Abstract
The cultivation of genetically modified (GM) crops has increased significantly over the last decades. However, concerns have been raised that some GM traits may negatively affect beneficial soil biota, such as arbuscular mycorrhizal fungi (AMF), potentially leading to alterations in soil functioning. Here, we test two maize varieties expressing the Bacillus thuringiensis Cry1Ab endotoxin (Bt maize) for their effects on soil AM fungal communities. We target both fungal DNA and RNA, which is new for AM fungi, and we use two strategies as an inclusive and robust way of detecting community differences: (i) 454 pyrosequencing using general fungal rRNA gene-directed primers and (ii) terminal restriction fragment length polymorphism (T-RFLP) profiling using AM fungus-specific markers. Potential GM-induced effects were compared to the normal natural variation of AM fungal communities across 15 different agricultural fields. AM fungi were found to be abundant in the experiment, accounting for 8% and 21% of total recovered DNA- and RNA-derived fungal sequences, respectively, after 104 days of plant growth. RNA- and DNA-based sequence analyses yielded most of the same AM fungal lineages. Our research yielded three major conclusions. First, no consistent differences were detected between AM fungal communities associated with GM plants and non-GM plants. Second, temporal variation in AMF community composition (between two measured time points) was bigger than GM trait-induced variation. Third, natural variation of AMF communities across 15 agricultural fields in The Netherlands, as well as within-field temporal variation, was much higher than GM-induced variation. In conclusion, we found no indication that Bt maize cultivation poses a risk for AMF.
INTRODUCTION
In recent decades, significant advances have been made in developing more productive crop varieties. One of these developments has been the generation of genetically modified (GM) crops, which has been advocated as a tool to help alleviate some of the major constraints on crop production, such as pest infestations and weed growth (22). However, because genetic modification alters the functioning of crop plants, there is a risk that these changes may have adverse effects on the plant's biotic environment (5, 19), implying that the potential effects of such modifications should be examined (4). This is particularly relevant given the forecasted increases in GM cultivation worldwide (7).
One important component of a plant's environment is the soil ecosystem, which plays an essential role in nutrient cycling and plant productivity (55). Within this system, arbuscular mycorrhizal fungi (AMF) intricately interact with plants; they engage in a symbiosis with over 80% of land plants, including major crops such as maize, wheat, potato, and soybean (62), and provide mineral nutrients in exchange for plant carbohydrates (45). Because of these ecological features, they have been identified as an important group of organisms to study for risk assessments of GM crops (25, 31).
Some studies have shown altered AM fungal development, such as spore production and root colonization, in association with GM versus non-GM plants (52, 60), including maize expressing the Bacillus thuringiensis Cry1Ab endotoxin (Bt maize) (8, 9), suggesting that there is a potential for adverse GM plant-induced effects on AM fungi. However, because these studies involved testing of only a single AM fungal species under otherwise nearly sterile conditions, we do not know whether these results hold for more natural conditions. In nature, soil and plant roots are inhabited by more diverse fungal communities, whose members may respond differently to GM plants. In general, studies where roots of GM plants (Bt or other traits) were subjected to a mixture of AM fungal species, either in microcosms or in the field, no effects of GM plants on AM fungal colonization were reported (12, 24, 28, 38, 49).
In addition to root colonization levels, it is important to investigate whether the community composition of AM fungi is altered by the cropping of GM plants. Several studies have shown that plant productivity depends on the identity and diversity of AM fungal species colonizing a plant (33, 61). Hence, if GM plants influence AM fungal communities, this can subsequently influence plant productivity. There are only a few studies (Hannula et al. [21] on GM potato and Tan et al. [49] on Bt maize) reporting whether GM cropping influences AM fungal communities in natural soils. Moreover, no studies to date have targeted RNA to detect changes in AM fungal communities in relation to Bt cropping. Targeting RNA, in addition to DNA, is thought to provide a more comprehensive picture of community dynamics, as RNA has a faster turnover rate than DNA-based assays, which may also recover DNA from dead or inactive AM fungal cells or spores (e.g., reference 29); targeting RNA might therefore better capture changes in active fungal communities (3, 23).
Here, we tested and compared the response of AM fungal communities to two Bt and two non-Bt maize varieties at two different time points by examining partial 18S rRNA sequences recovered from DNA and RNA, separately. The GM trait we studied was insect resistance, which was mediated by expression of an insecticidal protein (Bt) that can be present in Bt maize root exudates (43). This protein does not appear to directly affect AM fungi (15), but some Bt plants have nevertheless been reported to exhibit an altered interaction with AM fungal symbionts (e.g., reference 9). The maize plants were grown in a greenhouse in intact soil cores collected from the field to target natural AM fungal communities and to ensure that our results have high ecological relevance.
We utilized general fungal primers in combination with deep-sequencing technology to minimize bias toward specific AM fungal lineages (17, 26) and to get an estimate of AM fungal abundance compared to that of other fungi. This approach provided the necessary phylogenetic resolution and sufficient depth of sampling to potentially detect significant community changes. For additional confirmation of observed trends, and to estimate whether potential Bt-induced effects were comparable to the normal natural variation of AM fungal communities, we also assessed communities by a lower-resolution molecular profiling technique (terminal restriction fragment length polymorphism [T-RFLP]) targeting the nuclear 25S rRNA gene with AM fungus-specific primers. In earlier work, we used this technique to explore AMF community composition and diversity in a wide range of agricultural fields distributed throughout The Netherlands (58, 59). This enabled us to compare differences between communities in our greenhouse experiment to community variation of AM fungi found in the field. Hence, these comparisons allowed us to assess whether GM crops induce changes that exceed normal temporal community variation in the field and between fields and thus whether they represent a potential risk for soil ecological functioning.
Our aim was to answer the following questions. (i) What are the effects of two GM maize varieties on (active) AM fungal communities? (ii) Do these effects exceed normal seasonal and spatial variation of AM fungal communities in the field? (iii) How well do DNA-based and RNA-based measures of communities agree in regard to community trends?
MATERIALS AND METHODS
Pot experiment and sampling.
In order to compare AM fungal communities between GM and non-GM plants, seeds were sown into pots that contained soil from a field in which we previously characterized the AM fungal community for multiple years (organic field “Biezenmortel” in reference 58). Because AM fungal communities have previously been shown to be sensitive to experimental manipulation (47, 57), we collected intact pot-sized soil cores from the field and transferred them to pots in order to maintain natural stratification and mycelial integrity of AM fungi. Soil cores were collected randomly from within a homogeneous 10- by 10-m plot in September 2009. The crop at that time was a grass-clover mixture (Trifolium pratense L. and Lolium perenne L.) which had been sown after maize in fall 2007 and mown twice a year. Soil chemical properties were as follows: pH (CaCl2 extractable) of 5.8, P (CaCl2 extractable phosphate) of 5.1 mg kg−1, N (total nitrogen) of 1.36 g kg−1, and OM (organic matter) of 1%.
In each pot (containing approximately 6 kg of soil; diameter of 20 cm, height of 18 cm), one of four different maize (Zea Mays L.) cultivars was grown. The four different cultivars included two GM cultivars: (i) “event MON810,” cultivar Monumental MON810; and (ii) “event MON810,” cultivar DKC3421YG. These were matched with two non-GM cultivars (Monumental and DKC3420, respectively). The GM cultivars had both been transformed to express the cry1Ab gene (an insecticidal endotoxin produced by Bacillus thuringiensis that is active against, among others, the European corn borer Ostrinia nubilalis (35). The non-Bt varieties are nontransformed maize lines with background genetics similar to the Bt counterparts but do not contain the cry1Ab expression cassette. Similar background genetics between Bt and non-Bt varieties are ensured by traditional back-crossing breeding processes. Different cultivars are abbreviated by letters: M for Monumental and K for DKC (additionally, Bt varieties are indicated by “GM,” i.e., M-GM and K-GM).
