Abstract
We report the detection of Salmonella carrying blaCTX-M in U.S. livestock populations. We identified 12 of 2,034 (0.6%) Salmonella isolates originating from turkeys, horses, and pigs from at least 6 U.S. states, all carrying blaCTX-M-1, many on a pandemic sequence type 1 IncN plasmid.
TEXT
We have previously reported commensal Escherichia coli isolated from U.S. livestock harboring blaCTX-M on conjugative plasmids (15, 29) and subsequently hypothesized that Salmonella spp. bearing blaCTX-M are present but unrecognized in U.S. livestock populations, posing a potential public health risk. Thus, our objective was to identify and characterize Salmonella carrying blaCTX-M among veterinary diagnostic submissions to the USDA APHIS National Veterinary Services Laboratories (NVSL).
To accomplish this, we screened 2,034 clinical Salmonella isolates submitted for serotyping to the NVSL between October 2010 and June 2011 (Table 1). Salmonella isolates that had been previously serotyped were tested in June 2011 by streaking to Mueller-Hinton agar containing 8 μg/ml cefepime. We identified a total of 12 (0.6%) Salmonella isolates carrying blaCTX-M on transferable plasmids (Table 2). We did not detect other classes of β-lactamase resistance genes, including CMY, TEM, SHV, and OXA (11, 14, 22), by PCR. MICs for these Salmonella isolates generally displayed the expected phenotype (Fig. 1). Southern blot hybridization (10, 13, 20) using a blaCTX-M probe indicated the localization of the gene on the plasmids (Fig. 2).
Table 1.
Salmonella clinical isolates screeneda
| S. enterica serovar | No. of represented states | No. of isolates screened by species |
Total no. of isolates | |||||
|---|---|---|---|---|---|---|---|---|
| Cattle | Chickens | Horses | Swine | Turkeys | Otherb | |||
| Typhimurium var. 5− | 31 | 26 | 5 | 7 | 215 | 1 | 17 | 271 |
| Agona | 16 | 25 | 1 | 4 | 111 | 6 | 2 | 149 |
| Dublin | 21 | 116 | 0 | 0 | 2 | 0 | 3 | 121 |
| Typhimurium | 24 | 30 | 4 | 9 | 54 | 0 | 15 | 112 |
| Cerro | 16 | 100 | 1 | 1 | 5 | 0 | 0 | 107 |
| Derby | 16 | 1 | 0 | 0 | 98 | 1 | 3 | 103 |
| Montevideo | 20 | 61 | 0 | 3 | 6 | 4 | 6 | 80 |
| Infantis | 15 | 4 | 1 | 13 | 55 | 0 | 2 | 75 |
| Heidelberg | 16 | 8 | 1 | 1 | 53 | 10 | 1 | 74 |
| Newport | 21 | 31 | 0 | 17 | 4 | 0 | 18 | 70 |
| Anatum | 13 | 14 | 0 | 11 | 33 | 0 | 5 | 63 |
| Senftenberg | 13 | 1 | 2 | 2 | 34 | 15 | 8 | 62 |
| Enteritidis | 13 | 3 | 42 | 2 | 1 | 0 | 8 | 56 |
| Kentucky | 21 | 30 | 9 | 2 | 3 | 0 | 2 | 46 |
| Mbandaka | 13 | 17 | 2 | 1 | 12 | 1 | 4 | 37 |
| Worthington | 16 | 0 | 1 | 1 | 30 | 0 | 0 | 32 |
| Ouakam | 7 | 0 | 1 | 0 | 8 | 16 | 1 | 26 |
| Bredeney | 5 | 4 | 0 | 2 | 7 | 3 | 0 | 16 |
| Rough O:d:e,n,z15 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| All other serotypesc | 38 | 110 | 13 | 68 | 208 | 31 | 103 | 533 |
| Total | 41 | 581 | 83 | 144 | 940 | 88 | 198 | 2,034 |
Summary of results for 2,034 Salmonella clinical isolates originally submitted to the NVSL for serotyping between October 2010 and June 2011 that were screened for the blaCTX-M phenotype using selective media.
Includes isolates submitted from wild animals, zoo animals, cats, dogs, goats, and sheep and isolates of unknown origin.
Includes 134 additional serotypes that were present in this isolate set.
Table 2.
