Abstract
Before new, rapid quantitative PCR (qPCR) methods for assessment of recreational water quality and microbial source tracking (MST) can be useful in a regulatory context, an understanding of the ability of the method to detect a DNA target (marker) when the contaminant source has been diluted in environmental waters is needed. This study determined the limits of detection and quantification of the human-associated Bacteroides sp. (HF183) and human polyomavirus (HPyV) qPCR methods for sewage diluted in buffer and in five ambient, Florida water types (estuarine, marine, tannic, lake, and river). HF183 was quantifiable in sewage diluted up to 10−6 in 500-ml ambient-water samples, but HPyVs were not quantifiable in dilutions of >10−4. Specificity, which was assessed using fecal composites from dogs, birds, and cattle, was 100% for HPyVs and 81% for HF183. Quantitative microbial risk assessment (QMRA) estimated the possible norovirus levels in sewage and the human health risk at various sewage dilutions. When juxtaposed with the MST marker detection limits, the QMRA analysis revealed that HF183 was detectable when the modeled risk of gastrointestinal (GI) illness was at or below the benchmark of 10 illnesses per 1,000 exposures, but the HPyV method was generally not sensitive enough to detect potential health risks at the 0.01 threshold for frequency of illness. The tradeoff between sensitivity and specificity in the MST methods indicates that HF183 data should be interpreted judiciously, preferably in conjunction with a more host-specific marker, and that better methods of concentrating HPyVs from environmental waters are needed if this method is to be useful in a watershed management or monitoring context.
INTRODUCTION
Fecal indicator bacteria (FIB), including fecal coliforms, Escherichia coli, and enterococci, have been approved indicators of sewage contamination in recreational waters for decades (49, 51). Swimmers exposed to waters contaminated with sewage are at a greater risk of infection by human pathogens, and subsequent gastrointestinal or other illnesses, than those exposed to unimpacted waters (57). The use of FIB to detect sewage contamination, however, relies on the assumptions that elevated FIB concentrations are representative of pathogen presence and that the fate of these bacteria mimics that of pathogens. Unfortunately, previous studies have shown that concentrations of FIB do not necessarily correlate well with bacterial, protozoan, and viral pathogens (2, 20).
To address the limitations of FIB, microbial source tracking (MST) methods have been developed to identify human fecal contamination (e.g., sewage) in recreational waters (7, 32). These methods are culture independent and are therefore more rapid than FIB methodologies that require a culture step. The potential for same-day results provides the possibility of advisories that reflect current conditions, rather than day-old ones, which allows more-immediate action to protect against public health risks posed by recreational water use (12, 56). The selection of MST markers represents a significant concern for resource managers, which has led to the establishment of uniform performance characteristics by which MST markers are evaluated. These characteristics include the method's sensitivity to various forms of human fecal contamination (i.e., sewer and septic), specificity against nontarget fecal sources, and limit of detection in environmental waters (43). The performance of several human-associated MST markers has been evaluated previously using these criteria in coastal waters to determine which markers are most representative of sewage contamination (16, 18).
An initial limitation of MST methods was the use of conventional endpoint PCR, which allows for presence/absence determination but cannot estimate marker concentrations. Recently, quantitative real-time PCR (qPCR) assays which allow for more rapid detection of markers, as well as determination of their relative concentrations, have been developed (10, 33, 34). While the performance of qPCR methods has been evaluated to some extent with coastal waters (16), little work has been done with inland waters, which are considerably smaller in scale and are likely to have considerably different hydrological or physicochemical parameters that could affect qPCR method performance (11).
In this study, the HF183 marker for human-associated Bacteroides (34) and the marker for human polyomaviruses (HPyVs) (33) were evaluated to determine their specificity and limits of detection (LOD). These markers were selected as being among the most promising MST markers for evaluation in inland waters based on both the existing body of literature evaluating their use in coastal waters and a high level of specificity for human fecal contamination (19). The LOD for sewage spiked into samples was determined both under ideal conditions, in sterile buffered water, and also in a variety of water types, including lake, river, tannic, estuarine, and marine waters. These water types represent complex matrices that potentially contain substances, such as humic acids, which may prove inhibitory to the PCR, which could in turn affect detection limits and produce artifacts, such as artificially low estimates of DNA gene copies. Furthermore, a quantitative microbial risk assessment (QMRA) was conducted to estimate the risk of gastrointestinal (GI) illness for adults resulting from the ingestion of diluted sewage, which was then linked to levels of MST markers detected in diluted sewage.
MATERIALS AND METHODS
Limit terminology.
For both the HF183 and HPyVs assays, a limit of detection (LOD) and limit of quantification (LOQ) were determined. Three distinct types of LOD for the qPCR methods were determined in this study. The analytical limit of detection (ALOD) refers to the number of gene copies that can reliably be detected in one qPCR. The method limit of detection (MLOD) refers to the extent to which DNA extracted from sewage influent can be diluted and still be reliably detected by qPCR. MLOD was determined in sterile buffered water. The process limit of detection (PLOD) refers to the smallest volume of sewage that can be subjected to the entire sample preparation process, from dilution in water and filtration through DNA extraction (incorporating the loss of target associated with these manipulations), and still be reliably detected by qPCR. PLOD was determined for each of the ambient water types sampled for the study. The LOD was determined as the concentration at which the target in a sample can be detected (e.g., distinguished from a nondetect) with reasonable certainty. In this study, we used the 95% confidence interval to establish the analytical LOD and designated amplification in two of three replicates (66% detection) as the cutoff for method and process LOD. The LOQ was determined as the concentration at which a sample can be quantified with reasonable precision, in this case when amplification was observed in triplicate reactions with a threshold cycle (CT) standard deviation of <1 among all replicates.
Sample collection and processing for ALOD and MLOD testing.
Untreated influent (sewage) was collected from the Falkenburg Advanced Wastewater Treatment plant in Tampa, FL, on three separate dates for determination of MLOD and PLOD. Samples were collected in sterile, 500-ml bottles and transported to the laboratory on ice. PLOD values were determined over two consecutive days, and therefore the sewage sample was stored at 4°C overnight and used again the following day.
The MLOD and MLOQ of each qPCR assay were determined in a simple matrix. DNA was extracted in triplicate (three separate extractions) from 700 μl raw sewage, which was serially diluted 1:10 in sterile buffered water (1) over 6 orders of magnitude. Undiluted and diluted DNA was then used as the template for qPCRs to determine the dilution at which the signal became nonquantitative and undetectable (see below for evaluation of quantification and detection of samples).
Sample collection and testing for specificity.
Fecal samples for specificity testing were collected from individual birds (seagulls, ducks, and chickens), cattle, and dogs in the Tampa Bay area. Fresh fecal samples (still moist) were collected using sterile tongue depressors. Ten to twenty-five grams (wet weight) was collected from each dropping whenever possible for dogs and cattle, and whole bird droppings were placed individually in sterile, 50-ml conical tubes, which were transported to the laboratory on ice and stored at −80°C until composite preparation. Composite samples (n = 10 for birds; n = 11 for cattle and dogs) were prepared by combining approximately 0.3-g samples from five individuals in one conical tube. In total, feces of 50 birds, 55 cattle, and 55 dogs were represented in the samples.