We used three replicates for each cultivar, resulting in 12 pots. Two seeds were sown into each pot on 1 October 2009 and were kept in a greenhouse with a 16/8-h light/dark cycle. After 2 weeks, pots were thinned to one seedling each. In pots where no seedlings had emerged after 1 week (2 pots), new seeds were sown and thinned in the same way after an additional 2 weeks. Hoagland solution (1/2 strength P; 250 ml per pot) was applied twice during the first month of plant growth.
On 24 November, after 47 days of plant growth, soil samples were taken using the following protocol: one core (diameter of 1 cm) per pot was taken, and the part originating from the 5- to 11-cm-depth level was immediately stored on dry ice and transferred to −80°C. Cores were taken 5 to 6 cm from the edge, which was approximately halfway between the edge of the pot and the stem of the plant. On 20 January, after 104 days of growth, samples were taken as described above, but the position of cores was shifted 45° in relationship to the first core to minimize potential disturbing effects of the first sampling event. At the end of the experiment (plant age, 130 days; at full maturity of the ears), total above- and below-ground plant biomass was harvested and dried (7 days at 50°C), and total plant dry weight and ear (grain plus cob) dry-weight were determined. Of a random subset of roots in each pot, the percentage of root colonization by AM fungi was determined according to the magnified intersection method (34), based on 50 root intersections per plant.
Nucleic acid extraction and cDNA preparation.
From each sample, 2 g of soil was used for simultaneous RNA and DNA isolation using the RNA PowerSoil kit and the DNA Elution accessory kit (MO BIO Laboratories Inc., Carlsbad, CA). The DNA from total RNA-enriched samples was removed by DNase I (RNase-free DNase set 79254; Qiagen, Hilden, Germany) according to manufacturer recommendations. The total RNA was measured with a ND-1000 spectrophotometer (Nanodrop Technology, Wilmington, DE), and the quality of the total RNA was checked with Experion (Bio-Rad Laboratories Inc., Hercules, CA). The total RNA was cDNA synthesized using random hexamer primers and the superscript double-stranded cDNA synthesis kit (Life Technologies Corp., Carlsbad, CA). Resulting DNAs and cDNAs were used as the template in parallel analysis using 454 pyrosequencing and T-RFLP as described below.
454 pyrosequencing methodology.
For 454 pyrosequencing, general fungal primers FR1 and FF390 (53) were used, which amplify a region approximately 350 base pairs in length that includes the V7 and V8 hypervariable regions of the small-subunit (SSU) rRNA gene. In a recent analysis, this primer pair was most suitable for soil fungi based on specificity, coverage, and amplicon length (39). We made one small modification to primer FF390 (and now refer to it as FF390.1), where at the 5′ terminus the third position is now degenerate (see below) to accommodate detection of some Glomeraceae members as inferred from bioinformatic analysis of GenBank sequences. Thermocycling conditions were as follows: denaturing at 94°C for 30 s (after initial denaturation of 4 min), initial annealing temperature was 55°C (1 min), and every two cycles the annealing temperature was lowered by 2°C until 47°C was reached, which was the annealing temperature used for an additional 20 cycles (thus 29 PCR cycles in total). Extension conditions were 68°C for 2 min for all cycles. Reaction mixtures contained about 25 ng of DNA or RNA template added to a standard PCR mix. The 5′ terminus of primers contained an adaptor sequence and a multiplex identifier tag (MID; 12 different 10-bp-long tags), which resulted in the following primer constructs (adaptor in boldface): Forward (FF390.1), 5′-CTATGCGCCTTGCCAGCCCGCTCAG-(MID)-CGWTAACGAACGAGACCT-3′; Reverse (FR1), 5′-CGTATCGCCTCCCTCGCGCCATCAG-(MID)-AICCATTCAATCGGTAIT-3′.
Pyrosequencing was performed by Macrogen Inc. (Seoul, South Korea) using a GS FLX Titanium kit (Roche, Basel, Switzerland). Sequence analysis was done using QIIME 1.2.1 scripts (6) incorporated into the Galaxy interface (18). All reads were checked for the right forward and reverse MID tags and assigned to samples accordingly. Four samples out of 24 were excluded at this stage, because they contained the wrong tag combinations. As a result, M-GM and K treatments (where M and K indicate parental cultivars, and GM indicates the GM trait) at first sampling and M and K treatments at second sampling are represented by two, instead of three replicates (see also Table 1 for an overview of replicate number per treatment). Of all other samples, barcodes and tags were removed, and sequences were denoised using Denoiser 0.91 (42) and clustered at 97% similarity using the UCLUST 1.2.21 algorithm (14). The resulting operational taxonomic units (OTUs), represented by the most abundant sequence within each OTU, were assigned to eukaryote families through BLAST searches against the QIIME-compatible version of the Silva 104 release (41). After this, we sorted this data set into “nonfungal,” “fungal,” and “putative AMF,” of which the latter two sets were used for subsequent analysis.
Table 1.
Average AM fungal richness for each plant variety at first (47 days) and second sampling (104 days)a
| DNA or RNA | Variety | Pyrosequencing |
Reads | 104 days |
Reads | T-RFLP |
104 days |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 47 days |
47 days |
||||||||||||||
| Mean | SE | n | Mean | SE | n | Mean | SE | n | Mean | SE | n | ||||
| DNA | M | 2.7 | 0.9 | 3 | 9 | 6.0 | 0.0 | 2 | 137 | 6.2 | 0.2 | 3 | 6.5 | 0.5 | 3 |
| M-GM | 3.5 | 0.5 | 2 | 35 | 5.3 | 0.4 | 3 | 67 | 4.2 | 1.0 | 3 | 5.5 | 0.6 | 3 | |
| K | 2.0 | 2.0 | 2 | 3 | 6.5 | 0.5 | 2 | 37 | 4.3 | 1.3 | 3 | 6.3 | 1.5 | 3 | |
| K-GM | 2.3 | 0.9 | 3 | 8 | 5.3 | 0.3 | 3 | 146 | 5.0 | 1.0 | 3 | 5.2 | 0.8 | 3 | |
| RNA | M | 5.0 | 1.0 | 3 | 459 | 5.8 | 0.0 | 2 | 1159 | 4.0 | 1.0 | 3 | 6.0 | 1.2 | 3 |
| M-GM | 3.8 | 0.4 | 2 | 1109 | 5.9 | 0.0 | 3 | 718 | 4.5 | 0.8 | 3 | 6.3 | 0.4 | 3 | |
| K | 4.6 | 1.4 | 2 | 190 | 5.2 | 0.4 | 2 | 815 | 5.3 | 0.7 | 3 | 5.3 | 1.4 | 3 | |
| K-GM | 4.7 | 0.7 | 3 | 651 | 5.7 | 1.0 | 3 | 792 | 5.0 | 0.5 | 3 | 5.5 | 0.8 | 3 | |
Assessments by pyrosequencing and by T-RFLP (average of forward and reverse T-RF) are presented separately, assessed by DNA and RNA analysis. Different plant varieties are abbreviated by letters: M, Monumental; K, DKC. GM varieties are indicated with an additional identifier (i.e., M-GM and K-GM). For pyrosequencing, to account for differences in total AM fungal read numbers when more than 100 reads were present, AM fungal richness of samples was estimated by a rarefaction analysis estimate of 100 individual sequences (e.g., reference 1). SE, standard error of the mean; n, number of observations; Reads, the mean number of AM fungal reads obtained. In none of the instances was there a significant effect of the GM trait (GM versus non-GM), the cultivar (M versus K), or their interaction on AM fungal richness. For values in italics (pyrosequencing of DNA at both times and RNA at 47 days), not all replicates were represented by at least 100 AM fungal reads and thus no statistics are performed on richness estimates comparing GM with non-GM plants.
