Abstract
Herpes simplex virus 1 (HSV-1) immediate-early protein ICP0 localizes to cellular structures known as promyelocytic leukemia protein (PML) nuclear bodies or ND10 and disrupts their integrity by inducing the degradation of PML. There are six PML isoforms with different C-terminal regions in ND10, of which PML isoform I (PML.I) is the most abundant. Depletion of all PML isoforms increases the plaque formation efficiency of ICP0-null mutant HSV-1, and reconstitution of expression of PML.I and PML.II partially reverses this improved replication. ICP0 also induces widespread degradation of SUMO-conjugated proteins during HSV-1 infection, and this activity is linked to its ability to counteract cellular intrinsic antiviral resistance. All PML isoforms are highly SUMO modified, and all such modified forms are sensitive to ICP0-mediated degradation. However, in contrast to the situation with the other isoforms, ICP0 also targets PML.I that is not modified by SUMO, and PML in general is degraded more rapidly than the bulk of other SUMO-modified proteins. We report here that ICP0 interacts with PML.I in both yeast two-hybrid and coimmunoprecipitation assays. This interaction is dependent on PML.I isoform-specific sequences and the N-terminal half of ICP0 and is required for SUMO-modification-independent degradation of PML.I by ICP0. Degradation of the other PML isoforms by ICP0 was less efficient in cells specifically depleted of PML.I. Therefore, ICP0 has two distinct mechanisms of targeting PML: one dependent on SUMO modification and the other via SUMO-independent interaction with PML.I. We conclude that the ICP0-PML.I interaction reflects a countermeasure to PML-related antiviral restriction.
INTRODUCTION
Promyelocytic leukemia protein nuclear bodies (PML-NBs), also known as ND10, are dynamic punctuate structures within the nuclei of mammalian cells that harbor a large number of permanently or transiently localized proteins (8, 22, 44). ND10 have been associated with many cellular functions, including DNA repair (17), regulation of transcription (42, 60), chromatin assembly and modification (18, 32), apoptosis (1, 55), stress (39), senescence (3), the ubiquitin pathway (30, 35, 36,) and oncogenesis (47, 48; reviewed in reference 2). Increasing evidence also links ND10 with an intrinsic cellular defense against many DNA viruses, such as human cytomegalovirus (HCMV), herpes simplex virus 1 (HSV-1), varicella zoster virus, human adenovirus type 5, and the murine gammaherpesvirus 68 (reviewed in references 19, 56, and 57).
Very early after HSV-1 infection, the immediate-early (IE) protein ICP0 localizes to ND10 and disrupts their integrity. ICP0, which is a RING finger E3 ubiquitin ligase (7), induces the proteasome degradation of two major ND10 components, namely, PML, which is the ND10 organizer, and the small ubiquitin modifier (SUMO)-modified forms of Sp100 (5, 11, 21, 27, 41). In the absence of ICP0, PML and Sp100 are both recruited to sites associated with parental HSV-1 genomes and early replication compartments, and this behavior contributes to the repression of viral gene expression. Indeed, depletion of either PML or Sp100 increases the plaque formation efficiency of ICP0-null mutant HSV-1 (28, 29).
Due to alternative splicing, there are six major nuclear PML isoforms (PML.I to VI), that share the N-terminal region (exons 1 to 6) and differ in their C-terminal parts. Exons 1 to 3 encode a tripartite motif (TRIM), which comprises a RING finger, two additional zinc-stabilized domains known as B-boxes, and a coiled-coil motif (34). The TRIM domain and SUMO posttranslational modification of PML are required for proper ND10 formation (50). We have shown recently that no single PML isoform is able to establish fully functional ND10 structures, implying that PML isoforms act in collaboration (15). PML.I is the most abundant of the isoforms and is also the most highly conserved between humans and mice (13). In the context of HSV-1 infection, expression of PML.I or PML.II partially reverses the improved replication of an ICP0-null mutant virus observed in PML-depleted cells (15).
The questions of how ICP0 is targeted to ND10 and how it induces the degradation of PML early after infection have been raised for many years. Using cotransfection, one previous investigation into the characteristics of PML that contribute to its degradation by ICP0 found that PML.IV mutants that do not localize to ND10, have lost C-terminal sequences, or lack the SUMO-1 modification site at lysine residue 160 are degraded less efficiently by ICP0 (6). That study did not find evidence of a direct interaction of PML.IV with ICP0 or of ubiquitination of PML.IV by ICP0 in vitro.
Using a depletion/reintroduction approach, we recently reported that ICP0 induces the preferential degradation of the SUMO modified forms of all nuclear PML isoforms during wild-type (wt) HSV-1 infection (5). We also found that ICP0 contains sequences related to SUMO interaction motifs (SIMs) and can interact with SUMO-1 and SUMO-2, which might in part explain its accumulation at SUMO-rich foci, such as ND10. However, PML.I was also efficiently degraded by ICP0 independently of SUMO modification. During the course of the present study we noted that SUMO modification-deficient PML.I, but not an analogous mutant of PML.IV, colocalized with ICP0 extremely well during HSV-1 infection. These observations raised the questions of how ICP0 colocalizes preferentially with PML.I and induces its degradation in a SUMO-independent manner.
To address these questions, we introduced wt and mutant forms of PML.I into cells depleted of endogenous PML. We found that the isoform-specific C-terminal part of PML.I is responsible for colocalization with and degradation by ICP0. Using yeast-two hybrid and coimmunoprecipitation (Co-IP) assays, we found that PML.I and ICP0 interact via the isoform-specific PML.I sequences and the N-terminal half of ICP0. We also observed that other SUMO-modified PML isoforms are less efficiently degraded by ICP0 in cells depleted of PML.I. When considered along with the results of our recent studies on the role of SUMO modification in ICP0 activity, these data demonstrate that ICP0 targets PML and ND10 not only via a SUMO-modification-dependent mechanism but also by sequence-specific recognition of the most abundant PML isoform.
MATERIALS AND METHODS
Viruses and cells.
Wild-type (wt) HSV-1 strain 17 syn+, ICP0-null mutant derivative dl1403 (ΔICP0) (53), and mutant E52X, which expresses only the N-terminal two thirds of ICP0 (40) were propagated in BHK (baby hamster kidney) cells and titrated in U2OS cells, in which ICP0 is not required for efficient replication of HSV-1 (59). Human foreskin diploid fibroblasts (HFs), U2OS, Vero and HEK-293T cells were grown in Dulbecco modified Eagle medium containing 10% fetal calf serum. BHK cells were grown in Glasgow modified Eagle medium supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. HepaRG hepatocyte cells (31) were maintained in William's medium E supplemented with 10% fetal bovine serum Gold (PAA Laboratories), 2 mM glutamine, 5 μg of insulin/ml, and 0.5 μM hydrocortisone. All cell growth media contained 100 U of penicillin/ml and 100 μg of streptomycin/ml. Lentivirus-transduced cell lines were maintained with continuous antibiotic selection as specified.
Lentivirus expression vectors and cells.