DNA from cattle and dog fecal samples used for specificity testing was initially screened via conventional (presence/absence) PCR using the assay for general members of the Bacteroidales (6) and the bacterial 16S rRNA gene (25) to verify that sufficient DNA of amplifiable quality was present in the sample, as has been previously suggested (37). Bird fecal samples were tested in the same way using a conventional PCR assay targeting the 16S rRNA gene using the Eco8F-1492RC primer set (25), since members of the Bacteroidales are not commonly found at high densities in bird fecal samples (28, 29). Undiluted DNA, as well as 1:10 and 1:20 dilutions, was used as the template to ensure that negative outcomes were not the result of inhibition. No amplification of the general Bacteroidales or 16S rRNA product was observed in 36% of cattle fecal composite samples; however, those samples produced amplicons from the 1:10 dilution, which was used for subsequent testing. All bird and dog fecal samples yielded amplicons from undiluted template.
Ambient water sampling.
Sampling sites included the highly tannic Green Swamp (28°18′46.88″N, 82°3′21.17″W), Hillsborough River (28°4′11.37″N, 82°22′39.06″W), Lake Carroll (28°2′45.37″N, 82°29′6.75″W), the estuarine Bahia Beach (27°43′44.63″N, 82°28′35.63″W), and the marine site Fort DeSoto, located on the Gulf of Mexico (27°37′1.43″N, 82°44′13.91″W) (Fig. 1). Grab samples of water were collected in sterile 2-liter bottles (total, 6 liters per site) at each sampling site on two separate dates (sample events) 2 weeks apart. Due to the distance between sites, sites were split and sampled on consecutive dates for each sample event. The sampling order was reversed for the second sample event to capture differences in the limit of detection resulting from refrigeration of sewage. Samples were transported on ice to the laboratory within 4 h of sampling and were processed within 2 h of receipt in the laboratory. Physicochemical parameters were also measured at each site (see Table S1 in the supplemental material). In the field, temperature, salinity, and pH were measured using an Oakton waterproof multiparameter tester 35 (Oakton Instruments, Vernon Hills, IL), dissolved oxygen was measured using a traceable portable dissolved oxygen meter (Fisher Scientific, Waltham, MA), and turbidity was measured at the lab using the DRT-15CE turbidimeter (HF Scientific, Fort Myers, FL). When salinity exceeded 10‰ (the upper limit of the field instrument), it was also measured in the lab using a handheld refractometer (Fisher handheld salinity refractometer with automatic temperature compensation; Fisher Scientific, Waltham, MA).
Fig 1.
Sampling sites used for determination of PLOD. Sites include Green Swamp (GS) (tannic water), Hillsborough River (HR) (river water), Lake Carroll (LC6) (lake water), Bahia Beach (BB) (estuarine water), and Fort DeSoto (FD) (marine water). The most dilute PLOD values observed during the study are shown for HPyVs and HF183. The map was generated by use of the Tampa Bay Water Atlas website (http://www.tampabay.wateratlas.usf.edu/).
Process limits of detection.
The PLOD was determined to further describe method performance in environmental matrices and was established by adding dilutions of sewage influent (see above) to ambient water, which was then filtered, and DNA was extracted as described below. Sewage was diluted in 10-fold increments ranging from 100 to 10−7 in sterile buffered water (total volume, 50 ml). Five-hundred-milliliter aliquots of ambient water from each site were spiked with 5 ml of each point on the sewage dilution series. An unamended, 500-ml aliquot of ambient water was used to determine background levels of the markers. Samples were processed as described previously (18) in order to capture HPyVs and bacteria. Briefly, the pH of the sample was lowered to 3.5 using 20% HCl, concentrated on a 0.45-μM-pore-size, 47-mm-diameter nitrocellulose membrane (Thermo Fisher Scientific, Waltham, MA), and placed in a PowerBead tube (MoBio Laboratories Inc., Carlsbad, CA). For each set of samples, a field blank and method blank were also processed. The field blank was a 500-ml sample of sterile buffered water which was carried on the sampling trip. The method blank consisted of 500-ml sterile buffered water. Both blanks were subjected to all methodological steps. Filters were stored at −80°C for up to 1 week prior to DNA extraction (the procedure is described below). To account for differences in qPCR method performance resulting from differences among water types, the sample limits of detection and quantification were then determined using the DNA extracted from these sewage-spiked environmental water samples.
DNA extraction and purification.
Between 0.2 and 0.3 g of each composite fecal sample was transferred to a PowerBead tube (MoBio Laboratories, Inc., Carlsbad, CA). DNA was extracted using the MoBio PowerSoil DNA isolation kit with modification of the manufacturer's instructions, including the following: (i) bead beating for at 4.0 m · s−1 using a FastPrep FP120 cell disruptor (Thermo Savant, Waltham, MA) and (ii) increasing the volumes of solutions C3, C4, and C5 to 285 μl, 1.6 ml, and 750 μl, respectively, to allow a larger portion of the supernatant to be carried through each step of the extraction process. An extraction blank which was subjected to all steps in the DNA extraction but had no sample material added was included in each set of DNA extractions.
DNA was extracted from each of three sewage samples (collected during three sample events; see above) in triplicate by delivering 700 μl of sewage directly into three separate PowerBead tubes (MoBio Laboratories Inc., Carlsbad, CA) without filtration in order to determine the MLOD. The same DNA extraction procedure as that described for fecal samples was used, with one exception. For the raw sewage extraction, 62.5% of the total supernatant (1 ml) was carried through the extraction following addition of the C2 solution; for subsequent steps, all of the supernatant was carried forward. The inability to carry over all of the supernatant was taken into account when reporting results, such that measured concentrations were multiplied by 1.6, and therefore the data presented reflect estimated concentrations presuming 100% of the sample was carried through the extraction. An extraction blank was included whenever DNA extraction was performed. See below (“Inhibition control”) for the secondary purification procedure used on some samples in which the qPCR was inhibited.
PCR and qPCR.
Primers and probes used in this study are shown in Table 1 and were directed at genes encoding the following: (i) the 16S rRNA of human-associated Bacteroides HF183 or (ii) the conserved T antigen of human polyomaviruses BK and JC. Endpoint PCR methods for bacterial 16S rRNA and a general group of the order Bacteroidales were used as controls for inhibition for DNA extracted from fecal samples as described above (6, 25). Primers were synthesized by Integrated DNA Technologies (Coralville, IA) and rehydrated to a concentration of 100 μM in nuclease-free water, and probes (100 μM) were synthesized by Applied Biosystems (Carlsbad, CA). Primer-probe mixes, including 7.68 μM (each) primer and 0.62 μM probe, were used for all PCRs. Reaction mixtures were composed of 12.5 μl TaqMan 2× universal PCR master mix No AmpErase UNG, 200 μM bovine serum albumin (BSA), 3 μl of primer-probe mix, and 5 μl template (total volume, 25 μl). PCRs were carried out in 96-well plates using the Applied Biosystems 7500 real time PCR system (Carlsbad, CA). All samples were run in triplicate, and for each target, three no-template controls (NTCs) were included. Thermocycler settings were 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 s and 60°C for 1 min for all targets. The threshold for all targets was set at 0.05 ΔRn (the fluorescence of the reporter dye divided by that of a passive reference dye). The baseline was determined by the software for each reaction plate. Quantity estimates for samples were extrapolated from comparison to a standard curve (described below).
