343 [Google Scholar]
- 17. Ghosh R, Bachofen R, Hauser H. 1985. Structural changes accompanying the irreversible oxidation of the chromatophores membrane from Rhodospirillum rubrum G9. Biochim. Biophys. Acta 765:97–105 [Google Scholar]
- 18. Ghosh R, Hardmeyer A, Thoenen I, Bachofen R. 1994. Optimization of the Sistrom culture medium for large-scale batch cultivation of Rhodospirillum rubrum under semiaerobic conditions with maximal yield of photosynthetic membranes. Appl. Environ. Microbiol. 60:1698–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giuliano G, Pollock D, Scolnik PA. 1986. The gene crtI mediates the conversion of phytoene into colored carotenoids in Rhodopseudomonas capsulata. J. Biol. Chem. 261:12925–12929 [PubMed] [Google Scholar]
- 20. Grammel H, Gilles E-D, Ghosh R. 2003. Microaerophilic cooperation of reductive and oxidative pathways allows maximal photosynthetic membrane biosynthesis in Rhodospirillum rubrum. Appl. Environ. Microbiol. 69:6577–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harada J, et al. 2001. Phytoene desaturase, CrtI, of the purple photosynthetic bacterium, Rubrivivax gelatinosus, produces both neurosporene and lycopene. Plant Cell Physiol. 42:1112–1118 [DOI] [PubMed] [Google Scholar]
- 22. Hunter CN, et al. 1994. Introduction of new carotenoids into the bacterial photosynthetic apparatus by combining the carotenoid biosynthetic pathways of Erwinia herbicola and Rhodobacter sphaeroides. J. Bacteriol. 176:3692–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jensen SL, Cohen-Bazire G, Nakayama TOM, Stanier RY. 1958. The path of carotenoid synthesis in a photosynthetic bacterium. Biochim. Biophys. Acta 29:477–498 [DOI] [PubMed] [Google Scholar]
- 24. Kajiwara S, Fraser PD, Kondo K, Misawa N. 1997. Expression of an exogeneous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 324:421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kakitani Y, et al. 2007. Conjugation-length dependence of the T1 lifetimes of carotenoids free in solution and incorporated into the LH2, LH1, RC, and RC-LH1 complexes: possible mechanisms of triplet-energy dissipation. Biochemistry 46:2181–2197 [DOI] [PubMed] [Google Scholar]
- 26. Kang MJ, et al. 2005. Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. Biotechnol. Bioeng. 91:636–642 [DOI] [PubMed] [Google Scholar]
- 27. Komori M, et al. 1998. A null lesion in the rhodopin 3,4-desaturase of Rhodospirillum rubrum unmasks a cryptic branch of the carotenoid biosynthetic pathway. Biochemistry 37:8987–8994 [DOI] [PubMed] [Google Scholar]
- 28. Lang HP, Cogdell RJ, Gardiner AT, Hunter CN. 1994. Early steps in carotenoid biosynthesis: sequences and transcriptional analysis of the crtI and crtB genes of Rhodobacter sphaeroides and overexpression and reactivation of crtI in Escherichia coli and Rhodobacter sphaeroides. J. Bacteriol. 176:3859–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee PC, Schmidt-Dannert C. 2002. Metabolic engineering toward biotechnological production of carotenoids in microorganisms. Appl. Microbiol. Biotechnol. 60:1–11 [DOI] [PubMed] [Google Scholar]
- 30. Lee PC, Mijts BN, Schmidt-Dannert C. 2004. Investigation of factors influencing production of the monocyclic carotenoid torulene in metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 65:538–546 [DOI] [PubMed] [Google Scholar]
- 31. Linden H, et al. 1991. Functional complementation in Escherichia coli of different phytoene desaturase genes and analysis of accumulated carotenes. Z. Naturforsch. 46:1045–1051 [DOI] [PubMed] [Google Scholar]
- 32. Lupo D, Ghosh R. 2004. The reaction center H subunit is not required for high levels of light-harvesting complex 1 in Rhodospirillum rubrum mutants. J. Bacteriol. 186:5585–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matthews PD, Wurtzel ET. 2000. Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of deoxyxylulose phosphate synthase. Appl. Environ. Microbiol. 53:396–400 [DOI] [PubMed] [Google Scholar]
- 34. Misawa N, et al. 1990. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172:6704–6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Misawa N, Shimada H. 1998. Metabolic engineering for the production of carotenoids in noncarotenogenic bacteria and yeasts. J. Bacteriol. 59:169–181 [DOI] [PubMed] [Google Scholar]
- 36. Miura Y, et al. 1998. Production of the carotenoids lycopene, β-carotene, and astaxanthin in the food yeast Candida utilis. Appl. Environ. Microbiol. 64:1226–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neudert U, Martinez-Ferez IM, Fraser PD, Sandmann G. 1998. Expression of an active phytoene synthase from Erwinia uredovora and biochemical properties of the enzyme. Biochim. Biophys. Acta 1392:51–58 [DOI] [PubMed] [Google Scholar]