Control and PML-depleted HepaRG cells reconstituted with individual PML isoforms expressed at close to endogenous levels have been described previously (15). PML isoforms are named as described previously (34). Lentivirus vector plasmids expressing an shRNA against all PML isoforms, and vectors expressing enhanced yellow fluorescent protein (EYFP), EYFP-PML isoforms I to VI, mutants of PML.I with lesions in the RING finger (ΔRING; C57A and C60A), B-Box 1 (ΔBB1; C139A and C142A), B-Box 2 (ΔBB2; C211A and C214A), the coiled-coil motif (ΔCC, deletion of E239 to L322), mutations in the SIM motif (MSIM) at SUMO modification sites K160 and K490 (PML.I.KK) and at SUMO modification sites K65, K160, K490, and K616 (PML.I.4K) were as described previously (5, 14, 15). A lentivirus vector plasmid expressing a mutant of PML.IV with substitutions at SUMO modification sites K160 and K490 (EYFP-PML.IV.KK) has also been described (15). A lentivirus vector plasmid expressing PML.II with mutations at SUMO modification sites K160 and K490 (EYFP-PML.II.KK) was obtained by fragment transfer from a previously characterized plasmid (6). A lentivirus vector plasmid expressing a PML.I mutant with substitutions at SUMO modification sites K65, K160, K490, and K616 and a deletion of exon 9 (EYFP-PML.I.4K.Δexon9) was constructed by PCR splicing with appropriate oligonucleotides. A lentivirus vector plasmid expressing a fusion of EYFP with PML.I C-terminal sequences (EYFP-PML.I.C-term) was obtained after deletion of the N-terminal part of PML.I from pLNGY-PML.I (exon 1 to the first half of exon 8a; residues 1 to 592), by replacement of the SalI-Bsu36I fragment with annealed oligonucleotides of sequences TCGACCCCATGGGCCCCAGCACCC and TGAGGGTGCTGGGGCCCATGGGG.
PML-depleted HepaRG cells reconstituted with individual PML isoforms expressed at high level were isolated by use of lentivirus vector plasmids pLNDY-PML.I, pLNDY-PML.II (15), and pLNDY-PML.IV (obtained by replacement of the gD promoter in pLNGY-PML.IV with the 372-bp NdeI-AgeI fragment of pEGFP-C1 [Clontech] containing the proximal part of the HCMV IE promoter/enhancer). HepaRG cells expressing EYFP-linked proteins were isolated using lentivirus transduction and fluorescence-activated cell sorting (FACS), as described previously (15). HFs, expressing control or anti-PML shRNAs, and EYFP or the EYFP-PML.I.C-term fusion were isolated using the same methodology.
HepaRG cells specifically depleted of PML.I were isolated by use of a lentivirus vector expressing an shRNA with the sense strand DNA sequence of 5′-CCAGACCTACCTGGCGAGAAA, which corresponds to a sequence specific to PML isoform I in exon 9. The shRNA sequence was built into double-stranded oligonucleotides for cloning into a lentivirus plasmid vector based on pLKO.1puro, from which lentivirus stocks were derived as described previously (25, 28, 29). HepaRG cells expressing EYFP-PML.I (15) were also transduced with lentiviruses expressing control or anti-PML.I shRNAs.
Transfections.
Cells were seeded onto coverslips in 24-well plates at 8 × 104 cells per well and transfected the next day with the plasmid pCI-FXE, using GeneJuice, according to the manufacturer's guidelines (Novagen). The plasmid pCI-FXE expresses a RING finger deletion mutant ICP0 (23). After 24 h, the cells were fixed for immunofluorescence.
Virus infections.
Cells were seeded into 24-well plates at 105 cells per well and infected the following day for immunofluorescence and immunoblotting experiments. Viruses (wt HSV-1 or mutant E52X or ΔICP0) were allowed to absorb for 1 h in 100 μl of medium with frequent agitation and then overlaid with 400 μl of medium.
For degradation studies using immunoblotting (Fig. 2, 4, 5, and 6), the cells were infected at a multiplicity of infection (MOI) of 2, 5, or 10 with wt HSV-1 in the absence or presence of proteasome inhibitor (MG132, 3 μM; Calbiochem) or with ICP0-null mutant HSV-1 (ΔICP0) at an MOI of 10 and then overlaid with medium with or without MG132. Four hours later, the cells were harvested for Western blot analysis.
Fig 2.
ICP0 degrades SUMO-modified EYFP-PML.II but not EYFP-PML.II.KK in HepaRG cells depleted of endogenous PML. Cells were infected with wt HSV-1 (MOI = 2) or ICP0-null mutant HSV-1 (ΔICP0, MOI = 10) in the presence or absence of MG132 and harvested 5 h later, and extracts were analyzed for ICP4, ICP0, and EYFP fusion proteins. Tubulin was used as a loading control. Ni indicates mock-infected control.
Fig 4.
ICP0 induces the degradation of TRIM, RING finger, and SIM motif mutants of PML.I. (A) ICP0 degrades mutant versions of EYFP-PML.I in HepaRG cells depleted of endogenous PML. Cells expressing the indicated proteins were infected with wt HSV-1 (MOI = 2) or ICP0-null mutant HSV-1 (ΔICP0, MOI = 10) in the presence or absence of MG132 and harvested 5 h later. All of the samples were analyzed on the same gel, but different exposures of the EYFP-PML sections are shown so that the intensities of the EYFP-PML bands in the mock-infected samples are similar. (B) Analysis of the degradation of EYFP-PML.I.ΔCC after infection with wt HSV-1 (MOI = 2, 5, or 10) or ICP0-null mutant HSV-1 (ΔICP0, MOI = 10) in the presence or absence of MG132. Two different exposures of the EYFP-PML section of the filter are shown. Viral and cellular proteins were detected by Western blotting, as indicated. Ni indicates the mock-infected controls.
Fig 5.
PML.I specific exon 9 is required for SUMO-independent PML.I colocalization with and degradation by ICP0. (A) Maps of PML.I.4K and PML.I.4K.Δexon9. (B and C) Immunofluorescence analysis of colocalization of PML.I.4K.Δexon9 and ICP0 (in HepaRG cells depleted of endogenous PML). Experimental details were as described for Fig. 1, and data were quantified as in Fig. 1D. The original data are in Table 1. A typical example image of lack of colocalization of ICP0 with PML.I.4K.Δexon9 is shown in panel B. (D) Western blot analysis of expression of PML.I, PML.I.4K, and PML.I.4K.Δexon9 in shPML cells. (E) ICP0 degrades EYFP-PML.I.4K but not EYFP-PML.I.4K.Δexon9. Cells depleted of endogenous PML and expressing EYFP-PML.I, EYFP-PML.I.4K, or EYFP-PML.I.4K.Δexon9 were infected with wt HSV-1 (MOI = 2) or ICP0-null mutant HSV-1 (ΔICP0, MOI = 10) in the presence or absence of MG132 and harvested 5 h later. ICP4, ICP0, EYFP-PML, and tubulin were analyzed by Western blotting. All of the samples were analyzed on the same gel, but different exposures of the EYFP-PML sections are shown so that the intensities of the EYFP-PML bands in the mock-infected samples are similar.