Table 1.
Primers and probes used for qPCR assays
| Target | Primer | Sequence (5′–3′)a | Reference |
|---|---|---|---|
| Human-associated Bacteroides 16S rRNA | HF183 | ATCATGAGTTCACATGTCCG | 7 |
| SSHBacR | TACCCCGCCTACTATCTAATG | 34 | |
| SSHBac-PRB | (FAM)-TTAAAGGTATTTTCCGGTAGACGATGG-(TAMRA) | 19 | |
| HPyV conserved T-antigen | SM2 | AGT CTT TAG GGT CTT CTA CCT TT | 33 |
| P6 | GGT GCC AAC CTA TGG AAC AG | 3 | |
| KGJ3 | (FAM)-TCA TCA CTG GCA AAC AT-(MGBNFQ) | 33 | |
| General Bacteroidales (GenBac) | GenBacF3 | GGGGTTCTGAGAGGAAGGT | 30 |
| GenBacR4 | CCGTCATCCTTCACGCTACT | 10 | |
| GenBacP2 | (FAM)- CAATATTCCTCACTGCTGCCTCCCGTA -(TAMRA) | 10 | |
| IAC | UCP1 | (VIC)-CCTGCCGTCTCGTGCTCCTCA-(TAMRA) | 22 |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Standard curve.
Standard curves for all targets were constructed using synthesized plasmid DNA (pIDTSMART with ampicillin resistance; Integrated DNA Technologies, Coralville IA). Plasmids contained the target sequence for HF183 (5′-TACCCCGCCTACTATCTAATGGAACGCATCCCCATCGTCTACCGGAAAATACCTTTAATCATGCGGACATGTGAACTCATGATA-3′), HPyVs (5′-AGTCTTTAGGGTCTTCTACCTTTCTTTTTTTTTTGGGTGGTGTTGAGTGTTGAGAATCTGCTGTTGCTTCTTCATCACTGGCAAACATATCTTCATGGCAAAATAAATCTTCATCCCATTTTTCATTAAAGGAACTCCACCAGGACTCCCACTCTTCTGTTCCATAGGTTGGCACC-3′), or general Bacteroidales (GenBac) (for determination of inhibition) (55). DNA was serially diluted in AE buffer (Qiagen, Valencia, CA) to final concentrations ranging from 101 to 106 gene copies/reaction as determined by multiplying the DNA concentration by Avogadro's number and dividing by the product of the plasmid size and average weight of a base pair (60). Each concentration of the standard curve was run in triplicate on each reaction plate. The average r2 value for all reaction runs was 0.98 for both targets, with average reaction efficiencies of 83% and 81% for HF183 and HPyVs, respectively. These relatively low efficiency values likely resulted from use of circular rather than linear plasmid DNA, as has been previously suggested (23).
Inhibition control.
To monitor PCR inhibition from the water matrix, DNA extracted from ambient water samples was analyzed in a multiplex reaction which included a synthetic internal amplification control (IAC) (UCP1; Table 1) in place of water in the qPCR mix at a concentration of 50 target copies per reaction. The IAC UCP1 is a plasmid designed with primer sites complementary to the GenBac primers (Table 1), with a unique probe region which was synthesized by Integrated DNA Technologies (Coralville, IA) (40). The multiplex primer-probe mix consisted of primers for GenBac and probes for both GenBac and IAC, allowing simultaneous amplification of GenBac and IAC. The expected CT value for amplification of the IAC in uninhibited samples was determined as the mean for all blanks (35.5 ± 0.9), since they did not contain inhibitory compounds. Reactions were deemed inhibited if the CT value was greater than three standard deviations of the average IAC CT (>38.2). Only samples from Hillsborough River were deemed inhibited on both dates, and DNA was further purified using the MoBio PowerClean DNA cleanup kit (Carlsbad, CA) before it was used as a template for qPCR according to the protocol described above.
Enumeration of FIB.
Membrane filtration using 0.45-μM-pore-size 47 mm nitrocellulose filters was performed via standard methods for fecal coliforms (1), E. coli (53), and enterococci (52) on all sewage samples and environmental water samples. Sewage was diluted 10−3 and 10−4 (vol/vol) in sterile buffered water (1) before filtration. Ambient (not amended with sewage) environmental water samples were filtered at volumes of 1 ml, 10 ml, and 100 ml. All samples were processed in duplicate. Concentrations were reported as numbers of CFU per 100 ml.
Data evaluation.
The ALOD was calculated as the inverse log of the intercept minus the upper 95% confidence interval of the intercept divided by the slope based on the standard curve. The sample was determined to be positive (detectable) if amplification was observed in at least two of three replicates. A sample was considered quantifiable when amplification was observed in all three replicates with a CT standard deviation of <1 CT. Based on these criteria, no target was detected in blank samples (extraction blanks, field blanks, method blanks, or NTCs). To assess the ability of the secondary DNA cleanup step (see above) to quantitatively recover DNA, a theoretical data set was generated by using the measured concentration of HF183 (gene copies · 100 ml−1) in the 10−2 sewage spike (5 ml undiluted sewage spiked into 500 ml ambient water) as a starting point and estimating the concentration in 10-fold dilutions (assuming 100% DNA recovery compared to that for the first dilution). The HF183 gene copy concentration was also measured by qPCR in a dilution series of the sewage spike over a range in which the target was quantifiable (10−2 to 10−5). Gene copy concentrations were log transformed for both data sets and were compared for waters from four sites (Fort DeSoto, Green Swamp, Lake Carroll, and Hillsborough River) via Pearson correlation analysis using the GraphPad Instat software program, version 3.0 (San Diego, CA). Values from the Bahia Beach site were not used for this analysis because the LOQ was too high (HF183 could not be reliably quantified in dilute samples).
Quantitative microbial risk assessment.
The QMRA process used to estimate the risk associated with each sewage dilution followed the guidance described for recreational waters (58). Norovirus was selected as the reference pathogen since human enteric viruses are thought to cause the majority of swimming-related illnesses in human-impacted water bodies (39, 41) and result in the highest risk estimates compared to other reference pathogens for these waters (41, 42). Furthermore, norovirus has been detected in sewage in high densities (17, 24).
The probability of infection (Pinf) was modeled using the hypergeometric function with parameters α = 0.04 and β = 0.055 for norovirus doses measured by qPCR from the work of Teunis et al. (47) for nonaggregated virus suspensions. A Monte Carlo simulation was conducted to capture the variation in norovirus density in sewage and ingestion volume. The pathogen densities in sewage were not quantified in this study; therefore, the pathogen dose was estimated using norovirus densities from the literature. Norovirus density in raw sewage was modeled as a lognormal distribution with parameters (μ = 10.8; σ = 6 [genome copies liter−1]) corresponding to a median density of 4.94 × 104 genome copies liter−1 and censored at 108 genome copies liter−1 (46). The volume of water ingested was also modeled as a lognormal distribution with parameters (μ = 2.92 ml; σ = 1.43 ml) corresponding to a mean volume of 18.6 ml.