- 38. Novak MA. 2006. Evolutionary dynamics: exploring the equations of life, p 13 Harvard University Press, Cambridge, MA [Google Scholar]
- 39. Ouchane S, Picaud M, Vernotte C, Reiss-Husson F, Astier C. 1997. Pleiotropic effects of puf interposon mutagenesis on carotenoid biosynthesis in Rubrivivax gelatinosus. J. Biol. Chem. 272:1670–1676 [DOI] [PubMed] [Google Scholar]
- 40. Peterson GL. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83:346–356 [DOI] [PubMed] [Google Scholar]
- 41. Picorel R, Bélanger G, Gingras G. 1983. Antenna holochrome B880 of Rhodospirillum rubrum S1: pigment, phospholipid, and polypeptide composition. Biochemistry 22:2491–2497 [Google Scholar]
- 42. Pinta V, et al. 2003. Characterization of unusual hydroxyl- and ketocarotenoids in Rubrivivax gelatinosus: involvement of enzyme CrtF or CrtA. Arch. Microbiol. 179:354–362 [DOI] [PubMed] [Google Scholar]
- 43. Ruther A, Misawa N, Böger P, Sandmann G. 1997. Production of zeaxanthin in Escherichia coli transformed with different carotenogenic plasmids. Appl. Microbiol. Biotechnol. 48:162–167 [DOI] [PubMed] [Google Scholar]
- 44. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Sandmann G, Woods WS, Tuveson RW. 1990. Identification of carotenoids in Erwinia herbicola and in a transformed Escherichia coli strain. FEMS Microbiol. Lett. 71:77–82 [DOI] [PubMed] [Google Scholar]
- 46. Sandmann G, Albrecht M, Schnurr G, Knörzer O, Böger P. 1999. The biotechnological potential and design of novel carotenoids by gene combination in Escherichia coli. Tibtech 19:233–237 [DOI] [PubMed] [Google Scholar]
- 47. Schmidt K, Liaan-Jensen S, Schlegel HG. 1963. Die Carotinoide der Thiorhodaceae: I. Okenon als Hauptcarotinoid von Chromatium okenii Perty. Arch. Microbiol. 46:117–126 [PubMed] [Google Scholar]
- 48. Schmidt-Dannert C, Umeno D, Arnold FH. 2000. Molecular breeding of carotenoid biosynthetic pathways. Nat. Biotechnol. 18:750–753 [DOI] [PubMed] [Google Scholar]
- 49. Shimada H, et al. 1998. Increased carotenoid production by the food yeast Candida utilis through metabolic engineering of the isoprenoid pathway. Appl. Environ. Microbiol. 64:2676–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 51. Sistrom WR. 1960. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 22:778–785 [DOI] [PubMed] [Google Scholar]
- 52. Smith JHC, Benitez A. 1955. Chlorophyll: analysis in plant materials, p 142–160 In Paech K, Tracy MV. (ed), Modern methods of plant analysis, vol 4 Springer-Verlag, Berlin, Germany [Google Scholar]
- 53. Stahlberg H, Dubochet J, Vogel H, Ghosh R. 1998. Are the light-harvesting I complexes from Rhodospirillum rubrum arranged around the reaction centre in a square geometry? J. Mol. Biol. 282:819–831 [DOI] [PubMed] [Google Scholar]
- 54. Stickforth P, Sandmann G. 2007. Kinetic variations determine the product pattern of phytoene desaturase from Rubrivivax gelatinosus. Arch. Biochem. Biophys. 461:235–241 [DOI] [PubMed] [Google Scholar]
- 55. Takaichi S. 2009. Distribution and biosynthesis of carotenoids, p 97–117 In Hunter CN, Daldal F, Thurnauer MC, Beatty JT. (ed), The purple phototrophic bacteria. Springer Science/Business Media, New York, NY [Google Scholar]
- 56. Tyo KEJ, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat. Biotechnol. 27:760–765 [DOI] [PubMed] [Google Scholar]
- 57. Vadali RV, Fu Y, Bennett GN, San KY. 2005. Enhanced lycopene productivity by manipulation of carbon flow to isopentenyl diphosphate in Escherichia coli. Biotechnol. Prog. 21:1558–1561 [DOI] [PubMed] [Google Scholar]
- 58. Wang C-W, Oh M-K, Liao JC. 1999. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol. Bioeng. 62:235–241 [DOI] [PubMed] [Google Scholar]
- 59. Wang C-W, Liao JC. 2001. Alteration of the product specificity of Rhodobacter sphaeroides phytoene deaturase by directed evolution. J. Biol. Chem. 276:41161–41164 [DOI] [PubMed] [Google Scholar]
- 60. Wiggli M, Cornacchia L, Saegesser R, Bachofen R, Ghosh R. 1996. Characterization of Rhodospirillum rubrum ST2: a new Tn5-induced carotenoid-less mutant for functional studies. Microbiol. Res. 151:57–62 [Google Scholar]
- 61. Wolfe AJ. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamono S, Ishii T, Nakagawa M, Ikenaga H, Misawa N. 1994. Metabolic engineering for production of β-carotene and lycopene in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 58:1112–1114 [DOI] [PubMed] [Google Scholar]
- 63. Yen H-C, Marrs B. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yoon S-H, et al. 2006. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol. Bioeng. 94:1025–1032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.