Fig 6.
EYFP-tagged PML.I.C-term is recruited into ICP0 foci and degraded after HSV-1 infection. (A) Maps of PML.I and PML.I.C-term. (B) EYFP-PML.I.C-term colocalizes with ICP0 after wt-HSV-1 infection. Experimental details were as described for Fig. 1B. Noninfected controls are also shown in the first two rows. (C) Data were quantified as described in Fig. 1D. (D) Western blot analysis of the expression of EYFP and EYFP-PML.I.C-term in shPML cells. (E) ICP0 degrades EYFP-PML.I.C-term in cells depleted of endogenous PML. Cells expressing EYFP or EYFP-PML.I.C-term were infected with wt HSV-1 (MOI = 2) or ICP0-null mutant HSV-1 (ΔICP0, MOI = 10) in the presence or absence of MG132 and harvested 5 h later. ICP4, ICP0, EYFP-PML, and tubulin were analyzed by Western blotting. All of the samples were analyzed on the same gel, but different exposures of the EYFP sections are shown so that the intensities of the EYFP bands in the mock-infected samples are similar. (F and G) Colocalization of ICP0 and EYFP-PML.I.C-term is not dependent on virus infection. (F) ICP0, EYFP-PML.I.C-term, and hDaxx colocalize during wt infection. Cells were infected at an MOI of 2 in the presence of MG132 and stained 4 h later for ICP0 and hDaxx. (G) ICP0 and EYFP-PML.I.C-term colocalize independently of HSV-1 infection and of the ICP0 RING finger. PML-depleted cells expressing EYFP or EYFP-PML.I.C-term were transfected with a plasmid expressing ICP0ΔRING and stained 24 h later for ICP0 and hDaxx. These images are single-plane projections of short z-stacks.
For time course degradation studies using immunoblotting (Fig. 10B to E and 11D and E), the cells were infected at an MOI of 2 with wt HSV-1 or mutant E52X in the absence or presence of MG132 or with ICP0-null mutant HSV-1 (ΔICP0) at an MOI of 10 and then overlaid with medium with or without MG132 as relevant. The cells were harvested at hourly intervals for Western blot analysis.
Fig 10.
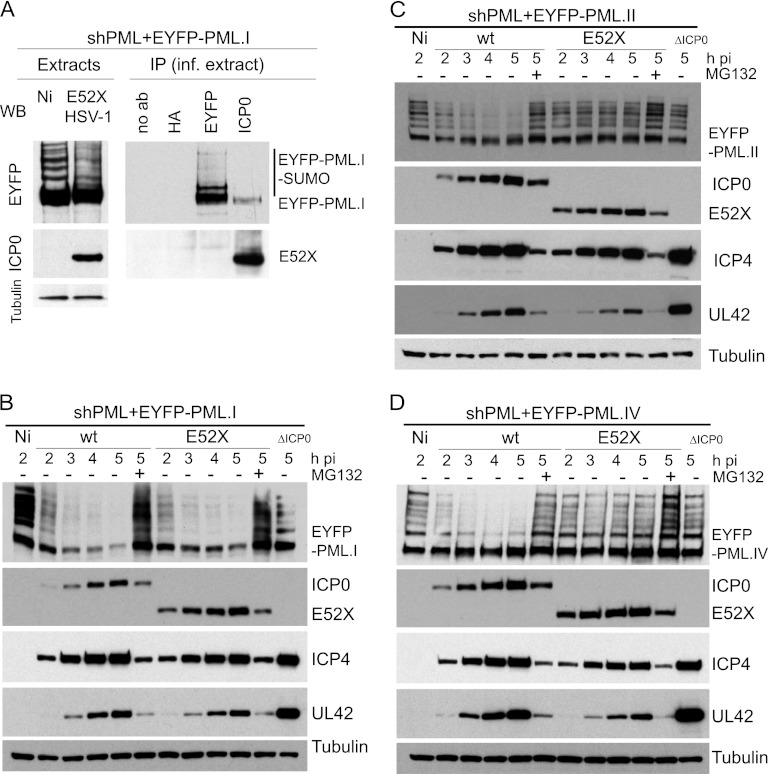
An ICP0 mutant lacking the C-terminal third of ICP0 (E52X) interacts with PML.I and induces its degradation. (A) Co-IP analysis of E52X and PML.I in HSV-1-infected PML-depleted HepaRG cells expressing EYFP-PML.I. Cells were infected with HSV-1 E52X mutant (MOI = 5) in the presence of MG132, and extracts were made 2 h later for immunoprecipitation with anti-EYFP, anti-ICP0, and anti-HA MAbs or with no antibody. The precipitates were analyzed by Western blotting for the EYFP-PML.I fusion protein and E52X. (B, C, and D) E52X induces degradation of PML.I but has no effect on PML.II or PML.IV. PML-depleted cells expressing PML.I (B), PML.II (C), or PML.IV (D) were infected with wt HSV-1 (MOI = 2) in the absence or presence of MG132, or with HSV-1 E52X mutant (MOI = 2) in the absence or presence of MG132, or with ICP0-null mutant HSV-1 (ΔICP0, MOI = 10) and harvested at hourly intervals until 5 h postinfection. Extracts were analyzed for ICP4, ICP0, UL42, and EYFP fusion proteins.
Fig 11.
Study of PML.I specific depletion. (A) Western blot analysis of HepaRG cells expressing EYFP-PML.I and control (shLuci) or PML.I-specific (shPML.I) shRNAs. (B) Western blot analysis of HepaRG cells expressing shLuci or shPML.I shRNAs using an antibody that detects all PML isoforms. (C) The relative plaque formation of wt and ICP0-null mutant HSV-1 in HepaRG cells expressing shLuci (control), shPML, or shPML.I shRNAs, as indicated. The data from several independent experiments were averaged and then plotted as means ± the standard deviations. Calculations were as described in Materials and Methods. (D and E) PML.I depletion delays the degradation of the other PML isoforms by ICP0. Control and shPML.I cells were infected with wt HSV-1 (MOI = 2) in absence or presence of MG132 or ICP0-null mutant HSV-1 (ΔICP0, MOI = 10) and harvested at hourly intervals until 7 h postinfection. Membranes were probed for PML, ICP4, ICP0, UL42, and tubulin.
For immunofluorescence, cells on coverslips were infected at an MOI of 2 with wt HSV-1 or E52X in the presence of MG132, overlaid 1 h later with medium that also contained MG132, and fixed 1 or 4 h later.
For immunoprecipitation studies, cells were seeded into 60-mm dishes at 1.5 × 106 cells per dish and infected the following day with wt HSV-1 or E52X at an MOI of 5 with 1 ml of medium containing MG132. One hour later, the cells were overlaid with 3 ml of medium with MG132, and the cells were harvested 2 h later.