The norovirus dose for each sewage dilution was estimated by multiplying the raw sewage density by the ingestion volume and dilution factor (point example: 3.86 × 10−4 genome copies liter−1 × 0.0186 liters × 10−2). The sewage dilutions were assumed to have a ratio of total to infectious virions equivalent to the inoculum used for the dose-response parameterization. Finally, the probability of illness was estimated by multiplying the probability of infection by a constant morbidity (fraction of infections resulting in illness) of 0.6 (42).
RESULTS
ALOD and positive reactions.
During this study, inconsistent amplification of replicate reactions (triplicate assays of a given DNA sample) was observed. An inability to detect the 101 gene copy standard (70% of standard curves for human polyomaviruses [HPyVs] [n = 10] and 33% of standard curves for HF183 [n = 12]) was also fairly common, even though initial evaluation of the ALOD determined it to be <10 gene copies for both methods. To address this issue, a sample was designated detectable (positive) for the target if amplification was observed in two of three replicates. Furthermore, samples in which the mean quantity of target copies estimated in the reaction was <102 frequently showed amplification in only two of three replicates or high standard deviations in CT values (>1 CT) for replicate reactions, which resulted in quantity estimates more than an order of magnitude different among triplicate reactions. This was observed in 64% of samples analyzed for HPyVs (n = 25) and 61% of samples analyzed for HF183 (n = 28). On the basis of these observations, amplification was considered to be quantitative only if it was observed in all replicates and the standard deviation of the replicates' CT values was <1.
Method specificity.
The HPyV assay showed 100% specificity, with no amplification observed for any of the 32 nontarget fecal composites tested. The HF183 assay, however, was less specific. Cross-reactivity (detectable target amplification) was seen with two dog, two chicken, and one duck fecal composite sample, giving the assay an overall specificity of 81.25%. The quantity of target determined for these samples was generally around 102 target copies · g−1 of fecal composite (wet weight); however, one chicken composite showed a much higher target concentration, 1.75 × 107 copies · g−1.
MLOD and MLOQ.
The culturable FIB concentrations in the sewage samples tested were on the order of 106 CFU · 100 ml−1 for E. coli and fecal coliforms, while the concentration of enterococci was on the order of 106 to 107 CFU · 100 ml−1. All sewage samples were positive for the HPyV and HF183 targets (on the order of 106 and 108 gene copies · 100 ml−1, respectively; Table 2). The MLOD for each method is shown as the greatest dilution of extracted sewage DNA that could be reliably detected, while the MLOQ was the greatest dilution that provided quantitative results. The HPyV assay had a higher MLOD (less dilution was required to dilute the signal to extinction) than the HF183 assay, i.e., HPyVs were detectable only in the 10−1 dilution of DNA from sewage, while the HF183 marker was still detectable after a 10−3 dilution (Table 2). Although the extent to which each target could be diluted and remain detectable differed by 2 orders of magnitude, the amount of each target estimated at the MLOQ was similar (on the order of 101 target copies per reaction). The MLOQ for HPyVs was in undiluted sewage (1.03 × 102 target copies per reaction), while the lowest quantifiable concentration of HF183 was observed at a 10−2 dilution (6.35 × 101 target copies per reaction). Generally, both targets could still be detected at approximately 1 order of magnitude below the MLOQ.
Table 2.
MLOD, MLOQ, and concentrations of DNA extracted from sewage samples and diluted in buffer for HPyVs and HF183a
| Target | Sample dateb | No. of gene copies per reaction (5 μl DNA) with indicated dilution |
MLODc | Estimated no. of gene copies/100 mld | |||||
|---|---|---|---|---|---|---|---|---|---|
| Undiluted (100) |
10−1 |
10−2 |
|||||||
| Mean | SD | Mean | SD | Mean | SD | ||||
| HPyVs | 8/30/10 | 5.91 × 102 | 2.12 × 102 | Not quantifiablee | Not quantifiable | 10−1 | 2.70 × 106 | ||
| 9/1/10 | 1.03 × 102 | 5.15 × 101 | Not quantifiable | Not quantifiable | 10−1 | 4.72 × 105 | |||
| 9/15/10 | 1.62 × 102 | 3.58 × 101 | Not quantifiable | Not quantifiable | 10−1 | 7.41 × 105 | |||
| HF183 | 8/30/10 | 7.43 × 104 | 1.31 × 104 | 4.98 × 103 | 5.49 × 102 | 5.74 × 102 | 9.26 × 101 | 10−3 | 3.40 × 108 |
| 9/1/10 | 2.25 × 104 | 1.02 × 104 | 1.20 × 103 | 3.82 × 102 | 9.62 × 101 | 4.22 × 101 | 10−3 | 1.03 × 108 | |
| 9/15/10 | 1.59 × 104 | 5.75 × 103 | 7.31 × 102 | 1.40 × 102 | 6.35 × 101 | 8.43 × 100 | 10−2 | 7.26 × 107 | |
Gene copies per reaction are a mean of qPCR results from three separate extractions. Bolded values represent the MLOQ for each marker in each sewage sample.
Month/day/year.
Lowest dilution in which target was amplified in at least two of three DNA extracts.
Mean target copies · 100 ml−1 of sewage were calculated as the mean concentrations in undiluted sewage samples.
Not quantifiable, target was detected but was not considered accurately quantifiable due to high standard deviations (>1 CT) among triplicate reactions.
PLOD and PLOQ.
Ambient water samples representing various water types common in Florida were used to determine the PLOD and PLOQ for sewage that was diluted and carried through membrane filtration and DNA extraction/purification (see Table S1 in the supplemental material for physical and chemical characteristics). The concentrations of FIB and MST markers present before the addition of sewage were determined (Table 3). FIB concentrations generally ranged from 101 to 102 CFU · 100 ml−1, while the sewage contained 106 to 107 CFU · 100 ml−1. The HPyV marker was not detected in any of the water samples collected prior to the addition of sewage. HF183 was detected at the Hillsborough River site on both sampling dates (1.41 × 103 and <4.52 × 102 copies · 100 ml−1) and at Green Swamp on the first sampling date (<2.15 × 102 copies · 100 ml−1) prior to spiking the water samples. Quantifiable concentrations of the HPyV marker were observed in all samples amended with undiluted sewage, representing a 10−2 dilution factor (5 ml sewage diluted in 500 ml water) (Table 4). The HPyV marker was also detectable but not quantifiable in most samples amended with further dilutions of sewage (10−3 and 10−4 dilution factors). Overall, the dilution at which detection was observed was inconsistent among sample sites and varied between sampling dates (Table 4).
Table 3.