Analysis of putative Glomeromycota sequences.
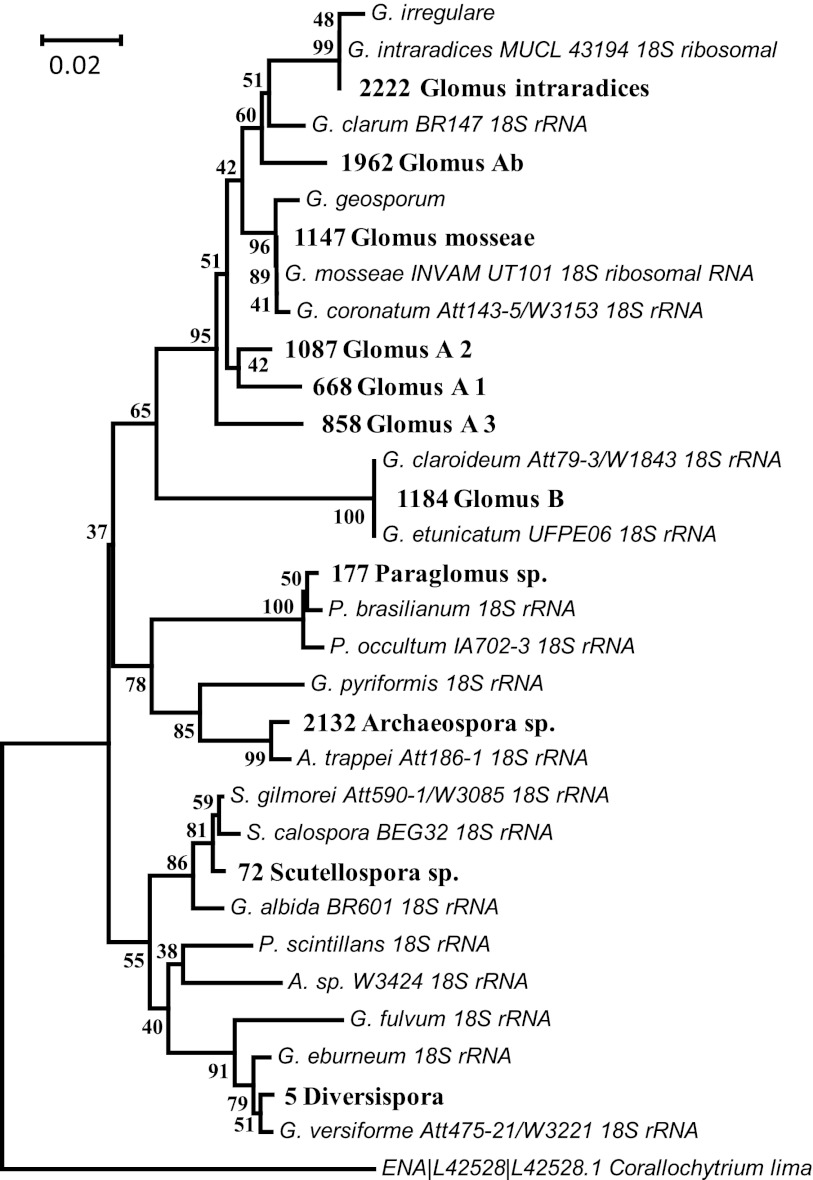
Assignment to eukaryote families yielded 185 OTUs having highest BLAST hits with families within the phylum of Glomeromycota (clustered at 97% similarity and after excluding singletons according to recommendations by Tedersoo et al. [50]). These were additionally blasted against the NCBI database for confirmation and were aligned with all Glomeromycota sequences (excluding those classified as “environmental”) within the SILVA database and, as the outgroup, the protist Corallochytrium limacisporum (GenBank accession no. L42528.1), which has been proposed to be an appropriate outgroup for fungal phylogenetic analysis (54). These analyses gave low confidence of membership of Glomeromycota for the majority (94%) of OTUs for the following reasons: (i) inclusion within the Glomeromycota had low bootstrap support, and many distinct clades were formed containing only sequences from this study and thus separated from all database sequences of known AM fungi; (ii) similarity of many of the obtained sequences to known AM fungal sequences was low (average of 93.9%) and often did not strongly exceed similarity to the highest non-AM fungal BLAST hits in the NCBI database (average of 92.4%; see Table S1 in the supplemental material for all putative AM fungal OTUs and BLAST scores). Therefore, as an operational definition, it was decided to consider OTUs to be conclusive members of the phylum of Glomeromycota only if similarity to known members exceeded 97%. Alignment, neighbor-joining (NJ) tree construction, and subsequent bootstrapping analysis (1,000 replicates) were performed using the program MEGA 4.1 (48). Nucleotide sequences were deposited in the European Nucleotide Archive under the accession no. HE970441 to HE970631.
AM fungal community analysis by T-RFLP.
AM fungal communities were also analyzed by T-RFLP. This was done to obtain additional information about the effects of Bt maize on AM fungal communities using a different molecular profiling technique. Moreover, the use of T-RFLP made it possible to compare potential GM-induced effects to the normal natural variation of AM fungal communities across 15 different agricultural fields, which we determined in earlier work (see Verbruggen et al. [58]). DNA and RNA derived from each of the pot soil samples at each time points were analyzed by T-RFLP after a nested PCR with primers LR1-FLR2 and FLR3-FLR4 (51, 56) with a standard PCR mix, respectively, and subsequent restriction digestion with TaqI. Cycling consisted of denaturing at 95°C for 5 min, 30 cycles of 15 s at 95°C, 30 s at 58°C, and 60 s at 72°C, with a final extension of 10 min. The product of the first PCR was diluted 500 times prior to second PCR. Reactions and electrophoresis conditions were as described in reference 58. The resulting electropherograms were analyzed using the program T-Rex (11) with a clustering threshold of 2.5 and exclusion of T-RFs less than 45 bp in length or contributing less that 0.5% of peak area per sample. As a reference for “normal” community variation, we analyzed samples from the same field from which soil was taken for the pot experiment (a maize field on sandy soil). These samples were taken in July 2007, September 2007, July 2008, September 2008, and at the time of the collection of experimental soil in September 2009. Freeze-dried roots (±100 mg) of six plants from each sampling date were homogenized and subjected to DNA isolation using the DNeasy plant minikit (Qiagen, Hilden, Germany). T-RFLP was performed using the same methods as the pot experiment to assess the AM fungal community present at each sampling time. Results were compared with AM fungal communities from 15 maize fields on sandy soil from earlier published work (58). However, because DNA was not available from all soil samples, we analyzed communities found in plant roots for these samples. Even though these might differ from those in soil, this procedure should still allow distinguishing main trends, especially since we previously found low systematic variation between root and soil AM fungal communities (59).