Salmonella isolates carrying blaCTX-Ma
| Isolate no. | Isolate ID | Serotype | Serogroup | Source | State | Date received (day/mo/yr) | PRTb |
|---|---|---|---|---|---|---|---|
| 1 | 11-13049 | Anatum | E | Equine | TX | 11/26/2010 | N |
| 2 | 11-13094 | Rough O:d:e,n,z15 | E | Swine | MN | 11/26/2010 | N, I1 |
| 3 | 11-13665 | Bredeney | B | Turkey | UNKc | 12/16/2010 | N |
| 4 | 11-13933 | Anatum | E | Equine | TX | 12/21/2010 | I1 |
| 5 | 11-104 | Ouakam | D | Turkey | AR | 1/6/2011 | N |
| 6 | 11-2362 | Ouakam | D | Turkey | MO | 3/7/2011 | N |
| 7 | 11-2945 | Anatum | E | Equine | TX | 3/25/2011 | nad |
| 8 | 11-2946 | Anatum | E | Equine | TX | 3/25/2011 | I1, HI1 |
| 9 | 11-3665 | Ouakam | D | Turkey | IN | 4/12/2011 | N |
| 10 | 11-4809 | Anatum | E | Equine | TX | 4/29/2011 | I1 |
| 11 | 11-4872 | Ouakam | D | Turkey | MO | 5/3/2011 | N |
| 12 | 11-6604 | Ouakam | D | Turkey | AR | 5/31/2011 | N |
| 13 | 11-9696e | Ouakam | D | Turkey | NC | 8/15/2011 | N |
Summary of results for Salmonella isolates carrying blaCTX-M identified from clinical isolates originally submitted to the NVSL for serotyping between October 2010 and June 2011.
PRT, plasmid replicon type. All IncN plasmids were identified as ST1 on pMLST. None of the IncI1 plasmids matched a reported ST.
Isolate 11-13665 state of origin unknown because the submission form was incomplete.
Isolate 11-2945 plasmid replicon type could not be identified using our typing procedure.
Isolate 11-9696 was not part of the original isolate set used for this study but was identified upon targeted screening of S. Ouakam isolates following completion of the original screening of 2,034 isolates.
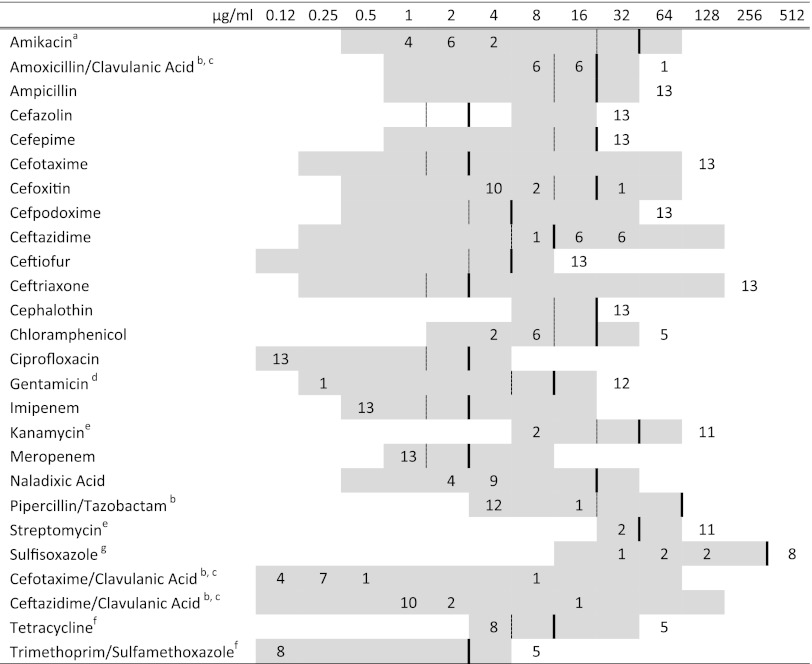
Fig 1.
MICs of 26 antimicrobial drugs for 13 Salmonella clinical isolates containing the blaCTX-M genetic element (numbers of isolates are shown in the body of the figure). Broken lines represent susceptible breakpoints, and solid lines represent resistant breakpoints when available. Corresponding to the concentration listed at the top of each column (μg/ml), the included range of each antimicrobial is shown in gray. a, amikacin MICs were not determined for isolate 11-9696. b, clavulanic acid and tazobactam were included at fixed concentrations of 4 μg/ml. c, isolate 11-13665 (S. Bredeney) was resistant to cefoxitin and the β-lactamase inhibitors. d, isolate 11-13094 (Salmonella with rough serotype O:d:e,n,z15) was susceptible to gentamicin. e, isolates 11-13094 (Salmonella with rough serotype O:d:e,n,z15) and 11-104 (S. Ouakam) were susceptible to kanamycin and streptomycin. f, the 5 S. Anatum isolates were resistant to chloramphenicol, tetracycline, and trimethoprim-sulfamethoxazole. g, the 5 isolates susceptible to sulfisoxazole were the S. Ouakam isolates except 11-9696.