FIB concentrations for ambient water and sewage samples
| Sample datea | Site | FIB concn (CFU · 100 ml−1) |
||
|---|---|---|---|---|
| E. coli | Fecal coliforms | Enterococci | ||
| 9/01/10 | Sewage sample 2 | 1.50 × 106 | 3.95 × 106 | 6.50 × 107 |
| Green Swampb | 9.57 × 101 | 1.57 × 102 | 9.57 × 101 | |
| Lake Carroll | 7.25 × 100 | 4.70 × 101 | 5.37 × 101 | |
| 9/02/10 | Hillsborough Riverb | 1.15 × 102 | 1.22 × 102 | 9.65 × 101 |
| Bahia Beach | 5.12 × 102 | 6.42 × 102 | 6.80 × 101 | |
| Fort DeSoto | 3.05 × 102 | 1.77 × 101 | 3.75 × 100 | |
| 9/15/10 | Sewage sample 3 | 3.20 × 106 | 2.25 × 106 | 1.15 × 107 |
| Bahia Beach | 8.80 × 100 | 2.53 × 101 | 3.20 × 101 | |
| Fort DeSoto | 1.00 × 100 | 1.00 × 100 | 1.55 × 101 | |
| 9/16/10 | Green Swamp | 1.62 × 102 | 1.42 × 102 | 3.45 × 102 |
| Lake Carroll | 1.82 × 102 | 2.62 × 102 | 3.66 × 102 | |
| Hillsborough Riverb | 9.57 × 102 | 3.02 × 103 | 2.12 × 102 | |
Sample date (month/day/year) corresponds to the sewage sample collection date.
HF183 was detected in ambient water samples prior to the addition of sewage.
Table 4.
PLOQ and PLOD of HPyV and HF183 assays using serially diluted sewage spiked into ambient waters
| Site | Datee | HPyV assay |
HF183 assay |
||||
|---|---|---|---|---|---|---|---|
|
PLOQ |
PLOD dilutionb |
PLOQ |
PLOD dilutionb | ||||
| No. of target copies · 5 μl−1a | Dilutionb | No. of target copies · 5 μl−1a | Dilutionb | ||||
| Bahia Beach | 9/2/10 | 3.23 × 102 | 10−2 | 10−3 | 8.31 × 102 | 10−4 | 10−6 |
| 9/15/10 | 1.06 × 101 | 10−3 | 10−3 | 6.63 × 101 | 10−3 | 10−3 | |
| Fort DeSoto | 9/2/10 | 2.05 × 101 | 10−4 | 10−4 | 8.81 × 101 | 10−5 | 10−6 |
| 9/15/10 | 3.07 × 101 | 10−3 | 10−3 | 9.07 × 101 | 10−5 | 10−5 | |
| Green Swampc | 9/1/10 | 3.91 × 102 | 10−2 | 10−3 | 1.66 × 102 | 10−5 | 10−5 |
| 9/16/10 | 1.64 × 102 | 10−2 | 10−2 | 1.31 × 101 | 10−5 | 10−6 | |
| Lake Carroll | 9/1/10 | 1.20 × 102 | 10−2 | 10−3 | 1.98 × 102 | 10−5 | 10−6 |
| 9/16/10 | 6.17 × 101 | 10−2 | 10−2 | 5.23 × 102 | 10−4 | 10−4 | |
| Hillsborough Riverd | 9/2/10 | 5.47 × 101 | 10−4 | 10−4 | 1.71 × 101 | 10−6 | 10−6 |
| 9/16/10 | 3.11 × 101 | 10−2 | 10−4 | 4.30 × 102 | 10−4 | 10−5 | |
Values represent the lowest number of target copies per reaction detected at the lowest sewage dilution quantifiable.
Sewage dilution corresponding to the limit of quantification or detection.
Results are estimated due to background contribution of HF183.
Extracted DNA was further purified due to detection of inhibition on both sample dates.
Sample date (month/day/year) corresponds to the collection date for the sewage sample used to spike each ambient water sample.
In general, the HF183 marker was quantifiable in sewage that was diluted approximately 100-fold more than for HPyVs (Table 4). All but two samples (the second sampling event for Bahia Beach and Lake Carroll) had detectable concentrations of HF183 at the 10−5 dilution factor (5 ml of sewage diluted 10−3 added to 500 ml water). Several samples had detectable concentrations of HF183 at the 10−6 dilution factor, and one of these samples was quantifiable (Table 4). For the two markers, the concentration of target measured at the ALOQ was similar, ranging from 101 to 102 target copies per qPCR.
Inhibition.
Samples collected from the Hillsborough River on both dates contained inhibitors of the qPCR, since the mean CT values for the IAC were 42.4 and undetermined, respectively, and a sample CT value over 40 was determined to be inhibited. Green Swamp samples collected on the first sample date may also have been inhibited and produced an average CT value of 40 for the IAC, but no action was taken to relieve inhibition for these samples. To relieve inhibition for Hillsborough River samples, DNA extracts from ambient and sewage-spiked waters from this site were further purified using a cleanup kit. Purified DNA extracts produced mean CT values for the IAC of ≤40 when the samples were reanalyzed (data not shown). The purified DNA showed detectable concentrations of HPyVs and HF183 in the 10−4 and ≥10−5 dilutions of sewage, respectively.
The effect of the secondary DNA cleanup procedure on recovery of DNA from sewage-amended samples was assessed by comparing idealized values for HF183 gene copies (assuming 100% DNA recovery) to measured values for each dilution of the sewage spiked into ambient waters (Fort DeSoto, Green Swamp, Lake Carroll, and Hillsborough River), which were carried through the entire filtration and DNA extraction procedure (Table 5). For sites at which DNA was not subjected to a secondary round of purification, correlations (r2 > 0.85) between observed and theoretical target concentrations were observed, with P values ranging from 0.0164 to 0.0785. The correlation coefficient for the cleaned-up Hillsborough River DNA samples was not large and did not approach statistical significance (r2 = 0.62; P = 0.42).
Table 5.
Correlation analysis of observed values on second sampling date for HF183 compared to a theoretical data set for the sewage spike dilution series
| Sewage dilution factora | Data and correlation analysis for siteb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fort DeSoto |
Green Swamp |
Lake Carroll |
Hillsborough Riverc |
|||||||||
| Observed quantity | Theoretical quantity | Correlation | Observed quantity | Theoretical quantity | Correlationn | Observed quantity | Theoretical quantity | Correlation | Observed quantity | Theoretical quantity | Correlation | |
| 10−2 | 5.13 | 5.11 | 5.59 | 5.59 | 5.08 | 5.08 | 3.91 | 3.91 | ||||
| 10−3 | 4.66 | 4.11 | 3.64 | 4.59 | 4.08 | 4.08 | 4.05 | 2.91 | ||||
| 10−4 | 3.28 | 3.11 | 2.04 | 3.59 | 3.34 | 3.08 | 3.10 | 1.91 | ||||
| 10−5 | 2.56 | 2.11 | 2.21 | 2.59 | NQd | NQ | NQ | NQ | ||||
| r2 value | 0.97 | 0.85 | 0.99 | 0.62 | ||||||||
| P value | 0.0164 | 0.0785 | 0.0548 | 0.4205 | ||||||||
Dilution factor of 10−2 represents 5 ml sewage inoculated into 500 ml water.
Quantities are expressed as log10 gene copies · 100 ml−1.
DNA extracted from Hillsborough River samples on both sample dates was further purified to relieve inhibition.
NQ, not quantifiable.
Risk assessment.
