Abstract
A novel targeted massive parallel sequencing approach identified genetic variation in eight known or predicted fimbrial adhesins for 46 Salmonella strains. The results highlight associations between specific adhesin alleles, host species, and antimicrobial resistance. The differentiation of allelic variants has potential applications for diagnostic microbiology and epidemiological investigations.
TEXT
Salmonella is one of the most common agents of food-borne diseases (7, 18). It infects a variety of host species and can cause different types of disorders, particularly typhoid fever and gastrointestinal illnesses (11, 19). Genetic variation within a Salmonella serovar and between different serovars reflects evolutionary forces that have shaped host and disease specificities. Most studies on bacterial pathogens focus on the presence or absence of virulence and housekeeping genes that are involved in the different adaptive phenotypes and benefit bacterial survival and transmission. In contrast, little is known about the extent of allelic variation among these genes and the role of allelic sequences in pathogenesis and host specificity.
Comparative genomic analysis has demonstrated that unrelated strains of the same Salmonella serovar are typically more similar to each other than to members of other serovars, suggesting serovar-specific evolution (20). Genomic studies of individual serovars have also uncovered an expanding number of open reading frames with single-nucleotide polymorphisms (SNPs) that give rise to allelic variants that may be involved in the adaptation of individual strains to new environments or hosts (1). Studies have shown how allelic variants of the Salmonella FimH adhesin can determine cell and host specificities (2, 8, 10, 12). Other virulence factors, particularly colonization factors, have genes with nonsynonymous substitutions, suggesting the existence of additional allele-specific effects that determine host adaptation and pathogenesis (pathotype). Here, we tested a targeted massive parallel sequencing (MPS) strategy to evaluate the extent of allelic variation in known and predicted fimbrial adhesins and correlated specific alleles with host species. Moreover, it was hypothesized that Salmonella strains with adhesin alleles that mediate efficient colonization of mammalian intestines, a propitious environment for horizontal gene transfer (6, 13, 14, 16), accumulate mobile genes that encode antimicrobial resistance factors. Accordingly, whether antibiotic resistance of a strain (antibiotype) could be associated to the presence of specific adhesin alleles was investigated. The focus of this study was Salmonella enterica serovar Newport, which is a leading and genetically diverse serovar that causes outbreaks in bovine, equine, and human populations (17).
A novel target enrichment strategy was used to sequence eight known or predicted fimbrial adhesin genes (21). Genomic DNA was prepared from S. Newport strains isolated independently from fecal samples or animal-associated environments between 1997 and 2003 (see Table S1 in the supplemental material). All of the bovine isolates were obtained from animals from dairy farms in Pennsylvania. Fecal samples from these animals were submitted for diagnostic testing to the Pennsylvania Animal Diagnostic Laboratory System (PADLS). Two previously sequenced S. Newport human isolates (SL317 and SL254) were used as reference controls (5). Primers were designed (see Table S2 in the supplemental material) and used to specifically amplify bcfD, stbD, fimH, lpfD, sthE, stiH, stjA, and stfH (21). A microfluidic device (Access Array system; Fluidigm, South San Francisco, CA) was used to independently amplify separately 48 DNA segments each from 48 DNA templates for a total of 2,304 PCR products. DNA was collected from the 48 amplicon libraries containing each of the 48 amplicons from one of the Salmonella genomic templates (Fig. 1). These libraries were purified, quantified, and pooled for MPS. Sequences from all the resulting amplicons were determined by pyrosequencing with a 454 GS FLX sequencer using Titanium chemistry on one-eighth of a PicoTiterPlate.
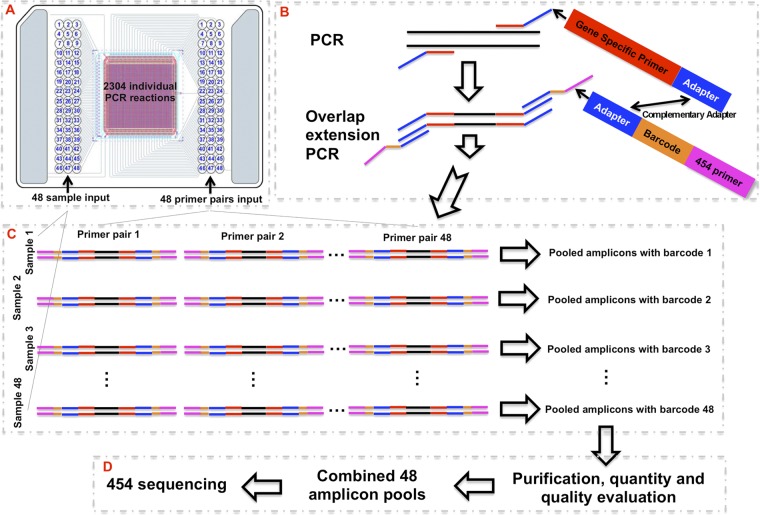
Fig 1.