Data analysis and statistics.
T-RFLP- and pyrosequencing-derived community data were analyzed in a complementary manner. T-RFLP was specifically used to compare AM fungal communities among pot treatments and to field communities. Pyrosequencing was used to compare communities between pot treatments, increase taxonomic information, and get an estimate of AM fungal abundances as related to other fungi. DNA- and RNA-derived communities were compared between GM and non-GM plants (factor GM) and their parental cultivars (factor cultivar), and the interaction between these two factors was examined in a crossed analysis using two-way nonparametric multivariate analysis of variance (NPMANOVA) (permutation with 10,000 replicates) with Bray-Curtis similarity indices. The Bray-Curtis similarity index is a commonly used similarity estimate in ecology (10) and was previously used for comparing AM fungal communities (44). We also estimated AM fungal richness through a two-way ANOVA with the same factors. Here, we used the average of forward and reverse T-RFs for T-RFLP (as each unique sequence produces both a forward and a reverse peak) and by individual rarefaction analysis for pyrosequencing based on 100 reads (as for the majority of samples, more than 100 reads were obtained), to account for variation in read numbers among samples. To assess whether the communities derived by analysis of DNA and RNA were correlated, we compared Bray-Curtis similarities between RNA and DNA derived from the same sample versus DNA-derived communities from all other samples with a nonparametric Mann-Whitney U test. Nonmetric multidimensional scaling (NMDS) (27) was performed to assess similarity of communities of pots at different plant stages for pyrosequencing and T-RFLP-derived community data. We analyzed the correlation between these two methods through a Mantel test of Bray-Curtis similarities (permutation with 10,000 replicates). These analyses were performed in the program PAST (20).
Usage of general fungal primers for pyrosequencing also enabled us to compare numbers of sequencing reads of AM fungi and those of all fungi and thus gave an indication of AM fungal relative abundance and its development through time. Relative abundance of each OTU at each time point, for DNA as well as RNA, was tested for significant differences between GM and non-GM plant-associated communities using a nonparametric Kruskal-Wallis test. Relative abundance of AM fungal reads to total fungi (pyrosequencing), AM fungal richness (T-RFLP and pyrosequencing), percentage root colonization, total plant dry weight, and ear dry weight were assessed using a one-way ANOVA comparing GM and non-GM plants. These analyses were performed using SPSS version 17.0.
RESULTS
Plant colonization and growth.
Roots of all maize plants were colonized by AM fungi, with percentage root length colonization varying from 12% to 64% (not shown). This variation was, however, not attributable to the GM trait, as colonization percentages did not differ significantly between plant varieties (F3,8 = 2.40; P = 0.14). Total plant dry-weight (average of 74.9 g ± 2.3 standard error [SE]) and ear dry weight (average of 25.5 g ± 3.9 SE) also did not differ between varieties (total dry weight of F3,8 = 0.22, P = 0.88, n = 12; ear dry weight of F3,8 = 0.17, P = 0.91, n = 12). Ear dry weight was significantly positively correlated with AM fungal root colonization (R2 = 0.46, P = 0.016, n = 12), but total dry weight was not (R2 = 0.07; P = 0.40, n = 12).
AM fungal taxa assessed by pyrosequencing.
The pyrosequencing approach yielded 222,401 reads after denoising, with a mean length of 351 bp (standard deviation [SD] = 7.7) and a minimum length of 284 bp. Of these, 155,206 reads belonged to OTUs assigned to fungal families, and 20,465 (∼9.2% of total, or 13.2% of fungi) were assigned to AM fungal families (excluding eight singleton OTUs). However, as the majority of these could not be confidently assigned to the phylum of Glomeromycota (see Materials and Methods), we have defined an OTU as belonging to AM fungi on the basis of having at least 97% similarity to sequences classified as AM fungi in the NCBI database. This conservative approach yielded 11 OTUs that could be unequivocally assigned to the Glomeromycota and showed good phylogenetic support of topology of the containing clades (Fig. 1). These OTUs represented a considerable portion of the putative AM fungal reads: 15,518 (75.8%) out of the 20,465 putative AM fungal sequencing reads. They represent most of the major AM fungal lineages, including Glomus group A (family Glomeraceae; six OTUs, comprising 58% of reads), Glomus group B (or Claroideoglomus; one OTU, 15%), Paraglomus (one OTU, 15%), Diversispora (one OTU, 11%), Scutellospora (one OTU, 0.2%), and Archaeospora (one OTU, 0.05%). The 11 AM fungal OTUs include all of the abundant putative AM fungal taxa, as the most abundant putative AM fungus not included in these is responsible for only 3% of all putative AM fungal reads.
Fig 1.
Neighbor-joining (NJ) tree showing topology of OTUs having at least 97% similarity to known AM fungal taxa (sequences obtained in this study are in bold and are preceded with a number, an identifier for storage purposes, as in Table S1 in the supplemental material). OTUs are named according to placement within the phylogenetic tree. Bootstrap values are given at the branch points.
AM fungal communities associated with GM and non-GM plants are not different.
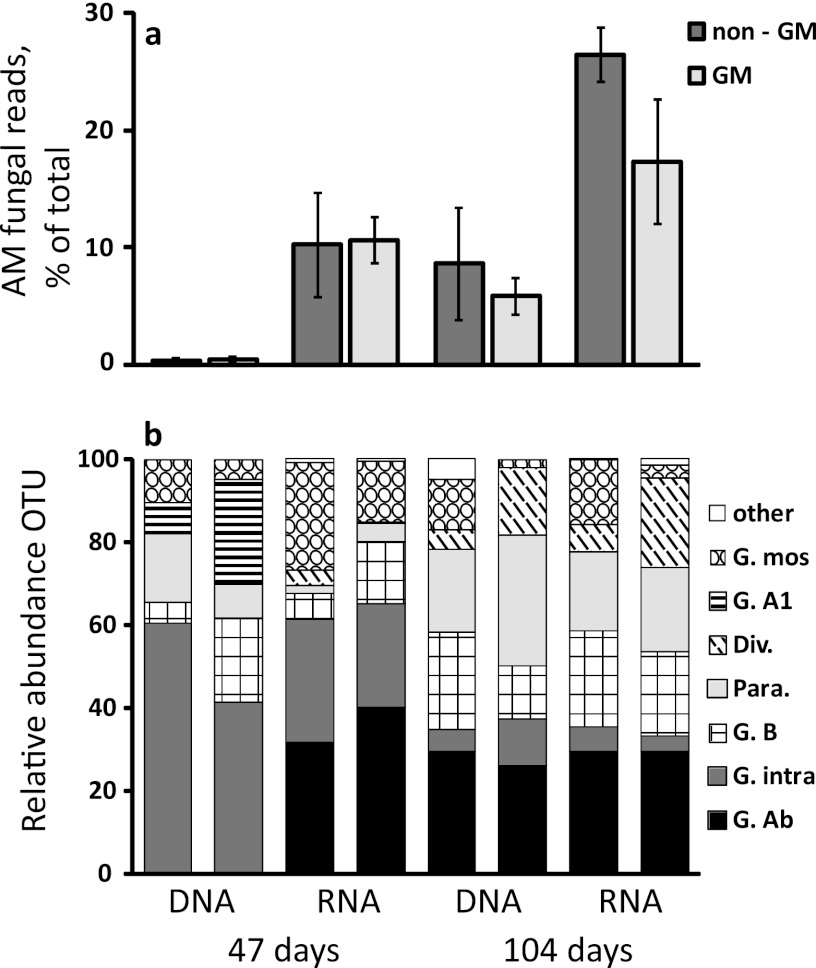
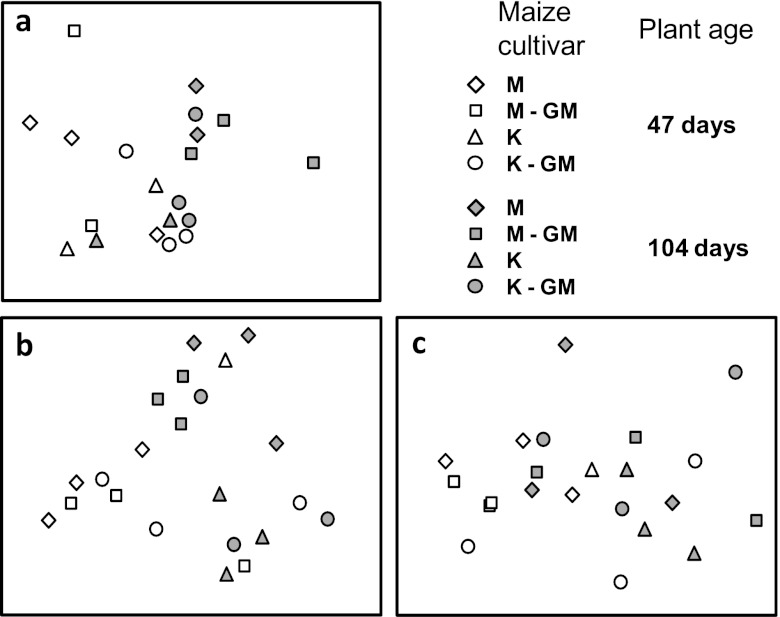