The objective of the risk assessment was to evaluate if the HF183 and HPyV markers could be detected in sewage dilutions that present a potential health hazard to swimmers if ingested. A QMRA was conducted to estimate the risk of gastrointestinal (GI) illness for adults resulting from the ingestion of diluted sewage; the sewage dilutions modeled in the QMRA were equivalent to those prepared for the PLOD analysis. Ideally, the PLOD for each marker will be a dilution with a low predicted human health risk (i.e., below the health benchmark); if this is the case, the markers will theoretically also be detectable if that same sewage is diluted less, resulting in a higher predicted human health risk. The health benchmark of 10 GI illnesses per 1,000 exposures (i.e., 0.01) was selected based on the 1986 Ambient Water Quality Criteria (42).
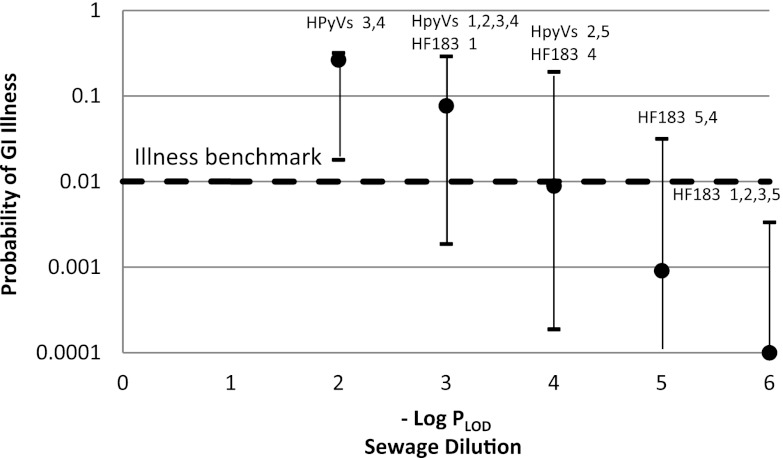
The benchmark illness risk of 10 GI illnesses per 1,000 exposures (i.e., 0.01) was compared to the predicted GI illness risk associated with norovirus for each marker's PLOD to evaluate whether HF183 and HPyVs could be detected in sewage dilutions corresponding to the lower range of risk levels that are still above the 0.01 benchmark (Fig. 2). Sewage dilutions that were 10−5 or more dilute had predicted median probabilities of illness below the benchmark risk (with a median pathogen dose less than 1 genome copy). Sewage dilutions that were less dilute had predicted illness risks at or above the benchmark risk using norovirus as the reference pathogen.
Fig 2.
Relationship of the PLOD for HF183 and HPyVs with gastrointestinal illness rates estimated for norovirus in sewage. Median probability of GI illness from norovirus (●) is plotted for accidental ingestion of water contaminated with each sewage dilution corresponding to a limit of detection. Error bars show the 25th and 75th percentile ranges for illness probability. Numbers beside each marker designate the water body corresponding to that LOD: 1, Bahia Beach; 2, Fort DeSoto; 3, Green Swamp; 4, Lake Carroll; 5, Hillsborough River. For source waters with a range of LOD, the maximum and minimum PLOD are included.
The HF183 marker could be detected in the 10−3 sewage dilution in Bahia Beach water but not in more dilute sewage dilutions, and at this level the median risk of illness was close to 0.1. The best detection performance for the HF183 marker was in Green Swamp water, where the marker was detected in a sewage dilution of 10−6. The predicted probability of illness from accidental ingestion of a sewage dilution of 10−6 was only 0.0001. For the majority of waters, the HF183 marker was detected in sewage dilutions with potential illness risks greater than or equal to the benchmark. In contrast, the PLOD for the HPyV marker was generally not sensitive enough to detect sewage dilutions near the benchmark level, except in one instance each at Fort DeSoto (2) and Hillsborough River (5) (Fig. 2).
DISCUSSION
Method sensitivity (ability to detect the target when the contamination source is known to be present) was assessed with respect to dilution of the contamination source (sewage) in order to directly relate LOD to the efficacy of the method for field application. The knowledge that one can detect x fg DNA may reflect a precise measurement, but it provides no context for interpreting the usefulness of the method for its intended purpose: detecting fecal contamination in environmental waters. This study advances the science of qPCR-based MST by doing the following: (i) assessing LOD on three levels, each of which is relevant to method performance (ALOD, MLOD, and PLOD), (ii) comparing method performances in various water types common to subtropical locales, and (iii) relating the limit of detection for the MST methods to quantitative estimates of the risk of gastroenteritis associated with exposure to sewage in recreational waters. Laboratory validation of the HPyV and HF183 markers in this study showed the HPyV target to be 100% specific against dog, cattle, and bird fecal samples while the HF183 marker cross-reacted with dog and bird fecal samples and was 81.25% specific to human fecal contamination. Previous studies have reported similar results for both markers (4, 18, 33). Higher concentrations of HF183 compared to HPyVs in sewage, however, resulted in an MLOD approximately 100-fold lower (more sensitive) for HF183 than for HPyVs in sterile buffered water. Resource managers typically have limited funds and must choose from a variety of markers to protect public health; here they are presented with the option of using a more conservative but highly specific marker for human contamination (HPyVs) or one that is more easily detected due to higher concentrations in human fecal contamination but less specific (HF183), or they may use both to maximize confidence in a positive or negative indication of sewage pollution.
Evaluation of these methods for a variety of inland and coastal water types revealed that PLODs and PLOQs varied up to 100-fold in water samples ranging from marine to freshwater that were inoculated using the same sewage sample. Furthermore, PLODs and PLOQs for water samples collected from the same sites but on different dates differed by 1 to 2 orders of magnitude for both MST markers. While different concentrations of the markers in sewage collected on different dates could have influenced the between-date variability, this factor was not an issue for the among-site variability on a given date. Inhibition of the PCR resulting from factors such as various concentrations of humic acids, fulvic acids, and other compounds may contribute to differences in LOD observed across different water types (48). By the method used here to assess inhibition, the UCP1 plasmid containing a target with GenBac primer sites and an engineered probe site, only the Hillsborough River samples were identified as inhibited. This control for inhibition may have been less useful than sample spikes and assays for the actual target or template dilution. IACs that can be multiplexed with primers and probes for target sequences are very attractive because they reduce the cost associated with controlling for inhibition; however, the magnitude of inhibition in a given sample may be method specific. In waters where inhibition is known to be an issue, the investment in spiked controls and/or dilutions of the template, both of which require multiple assays, may be worth the cost and effort.
The issue concerning how to interpret data in which replicate reactions perform differently (e.g., amplification in only one or two reactions out of triplicate reactions from the same DNA template) is of growing concern to scientists as well as resource managers when interpreting qPCR data. In this study, in samples with mean target copy concentrations of <102 target copies per reaction, triplicate reactions frequently (>60% of samples) returned variable results, including nondetects in replicates or quantity estimates over several orders of magnitude (100 to 102). We established criteria for a detectable (positive) sample as amplification in at least two of three replicates and for a quantifiable sample as amplification in all three replicates with a CT standard deviation of <1. These conditions represent conservative criteria for the interpretation of these data; however, further optimization of DNA extraction methodologies and/or qPCR preparation, such as automation and prepackaged reagents, may increase the sensitivity of these assays for accurate quantification of low target copy concentrations as well as increasing precision among replicate reactions.