Microfluidic PCR-based strategies for targeted sequencing. SNPs were detected by combining PCR target enrichment and barcoding with pyrosequencing. (A) The Fluidigm 48.48 Access Array system amplified 48 DNA segments each from 48 sample templates, for a total of 2,304 individual PCRs in a single run. (B) A unique overlapping PCR with two primer pairs that included complementary adapters resulted in sample-specific barcoded amplicons for each amplified gene (48 gene-specific primer pairs were each added to the 48 sample templates together with the 48 sample-specific barcoded primers for the PCR and overlap extension PCR, performed together in a single reaction compartment of the microfluidic system). (C) The 48 amplicons from each sample were collected as 48 sample-specific pools, each with its own barcode. (D) Amplicon pools from the 48 samples were purified, quantitated, and further pooled in an equimolar mixture for subsequent pyrosequencing with the 454 GS FLX sequencer.
The aim of the study was to correlate fimbrial adhesin alleles with host species and the presence of antimicrobial resistance. Investigating the S. Newport isolates (see Table S1 in the supplemental material) with uncharacterized genomic sequences identified SNPs leading to amino acid substitutions for the eight known or predicted adhesins examined. To identify major allele groups for correlation studies, phylogenic trees were constructed. Trees for the BcfD, FimH, and StfH proteins (see Fig. S1, S2, and S3 in the supplemental material), as well as for the SthE and StiH proteins (not shown), each detected the presence of two major groups of similar alleles (designated A and B) (see Fig. S1, S2, and S3, respectively) and a few minor groups (not shown). Group A of the BcfD, FimH, and StfH alleles overlapped for 22 strains, and reciprocally, 19 strains clustered as group B (see Fig. S4), highlighting significant allele associations in individual strains and suggesting convergent adaptation. Trees for the LpfD, StbD, and StjA alleles were more randomly distributed and did not form distinct groups (not shown). The data in Table 1 show that groups A and B of BcfD, FimH, or StfH correlated significantly with strains that were resistant or susceptible, respectively, to antimicrobial agents (P < 0.0001). There was also a significant association of groups A and B with the presence or absence of mobile DNA elements (as determined by the presence of plasmids or integrons) (see Table S1). Most of the strains with plasmids were resistant to antimicrobials (see Table S1), with several containing a previously characterized integron (3, 15), consistent with the typical location of Salmonella antimicrobial resistance genes on plasmids (4). However, only half of the plasmid-free strains were susceptible, suggesting that plasmids were lost after antimicrobial susceptibility testing was performed (done before plasmid profiling) or that the antimicrobial resistance genes of these strains were on the chromosome, possibly on integrative conjugative or mobilizable elements (9). Interestingly, groups A and B had different FimH alleles for strains from bovine or nonbovine sources (P < 0.05). No statistically significant associations were detected for the other adhesins studied (not shown). To summarize, the targeted sequencing system described here highlighted new associations of some bacterial adhesin alleles with host species and the presence of antibiotic resistance genes and plasmids.
Table 1.
Nine sets of 2-by-2 contingency tables correlating adhesin groups with antimicrobial resistance, mobile DNA, and host species
| Characteristic | No. of strains with: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| BcfD in: |
FimH in: |
StfH in: |
|||||||
| Groupa: |
P valueb | Group: |
P value | Group: |
P value | ||||
| A | B | A | B | A | B | ||||
| Resistant to antimicrobials | 24 | 3 | <0.0001 | 22 | 4 | <0.0001 | 22 | 4 | <0.0001 |
| Susceptible to antimicrobials | 2 | 16 | 1 | 18 | 1 | 16 | |||
| Mobile DNA elements | 15 | 2 | 0.0017 | 13 | 3 | 0.045 | 12 | 3 | 0.0231 |
| Nonmobile DNA elements | 11 | 17 | 10 | 19 | 11 | 17 | |||
| Bovine isolates | 10 | 3 | 0.1819 | 11 | 3 | 0.023 | 10 | 4 | 0.1191 |
| Nonbovine isolates | 16 | 16 | 12 | 19 | 13 | 16 | |||
Figure S4 in the supplemental material details how groups A and B of the three adhesins overlap.
Fisher's exact test was used to determine statistical significance.
In conclusion, we demonstrated that a targeted sequencing approach provides a time- and cost-effective means to identify SNPs in large numbers of bacterial isolates and highlight novel relationships by the detection of genotype-phenotype correlations (here, adhesin alleles and host-specific colonization or antibiotic resistance). The technology used is based on a simple PCR approach that prepares amplicons with barcodes and sequencing primers in one step on one array chip for direct MPS. The method bypasses the high cost and time-consuming approach of whole-genome sequencing and analysis. Such an approach could benefit translational and clinical research, including diagnostic microbiology and epidemiological investigations.