Comparing the number of AM fungal reads to the total number of fungal reads revealed a high presence of AM fungi, which increased from an average of 11% to 21% for RNA over the course of the experiment (Fig. 2a). AM fungi were represented at higher levels in RNA than in DNA, where they were very low at first sampling (Fig. 2a). There were no differences in AM fungal communities between GM and non-GM plants at either growth stage, as assessed by pyrosequencing (Table 2). Also, when analyzed separately, none of the AM fungal taxa occurred at significantly different relative abundances in GM or non-GM plant-associated soils (Fig. 2b). However, communities did differ between the two plant growth stages (Table 2; see also Fig. 2b), and for RNA-derived communities there was a significant cultivar effect at late growth stage (Table 2). Nonmetric multidimensional scaling of Bray-Curtis similarities of pyrosequencing data (Fig. 3a) indicated that AM fungal communities changed during the course of plant growth in a fairly similar direction for all treatments. In Table 1, estimated AM fungal taxon richness is presented for each plant variety. Also here, there was no significant effect of the GM trait, based upon RNA-derived sequences at the late plant stage. For RNA at early growth stage and for DNA-derived sequences, these statistics were not performed, because less than 100 AM fungal reads were obtained for some samples.
Fig 2.
(a) The total number of sequences derived from AM fungi is shown as a percentage of total fungal reads (mean ± SE). At both samplings, AM fungi were highly overrepresented in RNA compared to in DNA, and both increased from the first to the second sampling date. The relative abundances of AM fungi to total fungi were similar between GM and non-GM plants. (b) The mean relative abundance of each OTU (left and right bars of each panel representing non-GM and GM plants, respectively) also did not differ between GM and non-GM plants at any of the two time points or nucleic acid type (RNA or DNA; P > 0.05 for each OTU). For both panels, each column is represented by n = 5, except for GM at 104 days (both DNA and RNA) (n = 6) and non-GM at 104 days (both DNA and RNA) (n = 4).
Table 2.
Two-way NPMANOVA of Bray-Curtis similarities of plant-associated AM fungal communities, represented by DNA or RNA, sampled at either a plant age of 47 days or 104 daysa
| DNA or RNA | Factor | No. of days | Pyrosequencing |
T-RFLP |
||||
|---|---|---|---|---|---|---|---|---|
| F | P | n | F | P | n | |||
| DNA | GM vs non-GM | 47 | 2.73 | NS | 24 | |||
| Cultivar | 3.68 | 0.01 | ||||||
| GM vs non-GM | 104 | 0.83 | NS | 20 | 0.72 | NS | 24 | |
| Cultivar | 1.69 | NS | 0.81 | NS | ||||
| Time | 2.76 | 0.02 | 48 | |||||
| RNA | GM vs non-GM | 47 | 0.82 | NS | 20 | 2.86 | 0.04 | 24 |
| Cultivar | 0.13 | NS | 2.75 | NS | ||||
| GM vs non-GM | 104 | 0.49 | NS | 20 | 1.34 | NS | 24 | |
| Cultivar | 3.00 | 0.02 | 5.01 | 0.01 | ||||
| Time | 4.38 | 0.003 | 40 | 3.30 | 0.006 | 48 | ||
The crossed factors are GM versus non-GM and cultivar (K versus M parental cultivar; independent of the GM trait). There were no significant interactions between cultivar and GM versus non-GM in any of the analyses. The effect of the factor time is also presented, where a one-way NPMANOVA was performed contrasting communities at 104 days and 47 days. F, NPMANOVA statistic; P, chance to obtain result if null-hypothesis is true; n, number of observations. Bold numbers indicate significant effects at P < 0.05. No analyses were performed on pyrosequencing data for DNA at 47 days because of the low number of reads. Similarities between separate samples are presented in Table S2 in the supplemental material.
Fig 3.
NMDS biplots of Bray-Curtis similarities among communities found in the experimental treatments using pyrosequencing for RNA (stress = 0.14; n = 20 total samples) (a) and T-RFLP for RNA (stress = 0.16; n = 22) (b) and DNA (stress = 0.14; n = 22) (c). Note that for T-RFLP, two samples have been excluded; see the supplemental material for plots with these samples included.
T-RFLP also does not detect differences between GM and non-GM plants.
A total of 34 unique T-RF signals were found in the experimental pots, 17 of which contributed more than 1% to the total signal. This translates into an estimated total of 17 different taxa present across treatments (see Materials and Methods). There were no significant differences in AM fungal richness between GM and non-GM plants at either growth stage using either DNA or RNA (Table 1). Overall, richness estimates derived from DNA- and RNA-based community fingerprints were similar and tended to increase slightly with plant age. Using this method, there were also no significant differences found in community composition between GM and non-GM plant-associated AM fungal communities (Table 2). As with the pyrosequencing analysis, there was a significant effect of time on AM fungal communities assessed by RNA-based analysis, as well as cultivar (referring to parental variety, but not GM versus non-GM; see above) at second sampling for RNA (Table 2). Here, time was also found to have a significant effect for DNA-based analysis, as well as cultivar for AM DNA-based analysis at first sampling and GM versus non-GM for RNA at first sampling. This was caused by two replicates of the K variety, which were very different at the first sampling. Because these were the two pots where initially no seedling emerged, and communities converged afterward, this result may not be a true cultivar effect. An NMDS plot, including these two samples, is presented in Fig. S1 in the supplemental material, but these are excluded in Fig. 3b and c to show overall community trends. These two replicates did not strongly contribute to the measured time effect on RNA-based analysis (Table 1), because when these were excluded, there was still a significant difference between communities based on sampling time (not shown).