In addition to uncertainty about how to gauge certain performance characteristics (i.e., what constitutes a “detection”) and determining the most appropriate quality control strategies for accurate quantification (i.e., how outliers for a standard curve or sample should be determined), the issue of how to appropriately measure and handle inhibition in samples is crucial to all applications of qPCR in environmental waters. In the present study, an IAC (UCP1) was used at the level of the qPCRs, and inhibition was evaluated based on observation of a CT value higher than that expected for the IAC (35, 36). Chemical cleanup of DNA extracts from the Hillsborough River samples, which were strongly inhibitory to PCR amplification, helped to alleviate inhibition based on IAC CT values; however, the cleanup procedure did not result in consistent DNA recovery, and therefore accurate quantification of gene copies in the original sample was not possible. This breakdown in quantitative DNA recovery is likely due to more-extensive manipulation of the sample resulting in a variable loss of target between samples during the additional purification procedure. Dilution of samples has been previously suggested to reduce inhibition (54, 55, 59) and represents an acceptable alternative to reduce inhibitors without extensive sample manipulation.
The use of qPCR in field studies of environmental water quality is a relatively recent advancement in methodology. Initial studies at the beginning of the 21st century did not use controls for sample recovery during processing or inhibition (13, 14). As the field has matured, the need to account for sources of quantitative error has been increasingly recognized (11, 21, 38, 44, 54); however, there is little agreement in the literature on how this difficult goal should be accomplished. The tactics employed to assess and remove the effect of inhibitory compounds vary, ranging from simple template dilution (9) to addition of synthetic DNA sequences to reactions as internal controls (19, 38, 45, 54). Salmon sperm DNA has been used as a dual control for recovery through sample processing and inhibition (54). Cao et al. (9) compared the efficacy of four internal controls to that of sample dilution and concluded the following: (i) sample dilution provided the most reliable and readily interpretable evidence for inhibition, (ii) PCR chemistry, including the specific polymerase master mix used, had a major influence on the frequency of inhibited samples, and (iii) the choice of method to assess inhibition is not yet agreed upon and may vary by site or study objective.
Several methods were employed in this study to assess inhibition. Fecal samples were screened using conventional PCR targeting general Bacteroidales or bacterial 16S rRNA against both diluted and undiluted DNA; however, this was a qualitative rather than a quantitative assessment of the presence of the MST markers. We chose the use of internal amplification control UCP1 as a control for inhibition; however, in light of recent findings (9), the dilution approach may have been more appropriate. The IAC used here targeted a different sequence than the MST marker assays, which could cause false-negative results for inhibition (9).
DNA recovery following membrane filtration is inherently variable and inefficient (44); therefore, the LOD values presented here should be interpreted in light of this knowledge and with the understanding that a determination of minimal target concentrations (rather than sewage dilutions) would require the use of a sample processing control to account for target loss through filtration and DNA extraction (54). However, our previous work had shown that recovery of HPyVs from the DNA extraction protocol was ∼91% (see the supplemental material for reference 31) and that recovery, including the filtration protocol, was ∼79% when virus numbers were ∼105 per filter (the cell number recommended by the U.S. EPA for assessing recovery through filtration [50]).
The purpose of the QMRA was to evaluate whether the human sewage markers were detectable in sewage dilutions at the lowest levels that constitute an unacceptable level of health risk, i.e., 10 illnesses in 1,000 exposures. The PLOD of HF183 was generally sufficient to detect sewage in ambient water at dilutions that constituted a potential human health risk based on the QMRA assumptions, whereas, the HPyV marker was not. The reference pathogen, norovirus, provides a conservative risk estimate suited for comparing the general performance of each marker. The results should not be used to rule out the HPyV marker from the candidates of useful markers for human sewage. The decay rate of HPyVs in water strongly resembles that of adenoviruses, adding to its attractiveness as a surrogate for pathogenic human viruses (8, 33). Conversely, the decay of the Bacteroidales in the water has been demonstrated to occur rapidly under certain conditions, potentially faster than that of pathogenic human viruses (5, 15). The pathogen content of sewage varies (58), and therefore the HPyV marker may be detectable in sewage dilutions of concern for some populations. Likewise, the results shown here do not ensure that HF183 will be detectable in the environment when there is a human health risk.
This study represents a step toward closing the knowledge gaps as necessary to allow implementation of new rapid MST assays for the protection of human health in recreational waters. In the current work, the limits of detection and quantification of two human-associated markers in a variety of inland water types were established, and these results compared with QMRA data show that the HF183 marker can generally be detected and quantified at dilutions of sewage that allow it to be protective of human health, while current methods for HPyV detection are not effective at detecting these viruses at such sewage dilutions. More work on reducing the variability of DNA recovery and qPCR method performance in environmental waters is needed, as well as better methods for concentrating HPyVs without also concentrating inhibitors. The difference in limits of detection and quantification in various water types and their correlation to pathogen occurrence and risk will be an important consideration for regulators when adopting new water quality standards related to MST qPCR assays, such as the two studied here.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elizabeth Kennedy at the University of West Florida for the SSHBac-PRB sequence.
The funding for this work was provided by the Water Environment Research Foundation (PATH3C09), the Florida Stormwater Association, and the Florida Department of Environmental Protection.