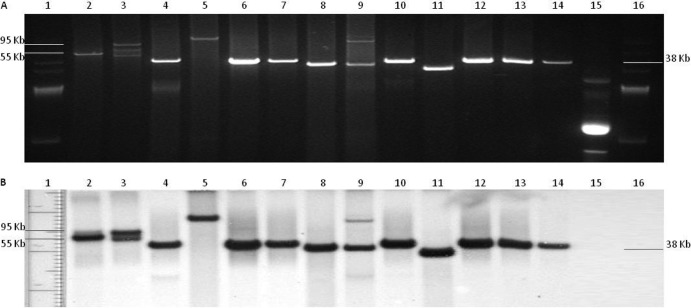
Fig 2.
(A) Plasmid content of 13 Salmonella clinical isolates containing the blaCTX-M genetic element originally submitted to the NVSL for serotyping. Lanes: 1, BAC-Tracker supercoiled DNA ladder; 2, 11-13049; 3, 11-13094; 4, 11-13665; 5, 11-13933; 6, 11-104; 7, 11-2362; 8, 11-2945; 9, 11-2946; 10, 11-3665; 11, 11-4809; 12, 11-4872; 13, 11-6604; 14, 11-9696; 15, control plasmid DNA containing blaCMY-2; 16, BAC-Tracker supercoiled DNA ladder. (B) Southern blot hybridization using CTX-M probe of plasmid content of 13 Salmonella clinical isolates containing the blaCTX-M genetic element originally submitted to the NVSL for serotyping. Lanes: 1, BAC-Tracker supercoiled DNA ladder; 2, 11-13049; 3, 11-13094; 4, 11-13665; 5, 11-13933; 6, 11-104; 7, 11-2362; 8, 11-2945; 9, 11-2946; 10, 11-3665; 11, 11-4809; 12, 11-4872; 13, 11-6604; 14, 11-9696; 15, control plasmid DNA containing blaCMY-2; 16, BAC-Tracker supercoiled DNA ladder.
We found that 6 of 88 (6.8%) turkey isolates carried blaCTX-M. Among these, one Salmonella enterica serovar Bredeney isolate was received December 2010 and carried blaCTX-M-1 on an IncN plasmid. The remaining five turkey isolates were all Salmonella enterica serovar Ouakam (Fig. 3) and were received between March and May 2011, originating from three U.S. states (Arkansas, Missouri, Indiana) that also carried blaCTX-M-1 on IncN plasmids. We screened an additional 48 S. Ouakam isolates submitted between January 2009 and October 2010 in an attempt to determine if earlier S. Ouakam isolates contained this gene, although none were identified. However, one additional S. Ouakam isolate, received in August 2011 from a turkey clinical diagnostic submission originating from North Carolina, carried blaCTX-M-1.
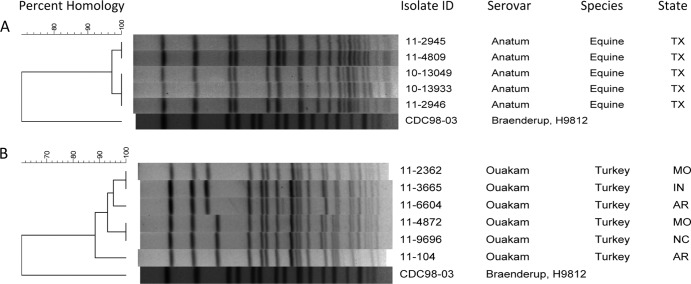
Fig 3.
(A) Dendrographic analysis of XbaI PFGE data for five Salmonella serovar Anatum clinical isolates containing the blaCTX-M genetic element. (B) Dendrographic analysis of XbaI PFGE data for six Salmonella serovar Ouakam clinical isolates containing the blaCTX-M genetic element.
Turkey production in the United States has consolidated such that there are few large centralized hatcheries that supply day-old turkey poults to grower-finisher operations throughout the United States. Ceftiofur is approved for the control of mortality in day-old turkey poults, and turkey grower-finisher operations can order poults from hatcheries that have been treated with ceftiofur prior to shipment. Turkey poults are maintained in high-population-density environments conducive to the exchange of enteric flora throughout the production system. The mass application of ceftiofur to large populations of day-old poults at the hatchery prior to shipment could provide the selection pressure required to support the emergence, dissemination, and maintenance of a ceftiofur-resistant strain of Salmonella at a central hatchery. The shipment of poults from the hatchery to grower-finisher operations in multiple states could then disseminate the resistant strain over a wide geographic area.