For plaque assays in HepaRG-derived lines, the cells were seeded into 24-well dishes at 105 cells per well and then infected the following day with appropriate sequential 3-fold dilutions of HSV-1 strain in1863 or dl1403/CMVlacZ that contain the lacZ gene under the control of the HCMV promoter/enhancer inserted into the tk gene. After virus adsorption, the cells were incubated in medium containing 1% human serum for 24 h before the plaques were identified by β-galactosidase staining (33). The relative efficiencies of plaque formation at a given virus input were calculated as described previously (29). All experiments were performed four times.
Immunofluorescence microscopy.
Cells on 13-mm glass coverslips were fixed using 5% (vol/vol) formaldehyde in phosphate-buffered saline (PBS) containing 2% sucrose and then treated with 0.5% Nonidet P-40 (NP-40) substitute (EuroClone) in PBS containing 10% sucrose. ICP0 was detected with mouse monoclonal antibody (MAb) 11060 (26), ICP4 with MAb 58S (51), hDaxx with rabbit polyclonal antibody (rAb) 07-471 (Upstate), and PML with rAb A301-167A (Bethyl Laboratories) MAb 5E10 (54). The secondary antibodies were fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Sigma), Alexa Fluor 633-conjugated goat anti-rabbit IgG (Molecular Probes), Alexa Fluor 633-conjugated goat anti-mouse IgG (Molecular Probes), and Cy3-conjugated goat anti-mouse IgG (GE Healthcare). A glycerol-based mounting medium was used (Citifluor AF1). The samples were examined using a Zeiss LSM 510 confocal microscope, using the 488-, 543-, and 633-nm laser lines and scanning each channel separately or under image capture conditions that eliminated channel overlap. The lens was an oil immersion ×63 Plan-Apochromat (NA 1.40). To ensure that all EYFP-PML and ND10 foci were recorded, the images were acquired as short z-stacks (three or four slices covering 1.5 to 2 μm), and then single-plane projections were produced for export as tiff files using LSM 510 software. Exported images were processed using Adobe Photoshop with minimal adjustment and then assembled for presentation using Adobe Illustrator. The quantification per nucleus of PML and ICP0 focus colocalization was done by eye using at least 10 single-plane projection images per cell line. The images were scored as positive only when clear colocalizing foci were present in both channels.
Immunoblotting.
Cells were seeded into 24-well dishes at 105 cells per well. The following day, the cells were washed twice with PBS before harvesting them in SDS-PAGE loading buffer. Proteins were resolved on 7.5 or 6% SDS gels and then transferred to nitrocellulose membranes by Western blotting. The following antibodies were used: anti-tubulin MAb (T4026; Sigma-Aldrich); anti-green fluorescent protein (anti-GFP) rAb ab290 (Abcam), which also detects EYFP; anti-ICP4 MAb 58S; anti-ICP0 MAb 11060; anti-UL42 MAb (Z1F11 [49]); anti-PML rAb A301-167A; and anti-Sp100 rAb SpGH (52).
Immunoprecipitation.
After 2 h of infection at an MOI of 5 in the presence of MG132, cell monolayers (1.5 × 106 cells) were harvested in 1 ml of immunoprecipitation lysis buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 1% NP-40, 10% glycerol, 2 mM EDTA, 2 mM dithiothreitol, and sodium fluoride [Sigma] at 4.7 mg/ml), supplemented with protease inhibitors (Roche) and iodoacetimide and N-ethylmaleimide (both from Sigma) at 1.25 mg/ml and 0.1 μg/ml, respectively, to inhibit SUMO proteases. Cells were incubated on ice for 30 min prior to centrifugation (10,000 × g for 25 min at 4°C). The lysates were precleared by incubation with protein G-beads supplemented with salmon sperm DNA (Millipore) for 30 min at 4°C (continuous mixing) and then centrifuged to remove beads and nonspecifically bound proteins (13,000 × g for 10 min at 4°C). The lysates were then incubated at 4°C with 0.5 μg of mouse antibodies per 900 μl of lysate and either anti-HA (Santa Cruz Biotechnology) as a negative control, anti-ICP0 11060, anti-GFP (ab1218; Abcam), or no antibody, utilizing continuous mixing. Immunocomplexes were captured by incubation with protein G-beads for 90 min at 4°C under continuous mixing and precipitated by centrifugation (2,000 × g for 5 min at 4°C). The beads were washed five times with 1 ml of immunoprecipitation lysis buffer (containing half the concentrations of salt and NP-40) at 4°C and then resuspended in 60 μl of gel loading buffer. Samples were boiled for 5 min and centrifuged twice (13,000 rpm, 2 min, room temperature) to remove the beads, and 40 μl of each sample was resolved on 6% polyacrylamide gels.
Y2H assays.
Yeast two-hybrid (Y2H) analysis was based on the Matchmaker 3 system (Clontech) utilizing yeast strains AH109 and Y187. The PML.I, PML.I.4K, and PML.I.4K.Δexon9 cDNAs were inserted into pGBK-T7 in frame with the GAL4 DNA binding domain (BD). Plasmids derived from pGBK-T7 containing USP7 cDNA, and pGAD-T7 plasmids encoding the GAL4 activation domain (AD) in frame with ICP0 cDNAs encoding wt ICP0.1-775, ICP0.1-593, ICP0.594-775, ICP0.1-388, ICP0.1-241, and ICP0.1-775ΔRING (FXE) or with SUMO-1 have been described previously (5). Transformed yeast colonies were picked and mated overnight into a final volume of 500 μl, and 7 μl of each of the diploid cultures was then plated out on selective medium (i) lacking leucine and tryptophan (−L−W), (ii) lacking leucine, tryptophan, and histidine (−L−W−H), or (iii) lacking leucine, tryptophan, and histidine and supplemented with 1 or 5 mM 3-amino-1,2,4-triazole (3AT). Colonies were grown for 3 days prior to image capture.
RESULTS
ICP0 colocalizes with and induces the degradation of SUMO modification deficient PML.I.
We previously reported that ICP0 can target PML.I, but not PML.IV, in a SUMO-modification-independent manner (5). In the present study, we set out to determine the molecular basis of the specific effects of ICP0 on PML.I. First, we tested whether ICP0 colocalizes with EYFP-tagged versions of various wt and mutant PML isoforms. To avoid complications caused by interactions between introduced PML proteins and endogenous PML (14, 15), we used cells expressing an shRNA that targets endogenous but not introduced PML isoforms. Cells were infected with wt HSV-1 in the presence of the proteasome inhibitor MG132 to inhibit ICP0-mediated PML degradation.