High similarity between present and active AM fungal communities.
T-RFLP community profiles derived from DNA and RNA were significantly related (Bray-Curtis similarities of RNA communities and DNA communities in the same sample versus DNA communities in other samples are 0.72 versus 0.47, respectively; Z (Mann-Whitney U statistic) = −5.72; P < 0.0001), indicating that the AM fungal community found in a sample through RNA-targeted analysis was relatively similar to DNA-targeted analysis of the same sample. For the pyrosequencing data, the relationship was less strong but also significant (0.56 versus 0.43; Z = −3.14; P < 0.001). However, when only communities at the late plant stage were analyzed, thus excluding the effect of the low number of DNA reads at sampling one (Fig. 2a; Table 1), similarity between communities represented by DNA and RNA as recovered by pyrosequencing increased (0.69 versus 0.53; Z = −3.30; P < 0.001). Also, community trends as derived from T-RFLP and pyrosequencing were significantly correlated (R = 0.21; P = 0.009), indicating that they partially detect similar AM fungal community compositional variation. For a more detailed account of community trends using T-RFLP and pyrosequencing, see Notes S1 in the supplemental material.
DISCUSSION
In this study, we compared AM fungal communities associated with two GM and two non-GM maize varieties using complementary approaches. Our analysis of RNA and DNA aimed to provide a sensitive and comprehensive assessment of community composition. DNA is much more stable than RNA, and thus it has been argued to have a strong historical component (2). In contrast, owing to its fast degradation, RNA is potentially more suitable for analysis of active communities at a given time point (40). Moreover, we analyzed DNA and RNA by pyrosequencing with general fungal primers and by T-RFLP using AM fungus-specific primers. Using this combined and sensitive approach, we did not find significant differences between AM fungal communities associated with GM and non-GM maize plants. Moreover, variation, including GM-induced variation, was much lower than the natural variation of AMF communities across a wide range of fields.
Our pyrosequencing-based assessment of total soil fungal communities provided us with novel information on AM fungal abundance and activity compared to other fungi (Fig. 2a). In most instances, relative abundance of AM fungal reads was very high, in particular in the RNA-derived assessments. This indicates that RNA-based analysis can provide a good estimate of AM fungal communities, which was in our case even superior to DNA-based analysis regarding contribution to total fungal reads. In our experiment, this contribution was lowest for DNA-derived assessment of young plants, where AM fungi accounted for only less than 1% of total fungal reads. A possible explanation is that the experimental procedure represented a strong disturbance, causing increased growth of fungi other than AM fungi or a decrease in the latter.
At plant maturity, however, AM fungal reads constituted an average of >20% of all fungal RNA reads and >10% of all fungal DNA reads. A similarly high contribution was observed recently by Jumpponen (23) in a North American tall-grass prairie system, where AM fungi were abundant in the rRNA pool (assessed on RNA), with a relative contribution ranging from about 15% to 35% of total fungal reads. In another study in a North American prairie, Miller and Kling (36) estimated AM fungi to contribute up to 23% of total microbial biomass. This may indicate that activity and abundance of AM fungi in our system is in the same order of magnitude as in these grassland systems. However, as rRNA is constitutively transcribed in living cells, it may not be a precise measure of taxon activity. Potentially, transcription of other genes which are indicative for specific physiological processes (e.g., phosphate transporters) can be assessed in future research as an approximate for specific activities (see Gamper et al. [16] for a discussion).
The high relative abundance of AM fungi observed here has important implications for studies on environmental risk of transgenic crops. In other studies on fungal community responses to GM plants (e.g., references 30 and 63), the primers used have been shown to display a bias toward other fungal groups over Glomeromycota and in some cases may not detect AM fungi at all. This means that these studies potentially underestimate a numerically significant and important group of soil fungi.
AM fungal community assessments by pyrosequencing and T-RFLP were significantly correlated, confirming that both methods are able to detect the same community trends. However, these methods did not always agree with respect to the relative abundances of individual taxa. These discrepancies are partially explained by the fact that our T-RFLP method highly underestimated the abundance of Paraglomus and Diversispora, and thus relative abundance of other taxa is consequently overestimated. Another factor that is likely to have contributed is the usage of two different rRNA regions and methods, SSU for pyrosequencing and LSU for T-RFLP. These regions differ in their phylogenetic resolutions (17), and the methods of T-RFLP versus full amplicon sequencing differ as well in this respect. For economic and practical reasons, the number of replicates per treatment was low in this study (2 or 3 replicates per treatment). An alternative would have been to include more replicates, thereby reducing the number of reads obtained per sample. However, because some AMF OTUs had very low abundances (e.g., Archaeospora represent 0.05% of AMF reads), this is likely to result in missing OTUs. Despite low replication, effects of time (see Table 2) on AM fungal communities were stronger than cultivar or GM-induced differences, indicating that our experiment had sufficient statistical power to measure community changes.
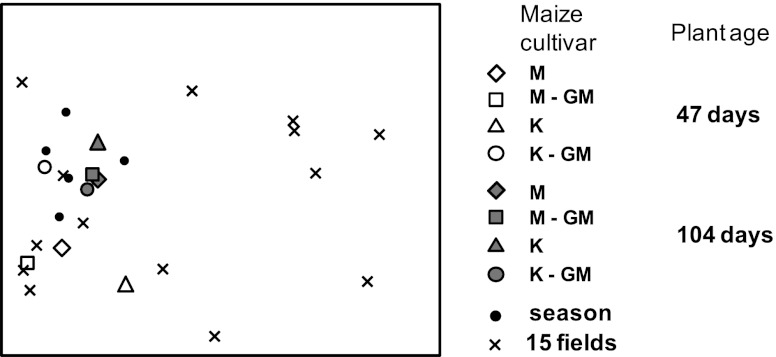
Additionally, we tested how community variation in our experiment relates to that in (i) the field where soil was derived from five samplings over the course of 3 years and (ii) 15 other agricultural maize fields (Fig. 4). As expected, we found that the variation of AM fungal communities was much larger among 15 different agricultural fields, and communities for each of the experimental varieties clustered within the seasonal variation of the donor field at the late sampling stage (Fig. 4). However, AM fungal communities at 47 days showed lower similarity to those obtained from the field but did not cluster outside the natural range found in other agricultural fields, nor did these show any systematic change in response to the GM trait. It should be noted that a variety of maize cultivars was cropped in the agricultural fields, and thus our assessment of natural variation includes cultivar-associated variation. Because our goal has been to sample a representative set of normal agricultural conditions, we think it is appropriate to include different cultivars in estimating baseline community variation. Therefore, our study suggests that the GM trait we assessed does not cause AM fungal communities to change outside baseline variation. This provides additional support to the conclusion that Bt maize poses no risk for the investigated soil fungi. For other GM traits, agricultural systems, or nontarget organism classes, such changes may occur. We therefore suggest that future studies on effects of GM plants on soil (microbial) communities address how effects, if any, scale against natural variation. This will provide important insight in the extent of GM-imposed disturbance and potential effects on soil ecological quality (25, 46). Moreover, it would be of great interest to extend this approach toward other soil microbes, such as bacteria and decomposing and pathogenic fungi.
Fig 4.