Pulsed-field gel electrophoresis (PFGE) analysis of the S. Ouakam isolates (Fig. 3) with XbaI (18) finds that they are similar but do not represent a single epidemic clone. Resistant Salmonella that disseminated clonally at the serotype level, but not the pulsotype level, have occurred previously, including S. Newport carrying blaCMY-2 on an IncA/C plasmid (6, 17) and Salmonella serovar Typhimurium DT104 expressing the characteristic ACSSuT resistance phenotype (resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline) (1, 5).
Of the 940 swine submissions, a single isolate (0.1%) was identified to be a Salmonella isolate, with a rough O:d:enz15 serotype, carrying blaCTX-M-1. As with turkey poults, ceftiofur is also commonly applied to large groups of piglets at weaning when they are processed into nurseries where they are maintained in a population-dense environment conducive to the spread of enteric pathogens. This mass application, typically with the extended-activity formulation of ceftiofur, may provide the selection pressure required for the emergence of a ceftiofur-resistant Salmonella strain in a population of pigs.
We found 5 of 143 (3.5%) equine isolates, all originating from Texas, were Salmonella enterica serovar Anatum (Fig. 3) carrying blaCTX-M-1 on multiple plasmid replicon types (Table 2). The PFGE similarity (>97%) of these isolates received over a relatively short time period suggests a common source exposure, as might occur at an equine event or facility. Ceftiofur is approved for the treatment of lower respiratory tract infections in horses, but the extended-activity formulation is applied at some breeding facilities to populations of mares as a postbreeding intrauterine infusion to improve conception rates (12). This application of ceftiofur therapy to mares while transiently housed in a population-dense environment may provide the population-level selection pressure required to support the emergence and spread of a ceftiofur-resistant strain of Salmonella.
None of the 581 cattle, 83 chicken, or 198 other source isolates were found to carry blaCTX-M. This result suggests that if Salmonella isolates carrying blaCTX-M are present in cattle or chicken populations in the United States, either they do not produce clinical disease sufficient to initiate a diagnostic investigation, including microbiological culture and serotyping at the NVSL, or their frequency in these populations is below the detection limits of our study.
Using plasmid multilocus sequence typing (MLST) (3, 4), we identified three unique I1 plasmid sequence types (ST) (Table 3). We also identified ST 1 IncN plasmids bearing blaCTX-M-1 in multiple Salmonella strains representing 4 serotypes from 3 animal species from diverse geographic origins. Sequence type 1 IncN plasmids carrying blaCTX-M-1 have been previously reported to be epidemic in humans, livestock, and food in Europe (16). The pandemic dissemination of multiresistant organisms, including S. Typhimurium DT104 (5, 28) and E. coli B2-O25:H4-ST131 (9, 26), has been previously reported. However, pandemic plasmid dissemination independent of clonal spread of organisms has not been reported.
Table 3.
pMLST allele variants of IncI1 plasmids from Salmonella spp. harboring blaCTX-M-1
| Isolate ID | Allele variant |
||||
|---|---|---|---|---|---|
| rep1 | ardA | trbA-pndC | sogS | pilL | |
| 11-13094 | 1 | 4 | 3 | 6 | 3 |
| 11-13933 | 1 | 4 | UNa | 9 | 3 |
| 11-2946 | 1 | 4 | UN | 9 | 3 |
| 11-4809 | 1 | 2 | UN | 9 | 3 |
UN, the allele variant has not been characterized in the pMLST database.
Salmonella bearing blaCTX-M have not been previously reported in livestock in the United States, although they have been recovered from multiple livestock species in Europe and Asia (2, 19, 27). In addition, Salmonella bearing blaCTX-M have been recovered sporadically from human cases of salmonellosis in the United States (23–25). The presence of these resistant isolates in U.S. livestock populations is important because most human salmonellosis cases in the United States result from food-borne zoonotic transmission in animal products (21). Empirical therapy of resistant infections by physicians without the benefit of microbiological culture and susceptibility results may result in treatment failure and thus increased health care costs (8) and higher risk of death (7).
ACKNOWLEDGMENTS
Partial funding for this project was provided by USDA NIFA award no. 2010-65201-20598.
We gratefully acknowledge the technical assistance of Edward Palmer, Tonya Mackie, Linda Cox, Ginger Harvey, and Brenda Morningstar-Shaw of the USDA APHIS NVSL and the technical support and expertise of Joshua B. Daniels of The Ohio State University. We also thank Alessandra Carattoli for generously providing positive-control strains for plasmid replicon typing.