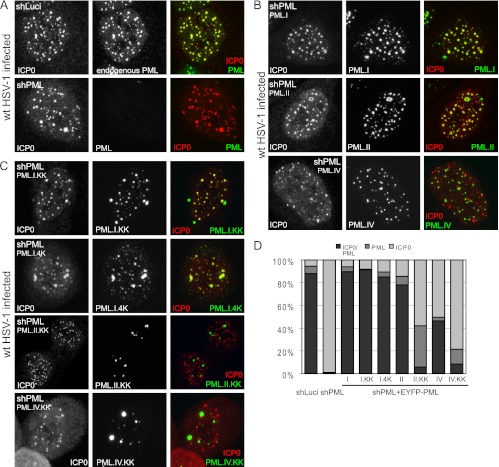
ICP0 precisely colocalizes with endogenous PML in the nucleus of control cells under these conditions (Fig. 1A), and this was also true of PML.I expressed in the absence of endogenous PML (Fig. 1B). Quantification of these and related data are presented in Fig. 1D and Table 1. In contrast, while both PML.IV and ICP0 formed foci in infected cells, half of the ICP0 foci did not contain PML.IV, and even when colocalization did occur, it occurred to a lesser extent (Fig. 1B, bottom row, compare the extent of red and green foci to the mainly yellow ones seen with PML.I). With the exception of PML.II (Fig. 1B), even higher proportions of ICP0 foci that were negative for PML were observed in cells expressing other individual PML isoforms in the absence of endogenous PML (Table 1; images not shown). Therefore, ICP0 has a marked preference for colocalization with PML isoforms I and II compared to the others.
Fig 1.
ICP0 colocalizes with SUMO-modification defective PML.I. (A) ICP0 and endogenous PML colocalize in control HepaRG cells expressing an anti-luciferase shRNA (shLuci). ICP0 also forms foci in PML depleted HepaRG cells (shPML). Cells were infected with wt HSV-1 at an MOI of 2 in the presence of MG132 and costained 4 h later for ICP0 and PML. (B) ICP0 colocalizes more efficiently with EYFP-PML.I than EYFP.PML.II or EYFP.PML.IV when these proteins are expressed in cells depleted of endogenous PML (shPML). Cells were infected as described above and stained 4 h later for ICP0 only. (C) PML.I SUMO-modification deficient mutants also colocalize with ICP0. Cells (shPML) expressing the indicated EYFP-PML.I isoform mutants were infected and stained as in panel B. All images are single-plane projections of short z-stacks. (D) Quantification of ICP0, PML, and colocalizing ICP0/PML foci. The ratios of colocalizing compared to noncolocalizing foci, as indicated by the shaded bars, are presented, based on the data of Table 1.
Table 1.
Analysis of PML colocalization with ICP0 during wt HSV-1 infection in the presence of MG132a
| Cell | Mean absolute no. ± SD |
% Foci |
|||
|---|---|---|---|---|---|
| PML | ICP0 | Co-loc | ICP0+PML dots/ICP0 total dots | ICP0+PML dots/PML total dots | |
| HA+shLuci (control) | 39.6 ± 9.5 | 39.2 ± 9.7 | 36.9 ± 9.5 | 94.13 | 93.18 |
| HA+shPML (HALP) | 0.4 ± 0.9 | 41.8 ± 12 | 0.4 ± 0.9 | 0.95 | 100 |
| HALP+PML.I | 29.1 ± 10.7 | 29.8 ± 11.5 | 27.9 ± 10.6 | 93.62 | 95.54 |
| HALP+PML.II | 42 ± 12.4 | 45.1 ± 11.8 | 38.1 ± 12.1 | 84.47 | 91.14 |
| HALP+PML.III | 8.2 ± 3.6 | 40 ± 10.2 | 6.8 ± 3 | 17 | 82.92 |
| HALP+PML.IV | 23.9 ± 4 | 49.7 ± 13.7 | 23.3 ± 4.2 | 46.88 | 93.57 |
| HALP+PML.V | 9.5 ± 4.7 | 34.5 ± 6 | 9.5 ± 5 | 27.53 | 100 |
| HALP+PML.VI | 8.1 ± 2.6 | 36.3 ± 9.6 | 7.2 ± 2.4 | 19.83 | 87.8 |
| HALP+PML.I.KK | 25.5 ± 11.2 | 27.6 ± 11.5 | 25.4 ± 11.2 | 92.03 | 99.5 |
| HALP+PML.I.4K | 26.8 ± 7.87 | 28.7 ± 9.2 | 25.5 ± 8.1 | 88.85 | 95.15 |
| HALP+PML.I.4K.Δexon9 | 7.7 ± 5.2 | 35.7 ± 14.5 | 2.4 ± 1.9 | 6.72 | 31.48 |
| HALP+PML.I.BB1 | 31 ± 8.4 | 31.6 ± 9 | 30.1 ± 8.5 | 95.25 | 97 |
| HALP+PML.I.BB2 | 22.8 ± 9.3 | 23.9 ± 9.8 | 22.2 ± 8.8 | 92.88 | 98.6 |
| HALP+PML.I.DCC | 12.5 ± 11 | 35 ± 12 | 12.5 ± 11 | 35.71 | 100 |
| HALP+PML.I.ΔRING | 30.11 ± 8.7 | 32.6 ± 9 | 30.11 ± 8.7 | 92.36 | 99.26 |
| HALP+PML.I.MSIM | 21.8 ± 5.8 | 23.5 ± 6.1 | 21 ± 5.8 | 89.36 | 96.33 |
| HALP+PML.II.KK | 26.1 ± 9 | 39.5 ± 9.1 | 3.6 ± 3.9 | 9.11 | 13.87 |
| HALP+PML.IV.KK | 5.6 ± 3 | 22.7 ± 9 | 2.2 ± 2 | 9.69 | 39.28 |
| HALP+EYFP | 0.2 ± 0.5 | 39 ± 9 | 0.2 ± 0.5 | 5.13 | 100 |
| HALP+EYFP-PML.I.C-term | 19.9 ± 6.1 | 23.7 ± 6.2 | 19.4 ± 6.1 | 81.85 | 97.48 |
| HA+shPML.I | 38.08 ± 10.6 | 59.33 ± 12.6 | 37.83 ± 10.6 | 63.76 | 99.34 |
Cells expressing control (shLuci), anti-PML.I (shPML.I), or anti-PML shRNAs (shPML) and shPML cells expressing EYFP-PML isoform I, II, III, IV, V, or VI fusion proteins, mutant forms of EYFP-PML.I, -PML.II, or -PML.IV, EYFP-PML.I.C-term, or EYFP on its own were treated with MG132 and infected at an MOI of 2. After 4 h, the control and shPML and shPML.I cells were costained for ICP0 and endogenous PML. EYFP-PML was detected in shPML cells expressing the various EYFP-PML fusion proteins by autofluorescence. Single plane projections of short z-stacks of at least 10 cells were used for quantification of the mean absolute number of PML, ICP0, and colocalizing (Co-loc) foci per nucleus. The percentage columns indicate (i) the proportions of ICP0 foci that are associated with PML compared to the total number of ICP0 nuclear dots observed and (ii) the percentage of PML foci that are associated with ICP0 dots compared to the total of nuclear PML dots observed.