Nucleotide sequence accession number.
The sequences produced by this study are available in GenBank under accession numbers JX026805 to JX026831.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jason McKinney, Fluidigm, as well as Kathleen O'Shea and Tapan Ganguly, for technical support.
This work was supported by a University of Pennsylvania Research Foundation grant and Research Initiative Funds from the University of Pennsylvania Veterinary Center for Infectious Disease.
Footnotes
Published ahead of print 10 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Allard MW, et al. 2012. High resolution clustering of Salmonella enterica serovar Montevideo strains using a next-generation sequencing approach. BMC Genomics 13:32 doi:10.1186/1471-2164-13-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boddicker JD, Ledeboer NA, Jagnow J, Jones BD, Clegg S. 2002. Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 45:1255–1265 [DOI] [PubMed] [Google Scholar]
- 3. Brown AW, Rankin SC, Platt DJ. 2000. Detection and characterisation of integrons in Salmonella enterica serotype enteritidis. FEMS Microbiol. Lett. 191:145–149 [DOI] [PubMed] [Google Scholar]
- 4. Foley SL, Lynne AM. 2008. Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86:E173–E187 [DOI] [PubMed] [Google Scholar]
- 5. Fricke WF, et al. 2011. Comparative genomics of 28 Salmonella enterica isolates: evidence for CRISPR-mediated adaptive sublineage evolution. J. Bacteriol. 193:3556–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia-Quintanilla M, Ramos-Morales F, Casadesus J. 2008. Conjugal transfer of the Salmonella enterica virulence plasmid in the mouse intestine. J. Bacteriol. 190:1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerner-Smidt P, Whichard JM. 2010. Foodborne disease trends and reports. Foodborne Pathog. Dis. 7:609–611 [DOI] [PubMed] [Google Scholar]
- 8. Grzymajlo K, Kuzminska-Bajor M, Jaworski J, Dobryszycki P, Ugorski M. 2010. The high-adhesive properties of the FimH adhesin of Salmonella enterica serovar Enteritidis are determined by a single F118S substitution. Microbiology 156:1738–1748 [DOI] [PubMed] [Google Scholar]
- 9. Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222 doi:10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo A, et al. 2009. FimH alleles direct preferential binding of Salmonella to distinct mammalian cells or to avian cells. Microbiology 155:1623–1633 [DOI] [PubMed] [Google Scholar]
- 11. Kingsley RA, Baumler AJ. 2000. Host adaptation and the emergence of infectious disease: the Salmonella paradigm. Mol. Microbiol. 36:1006–1014 [DOI] [PubMed] [Google Scholar]
- 12. Kisiela D, et al. 2006. Functional characterization of the FimH adhesin from Salmonella enterica serovar Enteritidis. Microbiology 152:1337–1346 [DOI] [PubMed] [Google Scholar]
- 13. Lester CH, Frimodt-Moller N, Hammerum AM. 2004. Conjugal transfer of aminoglycoside and macrolide resistance between Enterococcus faecium isolates in the intestine of streptomycin-treated mice. FEMS Microbiol. Lett. 235:385–391 [DOI] [PubMed] [Google Scholar]
- 14. Nijsten R, London N, van den Bogaard A, Stobberingh E. 1995. In-vivo transfer of resistance plasmids in rat, human or pig-derived intestinal flora using a rat model. J. Antimicrob. Chemother. 36:975–985 [DOI] [PubMed] [Google Scholar]
- 15. Rankin SC, et al. 2002. Molecular characterization of cephalosporin-resistant Salmonella enterica serotype Newport isolates from animals in Pennsylvania. J. Clin. Microbiol. 40:4679–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rowe-Magnus D, Mazel D. 2006. The evolution of antibiotic resistance, p 221–244 In Seifert HS, DiRita VJ. (ed), Evolution of bacterial pathogens. ASM Press, Washington, DC [Google Scholar]
- 17. Sangal V, et al. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J. Bacteriol. 192:6465–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scallan E, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schikora A, et al. 2011. Conservation of Salmonella infection mechanisms in plants and animals. PLoS One 6:e24112 doi:10.1371/journal.pone.0024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soyer Y, Orsi RH, Rodriguez-Rivera LD, Sun Q, Wiedmann M. 2009. Genome wide evolutionary analyses reveal serotype specific patterns of positive selection in selected Salmonella serotypes. BMC Evol. Biol. 9:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yue M, et al. 2012. Diversification of the Salmonella fimbriae: a model of macro- and microevolution. PLoS One 7:e38596 doi:10.1371/journal.pone.0038596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.