Detrended correspondence analysis (DCA; eigenvalue of first axis = 0.53, second axis = 0.21) of the soil AM fungal communities obtained in this study represented by DNA, together with season (seasonal variation in AM fungal communities at one field site, where soil was derived for this study), obtained during five field sampling events over 3 years, and 15 fields (variation of maize root AM fungal communities across 15 different agricultural fields sampled in September 2007) (data from reference 57). AM fungal community composition was assessed using T-RFLP for all data points. Note that the different maize varieties of the greenhouse study represent average communities of soil samples of three replicate plants (to accommodate comparison to pooled samples); season and 15 fields are pooled root samples of six plants. Three- to four-month-old maize roots were collected for 15 fields, while maize roots for season had variable age (from 2 to 4 months).
In our study, the majority of putative AM fungal OTUs (but not reads) had a low similarity to known AM fungal species. This is in contrast to previous studies where AM fungal communities were examined by pyrosequencing of rRNA genes. For these studies, recovered sequences typically had the highest BLAST hits with known AM fungal sequences (and thus putative AM fungal sequences) that exceeded 97% (13, 32, 37). One reason for this discrepancy may be that our primers target a different portion of the SSU rRNA gene, i.e., the V3-V4 and V7-V8 hypervariable regions, and that these portions differ in variability and/or phylum discrimination potential. Because other studies using the same primer set as used here rarely report the recovery of AM fungal sequences, it remains to be tested whether this is a common problem. We observed that affiliation of low-similarity OTUs with the Glomeromycota was not strongly supported through alignment and phylogenetic analysis, and therefore we have used very stringent similarity criteria to identify AMF. We acknowledge that this may have caused a significant underestimation of AMF diversity, although the OTUs included do represent the majority of putative AM fungal reads and should thus be sufficient to analyze major community trends. Because phylum assignment through BLAST of sequences obtained from soil material through 454 pyrosequencing can be problematic, at least for this SSU rRNA gene region, we recommend performing additional phylogenetic analysis to confirm affiliation of OTUs.
Conclusions.
In this study, we thoroughly analyzed community composition of important and beneficial soil fungi in association with transgenic and nontransgenic crops, targeting DNA as well as RNA at two plant growth stages. To our knowledge, assessing both DNA and RNA simultaneously has not previously been done for these fungi. Our assessment revealed that AM fungi represent a major portion of soil fungal communities, especially at the RNA (activity) level. We have not detected consistent changes in AM fungal community composition in response to the two GM maize cultivars compared to non-GM maize cultivars. Given the large contribution of AM fungi to the estimated total fungal activity we observed and the key ecological functions they perform, AM fungi should not be overlooked in risk-assessment of GM crops. Our approach of comparing GM-related changes to within and among field community variation can provide a suitable framework for scaling potential GM-imposed effects to natural community variation.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Ecology Regarding Genetically Modified Organisms (ERGO) grant 838.06.021. E.T.K. was supported by an NWO Vidi and Meervoud grant.
This is Netherlands Institute of Ecology (NIOO-KNAW) publication 5324.
We thank Monsanto for the provision of maize seeds used in the experiment.
Footnotes
Published ahead of print 10 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adrain JM, Westrop SR, Chatterton BDE, Ramskold L. 2000. Silurian trilobite alpha diversity and the end-Ordovician mass extinction. Paleobiology 26:625–646 [Google Scholar]
- 2. Anderson IC, Parkin PI. 2007. Detection of active soil fungi by RT-PCR amplification of precursor rRNA molecules. J. Microbiol. Meth. 68:248–253 [DOI] [PubMed] [Google Scholar]
- 3. Anderson IC, Parkin PI, Campbell CD. 2008. DNA- and RNA-derived assessments of fungal community composition in soil amended with sewage sludge rich in cadmium, copper and zinc. Soil Biol. Biochem. 40:2358–2365 [Google Scholar]
- 4. Andow DA, Zwahlen C. 2006. Assessing environmental risks of transgenic plants. Ecol. Lett. 9:196–214 [DOI] [PubMed] [Google Scholar]
- 5. Bruinsma M, Kowalchuk GA, van Veen JA. 2003. Effects of genetically modified plants on microbial communities and processes in soil. Biol. Fertil. Soils 37:329–337 [Google Scholar]
- 6. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carpenter JE. 2011. Impact of GM crops on biodiversity. GM Crops 2:7–23 [DOI] [PubMed] [Google Scholar]
- 8. Castaldini M, et al. 2005. Impact of Bt corn on rhizospheric and soil eubacterial communities and on beneficial mycorrhizal symbiosis in experimental microcosms. Appl. Environ. Microbiol. 71:6719–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheeke TE, Pace BA, Rosenstiel TN, Cruzan MB. 2011. The influence of fertilizer level and spore density on arbuscular mycorrhizal colonization of transgenic Bt 11 maize (Zea mays) in experimental microcosms. FEMS Microbiol. Ecol. 75:304–312 [DOI] [PubMed] [Google Scholar]
- 10. Clarke KR, Somerfield PJ, Chapman MG. 2006. On resemblance measures for ecological studies, including taxonomic dissimilarities and a zero-adjusted Bray-Curtis coefficient for denuded assemblages. J. Exp. Mar. Biol. Ecol. 330:55–80 [Google Scholar]
- 11. Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH. 2009. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics 10:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Vaufleury A, et al. 2007. Exposure and effects assessments of Bt-maize on non-target organisms (gastropods, microarthropods, mycorrhizal fungi) in microcosms. Pedobiologia 51:185–194 [Google Scholar]
- 13. Dumbrell AJ, et al. 2011. Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytol. 190:794–804 [DOI] [PubMed] [Google Scholar]
- 14. Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 15. Ferreira LHPL, Molina JC, Brasil C, Andrade G. 2003. Evaluation of Bacillus thuringiensis bioinsecticidal protein effects on soil microorganisms. Plant Soil 256:161–168 [Google Scholar]
- 16. Gamper HA, van der Heijden MGA, Kowalchuk GA. 2010. Molecular trait indicators: moving beyond phylogeny in arbuscular mycorrhizal ecology. New Phytol. 185:67–82 [DOI] [PubMed] [Google Scholar]
- 17. Gamper HA, Walker C, Schüssler A. 2009. Diversispora celata sp. nov.: molecular ecology and phylotaxonomy of an inconspicuous arbuscular mycorrhizal fungus. New Phytol. 182:495–506 [DOI] [PubMed] [Google Scholar]
- 18. Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hails RS. 2000. Genetically modified plants—the debate continues. Trends Ecol. Evol. 15:14–18 [DOI] [PubMed] [Google Scholar]
- 20. Hammer O, Harper DAT, Ryan PD. 2001. PAST: Palaeontological Statistics software package for education and data analysis. Pal. Elec. 4:9–17 [Google Scholar]
- 21. Hannula SE, de Boer W, van Veen JA. 2010. In situ dynamics of soil fungal communities under different genotypes of potato, including a genetically modified cultivar. Soil Biol. Biochem. 42:2211–2223 [Google Scholar]