We next investigated the SUMO-modification-deficient mutants PML.I.KK, PML.II.KK, and PML.IV.KK, in which the two major SUMO modification sites of PML (K160 and K490) are mutated. ICP0 colocalized very efficiently with PML.I.KK, and also with PML.I.4K, which has additional mutations in SUMO modification sites K65 and K616 (Fig. 1C). In contrast, ICP0 colocalized poorly with PML.II.KK and PML.IV.KK, although a minority of PML.II.KK and PML.IV.KK dots colocalized with low levels of ICP0 (Fig. 1C and D). Therefore, ICP0 colocalizes efficiently with PML.I, but not PML.II and PML.IV, independently of SUMO modification.
Interestingly, in uninfected cells the PML.I.KK, PML.I.4K, and PML.IV.KK foci are less abundant but larger than those of the wt proteins (14). This was also true of PML.II.KK (Fig. 1C). Notably, after infection of cells expressing PML.I.KK or PML.I.4K, we observed that these proteins were redistributed into a much larger number of foci that colocalized with ICP0. This did not occur with the analogous PML.II and PML.IV mutant proteins (Fig. 1C), indicating that the presence of ICP0 modifies the localization of the PML.I mutants specifically, with PML.I being recruited into the ICP0 foci.
Consistent with the colocalization studies, we previously reported that ICP0 preferentially induces the degradation of the SUMO-modified forms of all PML isoforms, expressed individually in cells depleted of endogenous PML (5, 15). We also reported that ICP0 can target PML.I but not PML.IV in a SUMO-modification-independent manner (5). In the present study we found that degradation of PML.II by ICP0 is also SUMO modification dependent (Fig. 2). Therefore, efficient colocalization with and degradation of PML that is not SUMO modified is specific for PML.I, implying that these properties are likely to be imparted by PML.I isoform-specific sequences.
Investigation of the involvement of the TRIM and SIM in ICP0-mediated PML.I degradation.
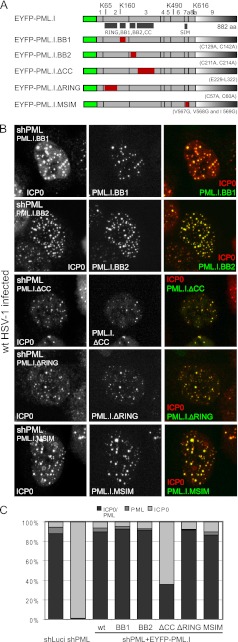
We next analyzed the characteristics of PML.I that are required for its colocalization with and degradation by ICP0. We previously reported the construction of cells depleted of endogenous PML and expressing mutants of PML.I with (i) substitutions of conserved cysteine residues in the RING finger, B-Box 1, or B-Box 2; (ii) a deletion of the coiled-coil element of the TRIM; and (iii) mutations in the SUMO interaction motif (SIM) (14, 15). These mutant proteins are named PML.I.ΔRING, PML.I.BB1, PML.I.BB2, PML.I.ΔCC, and PML.I.MSIM, respectively (Fig. 3A).
Fig 3.
ICP0 colocalizes with TRIM, RING finger, and SIM motif mutants of PML.I. (A) Maps of the relevant PML.I mutant EYFP-fusion proteins. (B) Immunofluorescence analysis of colocalization of the PML mutant proteins (in HepaRG cells depleted of endogenous PML) after infection at an MOI of 2 with wt HSV-1 in the presence of MG132. Cells were stained for ICP0 at 4 h after infection. The images are single-plane projections of short z-stacks. (C) Quantification of colocalization of ICP0 and the PML.I mutants, as described for Fig. 1D. The original data are in Table 1.
We first studied the colocalization of ICP0 with these PML.I mutants during wt HSV-1 infection in the presence of MG132. PML.I.BB1, PML.I.BB2, and PML.I.MSIM formed foci in uninfected cells (15) and, after infection, ICP0 colocalized as efficiently with these three mutants as with wt PML.I (Fig. 3 and Table 1). PML.I.ΔCC and PML.I.ΔRING mutants showed a diffuse expression pattern prior to infection (15) but, after infection, these mutants were also recruited into ICP0 foci (the latter more efficiently than the former) (Fig. 3B and C). It is striking that PML.I mutants with a diffuse nuclear expression pattern can be relocalized into ICP0 foci during HSV-1 infection.
We next analyzed ICP0-dependent degradation of these PML.I mutants. All were degraded 5 h after infection with wt HSV-1 at an MOI of 2, but to variable extents: PML.I.BB2, PML.I.ΔRING, and PML.I.MSIM were very efficiently degraded, whereas PML.I.BB1 was less sensitive (Fig. 4A). PML.I.ΔCC was degraded less efficiently than the other mutants at an MOI of 2, but the extent of its degradation was increased at higher MOIs (Fig. 4B). The reduced efficiency of degradation of the coiled-coil deletion mutant correlates with its reduced levels of colocalization with ICP0 (Fig. 3B and C).
PML.I specific exon 9 is required for PML.I colocalization with and degradation by ICP0.
At the sequence level, the six nuclear PML isoforms differ only in their C-terminal regions. Given that none of the elements of the TRIM, or the SIM, are absolutely required for PML.I colocalization with and degradation by ICP0 (Fig. 3 and 4), we investigated whether PML.I-specific sequences encoded by exon 9 are required for these effects. A mutant of PML.I that lacks all possible SUMO modification sites and also sequences encoded by exon 9 (PML.I.4K.Δexon9, Fig. 5D) colocalized poorly with ICP0 (Fig. 5B and C and Table 1) and was not sensitive to ICP0-induced degradation during wt HSV-1 infection (Fig. 5E). These features contrast strongly with the behavior of PML.I.4K, and the data therefore implicate sequences encoded by exon 9 as being required for the SUMO-modification-independent effects of ICP0 on PML.I.
EYFP-tagged PML.I.C-term is recruited into ICP0 foci and degraded.
We next investigated whether the sequences encoded by the C-terminal sequences of PML.I by themselves confer colocalization with and degradation by ICP0. We constructed a vector expressing the PML.I-specific sequences encoded by exon 9, plus part of exon 8a (which is also included in PML.IV), linked to EYFP. This EYFP fusion protein (EYFP-PML.I.C-term), which includes the putative exonuclease III domain defined previously (12), was expressed in PML-depleted cells and analyzed in comparison to analogous cells expressing EYFP alone (Fig. 6A and D). Both EYFP and EYFP-PML.I.C-term were diffusely localized throughout the cell (Fig. 6B, top rows). After infection with wt HSV-1, EYFP-PML.I.C-term, but not EYFP, was found in foci that colocalized with ICP0 (Fig. 6B and C and Table 1). Similar results were found using PML-depleted HFs expressing EYFP-PML.I.C-term (data not shown). EYFP-PML.I.C-term was partially degraded during wt HSV-1 infection in an ICP0-dependent and proteasome-dependent manner, whereas EYFP was not (Fig. 6E). EYFP-PML.I.C-term degradation was also observed in PML-depleted HFs (data not shown).