Footnotes
Published ahead of print 10 August 2012
REFERENCES
- 1. Besser TE, et al. 1997. Salmonellosis associated with S. Typhimurium DT104 in the USA. Vet. Rec. 140:75. [PubMed] [Google Scholar]
- 2. Cloeckaert A, et al. 2010. IncI1 plasmid carrying extended-spectrum-β-lactamase gene blaCTX-M-1 in Salmonella enterica isolates from poultry and humans in France, 2003 to 2008. Antimicrob. Agents Chemother. 54:4484–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. García-Fernández A, et al. 2011. Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 66:1987–1991 [DOI] [PubMed] [Google Scholar]
- 4. García-Fernández A, et al. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 61:1229–1233 [DOI] [PubMed] [Google Scholar]
- 5. Glynn MK, et al. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333–1338 [DOI] [PubMed] [Google Scholar]
- 6. Gupta A, et al. 2003. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188:1707–1716 [DOI] [PubMed] [Google Scholar]
- 7. Helms M, Vastrup P, Gerner-Smidt P, Mølbak K. 2002. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium. Emerg. Infect. Dis. 8:490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holmberg SD, Solomon SL, Blake PA. 1987. Health and economic impacts of antimicrobial resistance. Clin. Infect. Dis. 9:1065. [DOI] [PubMed] [Google Scholar]
- 9. Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 10. Kado C, Liu S. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koeck JL, et al. 1997. A plasmid-mediated CMY-2 β-lactamase from an Algerian clinical isolate of Salmonella Senftenberg. FEMS Microbiol. Lett. 152:255–260 [DOI] [PubMed] [Google Scholar]
- 12. LeBlanc M. 2009. The current status of antibiotic use in equine reproduction. Equine Vet. Ed. 21:156–167 [Google Scholar]
- 13. Lewis JS, II, Herrera M, Wickes B, Patterson JE, Jorgensen JH. 2007. First report of the emergence of CTX-M-type extended-spectrum β-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma L, et al. 2005. Variety of TEM-, SHV-, and CTX-M-type beta-lactamases present in recent clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae from Taiwan. Microb. Drug Resist. 11:31–39 [DOI] [PubMed] [Google Scholar]
- 15. Mollenkopf DF, et al. 2012. Diversity of CTX-M cephalosporinase-bearing Escherichia coli isolated from dairy cattle is variable within and between herds. Appl. Environ. Microbiol. 78:4552–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naseer U, Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17:83–97 [DOI] [PubMed] [Google Scholar]
- 17. Rankin SC, et al. 2002. Molecular characterization of cephalosporin-resistant Salmonella enterica serotype Newport isolates from animals in Pennsylvania. J. Clin. Microbiol. 40:4679–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 19. Rodriguez I, et al. 2009. Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J. Antimicrob. Chemother. 64:301–309 [DOI] [PubMed] [Google Scholar]
- 20. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed, p 2.82–2.98 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Scallan E, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siu L, et al. 2000. Beta-lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like beta-lactamase, OXA-30. Antimicrob. Agents Chemother. 44:2034–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sjölund M, et al. 2008. Human Salmonella infection yielding CTX-M beta-lactamase, United States. Emerg. Infect. Dis. 14:1957–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sjölund-Karlsson M, et al. 2011. CTX-M–producing non-Typhi Salmonella spp. isolated from humans, United States. Emerg. Infect. Dis. 17:97–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sjölund-Karlsson M, et al. 2010. Salmonella isolates with decreased susceptibility to extended-spectrum cephalosporins in the United States. Foodborne Pathog. Dis. 7:1503–1509 [DOI] [PubMed] [Google Scholar]
- 26. Smet A, et al. 2010. Characterization of extended-spectrum beta-lactamases produced by Escherichia coli isolated from hospitalized and nonhospitalized patients: emergence of CTX-M-15-producing strains causing urinary tract infections. Microb. Drug Resist. 16:129–134 [DOI] [PubMed] [Google Scholar]
- 27. Tamang MD, et al. 2011. Emergence of extended-spectrum β-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J. Clin. Microbiol. 49:2671–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Threlfall EJ. 2000. Epidemic Salmonella Typhimurium DT 104—a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7–10 [DOI] [PubMed] [Google Scholar]
- 29. Wittum TE, et al. 2010. CTX-M-type extended-spectrum β-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog. Dis. 7:1575–1579 [DOI] [PubMed] [Google Scholar]