Footnotes
Published ahead of print 10 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. American Public Health Association 1995. Standard methods for the examination of water and wastewater, 19th ed American Public Health Association, Washington, DC [Google Scholar]
- 2. Anderson KL, Whitlock JE, Harwood VJ. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arthur RR, Dagostin S, Shah KV. 1989. Detection of BK virus and JC virus in urine and brain tissue by the polymerase chain reaction. J. Clin. Microbiol. 27:1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balleste E, Bonjoch X, Belanche LA, Blanch AR. 2010. Molecular indicators used in the development of predictive models for microbial source tracking. Appl. Environ. Microbiol. 76:1789–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bell A, et al. 2009. Factors influencing the persistence of fecal Bacteroides in stream water. J. Environ. Qual. 38:1224–1232 [DOI] [PubMed] [Google Scholar]
- 6. Bernhard AE, Field KG. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bofill-Mas S, et al. 2006. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72:7894–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao Y, Griffith JF, Dorevitch S, Weisberg SB. 2012. Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters. J. Appl. Microbiol. 113:66–75 [DOI] [PubMed] [Google Scholar]
- 10. Dick LK, Field KG. 2004. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dorevitch S, et al. 2010. Meeting report: knowledge and gaps in developing microbial criteria for inland recreational waters. Environ. Health Perspect. 118:871–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Field KG, Samadpour M. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 41:3517–3538 [DOI] [PubMed] [Google Scholar]
- 13. Foulds IV, et al. 2002. Quantification of microcystin-producing cyanobacteria and E. coli in water by 5′-nuclease PCR. J. Appl. Microbiol. 93:825–834 [DOI] [PubMed] [Google Scholar]
- 14. Foulds IV, et al. 2002. Application of quantitative real-time PCR with dual-labeled hydrolysis probes to microbial water quality monitoring. J. Biomol. Tech. 13:272–276 [PMC free article] [PubMed] [Google Scholar]
- 15. Green HC, Shanks OC, Sivaganesan M, Haugland RA, Field KG. 2011. Differential decay of human faecal Bacteroides in marine and freshwater. Environ. Microbiol. 13:3235–3249 [DOI] [PubMed] [Google Scholar]
- 16. Griffith JF, Cao YP, McGee CD, Weisberg SB. 2009. Evaluation of rapid methods and novel indicators for assessing microbiological beach water quality. Water Res. 43:4900–4907 [DOI] [PubMed] [Google Scholar]
- 17. Haramoto E, et al. 2006. Seasonal profiles of human noroviruses and indicator bacteria in a wastewater treatment plant in Tokyo, Japan. Water Sci. Technol. 54:301–308 [DOI] [PubMed] [Google Scholar]
- 18. Harwood VJ, et al. 2009. Validation and field testing of library-independent microbial source tracking methods in the Gulf of Mexico. Water Res. 43:4812–4819 [DOI] [PubMed] [Google Scholar]
- 19. Harwood VJ, Gordon KV, Staley C. 2011. Validation of rapid methods for enumeration of markers for human sewage contamination in recreational water. Project no. PATH3C09 Water Environment Research Foundation, Alexandria, VA [Google Scholar]
- 20. Harwood VJ, et al. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71:3163–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harwood VJ, Stoeckel DM. 2011. Performance criteria, p 7–30 In Hagedorn C, Blanch AR, Harwood VJ. (ed), Microbial source tracking: methods, applications, and case studies. Springer, New York, NY [Google Scholar]
- 22. Haugland RA, et al. 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst. Appl. Microbiol. 33:348–357 [DOI] [PubMed] [Google Scholar]
- 23. Hou YB, Zhang H, Miranda L, Lin SJ. 2010. Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene. PLoS One 5:e9545 doi:10.1371/journal.pone.0009545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katayama H, et al. 2008. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 42:1441–1448 [DOI] [PubMed] [Google Scholar]
- 25. Lane D. 1991. 16S/23S rRNA sequencing, p 115–148 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom [Google Scholar]
- 26. Reference deleted.
- 27. Reference deleted.
- 28. Lu J, Santo Domingo JS. 2008. Turkey fecal microbial community structure and functional gene diversity revealed by 16S rRNA gene and metagenomic sequences. J. Microbiol. 46:469–477 [DOI] [PubMed] [Google Scholar]
- 29. Lu J, Santo Domingo JW, Hill S, Edge TA. 2009. Microbial diversity and host-specific sequences of Canada goose feces. Appl. Environ. Microbiol. 75:5919–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ludwig W, Schleifer KH. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556–562 [DOI] [PubMed] [Google Scholar]
- 31. McQuaig S, Griffith J, Harwood VJ. 2012. The association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Appl. Environ. Microbiol. 78:6423–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McQuaig SM, Scott TM, Harwood VJ, Farrah SR, Lukasik JO. 2006. Detection of human-derived fecal pollution in environmental waters by use of a PCR-based human polyomavirus assay. Appl. Environ. Microbiol. 72:7567–7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 75:3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seurinck S, Defoirdt T, Verstraete W, Siciliano SD. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol. 7:249–259 [DOI] [PubMed] [Google Scholar]
- 35. Shanks OC, et al. 2008. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 74:745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA. 2009. Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol. 75:5507–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shanks OC, Santo Domingo JW, Lamendella R, Kelty CA, Graham JE. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siefring S, Varma M, Atikovic E, Wymer L, Haugland RA. 2008. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J. Water Health 6:225–237 [DOI] [PubMed] [Google Scholar]
- 39. Sinclair RG, Jones EL, Gerba CP. 2009. Viruses in recreational water-borne disease outbreaks: a review. J. Appl. Microbiol. 107:1769–1780 [DOI] [PubMed] [Google Scholar]
- 40. Sivaganesan M, Seifring S, Varma M, Haugland RA, Shanks OC. 2008. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinformatics 9:120 doi:10.1186/1471-2105-9-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soller JA, Bartrand T, Ashbolt NJ, Ravenscroft J, Wade TJ. 2010. Estimating the primary etiologic agents in recreational freshwaters impacted by human sources of faecal contamination. Water Res. 44:4736–4747 [DOI] [PubMed] [Google Scholar]
- 42. Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 44:4674–4691 [DOI] [PubMed] [Google Scholar]
- 43. Stoeckel DM, Harwood VJ. 2007. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73:2405–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stoeckel DM, Stelzer EA, Dick LK. 2009. Evaluation of two spike-and-recovery controls for assessment of extraction efficiency in microbial source tracking studies. Water Res. 43:4820–4827 [DOI] [PubMed] [Google Scholar]
- 45. Stoeckel DM, Stelzer EA, Stogner RW, Mau DP. 2011. Semi-quantitative evaluation of fecal contamination potential by human and ruminant sources using multiple lines of evidence. Water Res. 45:3225–3244 [DOI] [PubMed] [Google Scholar]
- 46. Teunis PF, et al. 2010. Enteric virus infection risk from intrusion of sewage into a drinking water distribution network. Environ. Sci. Technol. 44:8561–8566 [DOI] [PubMed] [Google Scholar]
- 47. Teunis PFM, et al. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468–1476 [DOI] [PubMed] [Google Scholar]
- 48. Tsai YL, Olson BH. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. U.S. Environmental Protection Agency 1986. Ambient water quality criteria for bacteria—1986. U.S. Environmental Protection Agency, Washington, DC: http://www.epa.gov/waterscience/beaches/files/1986crit.pdf [Google Scholar]
- 50. U.S. Environmental Protection Agency 2011. (EPA-OW-2011-0466; FRL-9609-3) Notice of availability of draft recreational water quality criteria and request for scientific views. Fed. Regist. 76:79176–79177 [Google Scholar]
- 51. U.S. Environmental Protection Agency 2002. Implementation guidance for ambient water quality criteria for bacteria. EPA-823-B-02-003 U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 52. U.S. Environmental Protection Agency 2002. Method 1600: enterococci in water by membrane filtration using membrane-enterococcus indoxyl-B-d-glucoside agar (mEI). U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 53. U.S. Environmental Protection Agency 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 54. U.S. Environmental Protection Agency 2010. Method A: enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay. EPA-821-R-10-004 U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 55. U.S. Environmental Protection Agency 2010. Method B: Bacteroidales in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay. EPA-822-R-10-003 U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 56. Wade TJ, et al. 2006. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ. Health Perspect. 114:24–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wade TJ, Pai N, Eisenberg JNS, Colford JM. 2003. Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 111:1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Water Environment Research Foundation 2011. Quantification of pathogens and sources of microbial indicators for QMRA in recreational waters, report PATH2R08. Water Environment Research Foundation, Alexandria, VA [Google Scholar]
- 59. Wilson IG. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yun JJ, et al. 2006. Genomic DNA functions as a universal external standard in quantitative real-time PCR. Nucleic Acids Res. 34:e85 doi:10.1093/nar/gkl400 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.