ND10 proteins, such as Sp100 and hDaxx, are mostly diffusely localized in the nucleus of cells lacking or depleted of endogenous PML (15, 28, 32, 60). During wt HSV-1 infection of PML-depleted HFs, however, Sp100 and hDaxx are transiently recruited into ICP0 nuclear foci that are likely to be associated with viral genomes (29). Consistent with this earlier result, we found that hDaxx and EYFP-PML.I.C-term colocalized with ICP0 at early times of infection in cells depleted of endogenous PML (Fig. 6F). Recruitment of EYFP-PML.I.C-term into ICP0 foci, however, was not dependent of HSV-1 infection and the presence of the viral genome, since it occurred in cells transfected with a plasmid expressing RING finger mutant ICP0 (ICP0ΔRING) (Fig. 6G, lower row). Neither EYFP nor hDaxx was recruited into these ICP0ΔRING foci in transfected cells (Fig. 6G). Similarly, Sp100 was not found in these foci (data not shown). Therefore, colocalization between ICP0 and the C-terminal region of PML.I occurs independently of infection and of other major ND10 components.
ICP0 interacts specifically with PML.I.
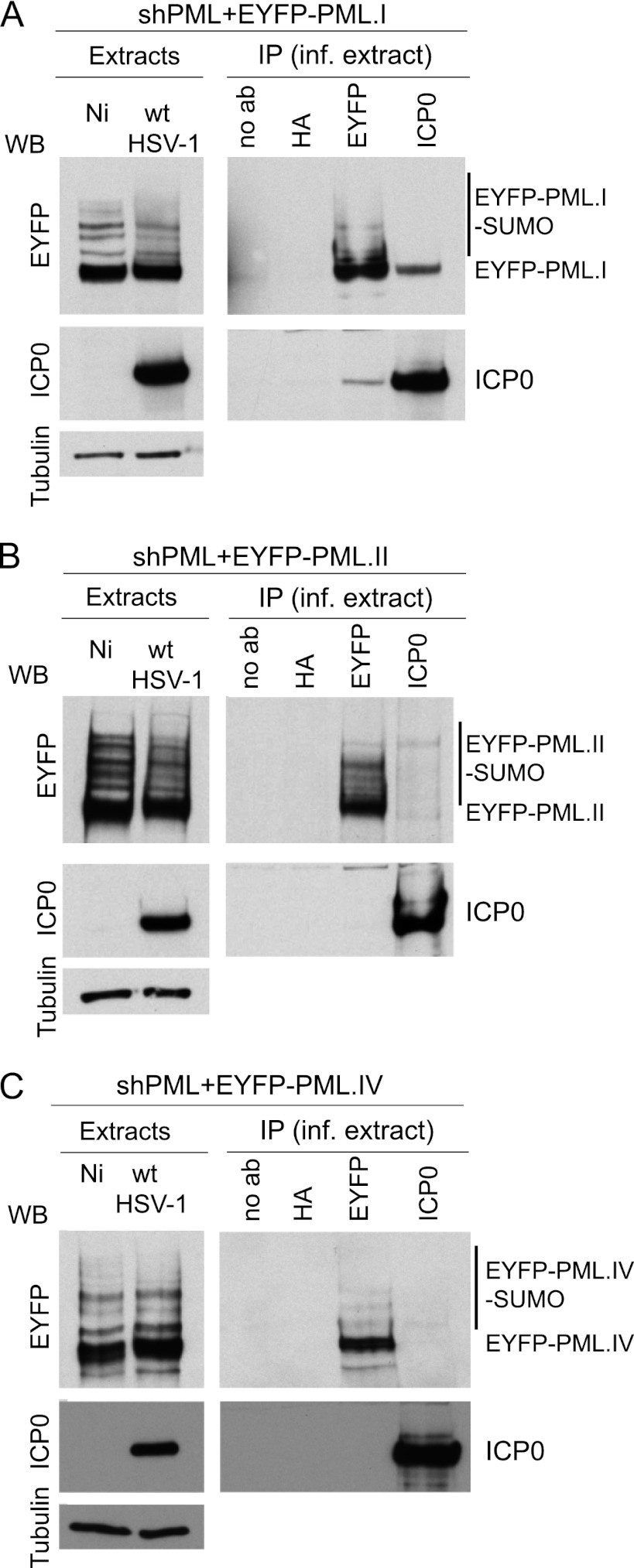
We next tested whether ICP0 and PML.I interact in Co-IP assays using PML-depleted cells expressing high levels of EYFP-linked PML isoform I, II, or IV (15). These three cell lines were infected with wt HSV-1 in the presence of MG132, and then at 2 h after infection extracts were prepared for analysis. Both unmodified EYFP-PML.I and ICP0 were detected in immune precipitates prepared from infected cells expressing EYFP-PML.I, using either anti-EYFP or anti-ICP0 antibodies (Fig. 7A). However, EYFP-PML.II and PML.IV were detected only in immune precipitates using the anti-EYFP antibody (Fig. 7B and C). Therefore, reciprocal immunoprecipitation of ICP0 with PML.I, but not PML.II or PML.IV, was observed.
Fig 7.
ICP0 interacts with PML.I. Co-IP analysis of ICP0 and PML isoforms in HSV-1-infected PML-depleted HepaRG cells expressing PML.I (A), PML.II (B), or PML.IV (C). Cells were infected with wt HSV-1 (MOI = 5) in the presence of MG132, and extracts were made 2 h later for immunoprecipitation with anti-EYFP, anti-ICP0, and anti-HA MAbs or with no antibody. The precipitates were analyzed by Western blotting for EYFP-PML fusion proteins and ICP0.