- 22. Jones JDG. 2011. Why genetically modified crops? Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 369:1807–1816 [DOI] [PubMed] [Google Scholar]
- 23. Jumpponen A. 2011. Analysis of ribosomal RNA indicates seasonal fungal community dynamics in Andropogon gerardii roots. Mycorrhiza 21:453–464 [DOI] [PubMed] [Google Scholar]
- 24. Knox OGG, Nehl DB, Mor T, Roberts GN, Gupta V. 2008. Genetically modified cotton has no effect on arbuscular mycorrhizal colonisation of roots. Field Crop. Res. 109:57–60 [Google Scholar]
- 25. Kowalchuk GA, Bruinsma M, van Veen JA. 2003. Assessing responses of soil microorganisms to GM plants. Trends Ecol. Evol. 18:403–410 [Google Scholar]
- 26. Krüger M, Stockinger H, Kruger C, Schüssler A. 2009. DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol. 183:212–223 [DOI] [PubMed] [Google Scholar]
- 27. Kruskal JB. 1964. Nonmetric multidimensional scaling—a numerical method. Psychometrika 29:115–129 [Google Scholar]
- 28. Landi L, Capocasa F, Costantini E, Mezzetti B. 2009. ROLC strawberry plant adaptability, productivity, and tolerance to soil-borne disease and mycorrhizal interactions. Transgenic Res. 18:933–942 [DOI] [PubMed] [Google Scholar]
- 29. Lanzen A, et al. 2011. Exploring the composition and diversity of microbial communities at the Jan Mayen hydrothermal vent field using RNA and DNA. FEMS Microbiol. Ecol. 77:577–589 [DOI] [PubMed] [Google Scholar]
- 30. Lee SH, Kim CG, Kang H. 2011. Temporal dynamics of bacterial and fungal communities in a genetically modified (GM) rice ecosystem. Microb. Ecol. 61:646–659 [DOI] [PubMed] [Google Scholar]
- 31. Liu WK. 2010. Do genetically modified plants impact arbuscular mycorrhizal fungi? Ecotoxicology 19:229–238 [DOI] [PubMed] [Google Scholar]
- 32. Lumini E, Orgiazzi A, Borriello R, Bonfante P, Bianciotto V. 2010. Disclosing arbuscular mycorrhizal fungal biodiversity in soil through a land-use gradient using a pyrosequencing approach. Environ. Microbiol. 12:2165–2179 [DOI] [PubMed] [Google Scholar]
- 33. Maherali H, Klironomos JN. 2007. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316:1746–1748 [DOI] [PubMed] [Google Scholar]
- 34. McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. 1990. A new method which gives an objective measure of colonization of roots by vesicular arbuscular mycorrhizal fungi. New Phytol. 115:495–501 [DOI] [PubMed] [Google Scholar]
- 35. Meissle M, Romeis J, Bigler F. 2011. Bt maize and integrated pest management—a European perspective. Pest Manag. Sci. 67:1049–1058 [DOI] [PubMed] [Google Scholar]
- 36. Miller RM, Kling M. 2000. The importance of integration and scale in the arbuscular mycorrhizal symbiosis. Plant Soil 226:295–309 [Google Scholar]
- 37. Öpik M, Metsis Daniell M, Zobel TJ, Moora M. 2009. Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol. 184:424–437 [DOI] [PubMed] [Google Scholar]
- 38. Powell JR, et al. 2007. Mycorrhizal and rhizobial colonization of genetically modified and conventional soybeans. Appl. Environ. Microbiol. 73:4365–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prévost-Bouré NC, et al. 2011. Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS One 6:13 doi:10.1371/journal.pone.0024166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prosser JI. 2002. Molecular and functional diversity in soil micro-organisms. Plant Soil 244:9–17 [Google Scholar]
- 41. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7:668–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saxena D, Flores S, Stotzky G. 2002. Bt toxin is released in root exudates from 12 transgenic corn hybrids representing three transformation events. Soil Biol. Biochem. 34:133–137 [Google Scholar]
- 44. Schechter SP, Bruns TD. 2008. Serpentine and non-serpentine ecotypes of Collinsia sparsiflora associate with distinct arbuscular mycorrhizal fungal assemblages. Mol. Ecol. 17:3198–3210 [DOI] [PubMed] [Google Scholar]
- 45. Smith SE, Read D. 2008. Mycorrhizal symbiosis, 3rd ed Academic Press, London, United Kingdom [Google Scholar]
- 46. Snow AA, et al. 2005. Genetically engineered organisms and the environment: current status and recommendations. Ecol. Appl. 15:377–404 [Google Scholar]
- 47. Sykorova Z, Ineichen K, Wiemken A, Redecker D. 2007. The cultivation bias: different communities of arbuscular mycorrhizal fungi detected in roots from the field, from bait plants transplanted to the field, and from a greenhouse trap experiment. Mycorrhiza 18:1–14 [DOI] [PubMed] [Google Scholar]
- 48. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 49. Tan F, et al. 2011. Assessment of the arbuscular mycorrhizal fungal community in roots and rhizosphere soils of Bt corn and their non-Bt isolines. Soil Biol. Biochem. 43:2473–2479 [Google Scholar]
- 50. Tedersoo L, et al. 2010. 454 pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 188:291–301 [DOI] [PubMed] [Google Scholar]
- 51. Trouvelot S, van Tuinen D, Hijri M, Gianinazzi-Pearson V. 1999. Visualization of ribosomal DNA loci in spore interphasic nuclei of glomalean fungi by fluorescence in situ hybridization. Mycorrhiza 8:203–206 [Google Scholar]
- 52. Turrini A, Sbrana C, Nuti MP, Pietrangeli BM, Giovannetti M. 2004. Development of a model system to assess the impact of genetically modified corn and aubergine plants on arbuscular mycorrhizal fungi. Plant Soil 266:69–75 [Google Scholar]
- 53. Vainio EJ, Hantula J. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104:927–936 [Google Scholar]
- 54. Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JPW. 2002. Evolution—extensive fungal diversity in plant roots. Science 295:2051–2051 [DOI] [PubMed] [Google Scholar]
- 55. van der Heijden MGA, Bardgett RD, van Straalen NM. 2008. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11:296–310 [DOI] [PubMed] [Google Scholar]
- 56. van Tuinen D, Jacquot E, Zhao B, Gollotte A, Gianinazzi-Pearson V. 1998. Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol. Ecol. 7:879–887 [DOI] [PubMed] [Google Scholar]
- 57. Verbruggen E, Kiers ET, Bakelaar PNC, Röling WFM, van der Heijden MGA. 2012. Provision of contrasting ecosystem services by soil communities from different agricultural fields. Plant Soil 350:43–55 [Google Scholar]
- 58. Verbruggen E, et al. 2010. Positive effects of organic farming on below-ground mutualists: large-scale comparison of mycorrhizal fungal communities in agricultural soils. New Phytol. 186:968–979 [DOI] [PubMed] [Google Scholar]
- 59. Verbruggen E, van der Heijden MGA, Weedon JT, Kowalchuk GA, Röling WFM. 2012. Community assembly, species richness and nestedness of arbuscular mycorrhizal fungi in agricultural soils. Mol. Ecol. 21:2341–2353 [DOI] [PubMed] [Google Scholar]
- 60. Vierheilig H, et al. 1995. Colonization of transgenic tobacco constitutively expressing pathogenesis-related proteins by the vesicular-arbuscular mycorrhizal fungus Glomus mosseae. Appl. Environ. Microbiol. 61:3031–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wagg C, Jansa J, Schmid B, van der Heijden MGA. 2011. Belowground biodiversity effects of plant symbionts support aboveground productivity. Ecol. Lett. 14:1001–1009 [DOI] [PubMed] [Google Scholar]
- 62. Wang B, Qiu YL. 2006. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16:299–363 [DOI] [PubMed] [Google Scholar]
- 63. Weinert N, et al. 2009. Rhizosphere communities of genetically modified zeaxanthin-accumulating potato plants and their parent cultivar differ less than those of different potato cultivars. Appl. Environ. Microbiol. 75:3859–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.