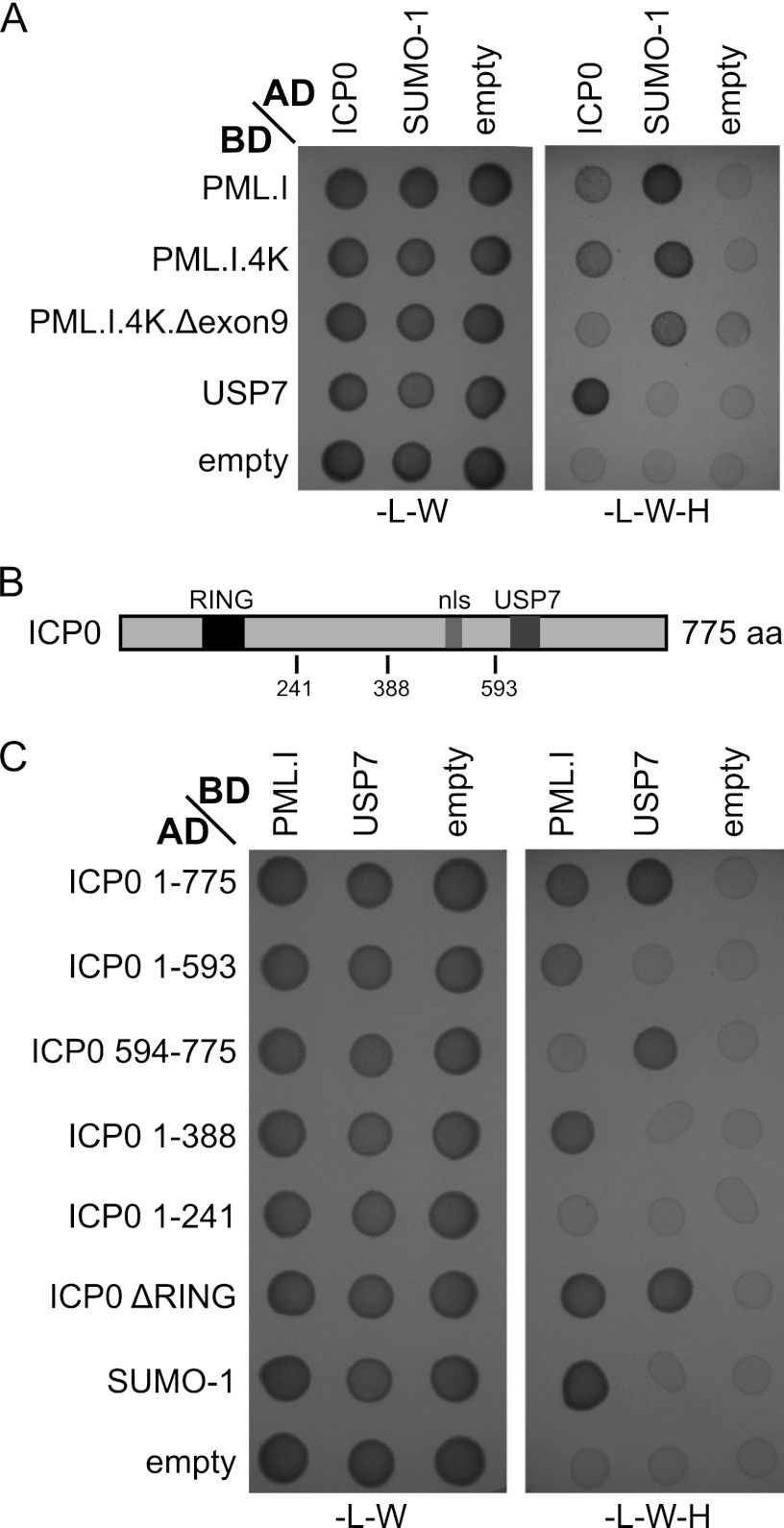
Consistent with the Co-IP results, Y2H assays demonstrated that ICP0 interacts with PML.I (Fig. 8A). We used USP7 and SUMO-1 as positive controls for interactions with ICP0 (24) and PML.I (4), respectively. SUMO-1 interacted with PML.I in a SIM-dependent manner and also with PML.I.4K and PML.I.4K.Δexon9 in this system. ICP0 did not interact with PML.I.4K.Δexon9, a finding consistent with the hypothesis that the PML.I-ICP0 interaction occurs in a PML.I exon 9-dependent manner (Fig. 8A).
Fig 8.
Y2H analysis of the ICP0-PML.I interaction. The same volume of each mated diploid culture was plated on media lacking either leucine and tryptophan (−L−W) (indicating the presence of both plasmids) or lacking leucine, tryptophan, and histidine (−L−W−H) (indicating positive interactions). The presence of 3AT (5 mM) decreases false-positive interactions. AD and BD indicate whether the proteins were linked to the GAL4 activation or DNA binding domains. SUMO-1 and USP7 are positive controls for PML.I and ICP0 interactions, respectively, and empty vectors provide the negative controls. Residual growth of the PML.I.4K.Δexon9 cells in the empty vector control was slightly higher than in the other instances. Growth in the presence of ICP0 was no greater than in the control. (B) A map of the locations of the RING finger (black box), nuclear localization sequence (nls, gray box), and USP7 binding domains (dark gray box) and other significant coordinates within ICP0. (C) Y2H analysis of the region of ICP0 that is required for interaction with PML.I. Experimental details were as described for panel A, and 3AT was used here at 1 mM.
The N-terminal half of ICP0 is required for interaction with PML.I.
We next investigated the region of ICP0 that is important for its interaction with PML.I. The C-terminal part of ICP0 is responsible for efficient ICP0 localization at ND10 (38). Surprisingly, the C-terminal third of ICP0 is not required for interaction with PML.I in the Y2H assay, since ICP0 mutant (residues 1 to 593) [ICP0(1-593)] still interacted with PML.I, whereas the C-terminal third itself (residues 594 to 775) did not (Fig. 8C). The Y2H assays also suggested that the ICP0 RING finger is unlikely to be involved in the interaction, which is consistent with the transfection assay data with RING finger mutant ICP0 (Fig. 6G), since ICP0(1-388) still interacted with PML.I, unlike ICP0(1-241) (Fig. 8C). Therefore, it appears that sequences in the region from residues 241 to 388 are required for ICP0 to interact with PML.I, assuming that ICP0 residues 1 to 241 are properly folded in yeast. This qualification is necessary because, although this segment of ICP0 has a functional interaction with UBE2D1 (UbcH5a) in vitro (7), longer versions of the protein are required for a detectable Y2H assay interaction (E. Vanni and C. Boutell, unpublished data). Further analysis of specific motifs within the 241-388 region was conducted using a variety of assays but, as discussed below, these were not informative.
Influence of PML.I on ICP0 localization.
Efficient ND10 localization of ICP0 requires its C-terminal third (38, 40). Given that the ICP0 regions responsible for PML.I interaction and localization at ND10 differ (Fig. 9A), it appears that interaction with PML.I does not have a dominant effect on ICP0 localization. To investigate this in more detail, we reanalyzed the localization of an ICP0 mutant (E52X) that lacks the C-terminal third of the protein (Δ593-775). As expected, E52X was mainly diffuse throughout the nucleus of HepaRG cells during infection, although there were minor accumulations in some ND10 at early times (data not shown). The accumulations at ND10 were more pronounced in the presence of MG132 (Fig. 9B, top row), but they were not observed in cells specifically depleted of PML.I (Fig. 9B, row 2; see Fig. 11 for characterization of these cells). In contrast, E52X was efficiently recruited into ND10 in cells expressing increased amounts of PML.I (Fig. 9B, row 3). In cells depleted of endogenous PML, the expression of PML.I, but not PML.II or PML.IV, also resulted in some accumulation of E52X at the PML foci (Fig. 9B, rows 4 to 6). In addition, PML.I coimmunoprecipitated with E52X (Fig. 10A) and was degraded by E52X as efficiently as by wt ICP0 (Fig. 10B), but E52X failed to degrade PML.II or PML.IV (Fig. 10C and D). Although we did not detect E52X in the reciprocal PML.I Co-IP (Fig. 10A), we suspect that this was because the Co-IP was always less efficient in this orientation (see Fig. 7A) and because of reduced levels of the mutant protein at the early time point of infection used. We conclude that ICP0 can localize to ND10 independently of its C-terminal third in a PML.I-dependent manner, but this is less distinct than with full-length ICP0 unless PML.I is overexpressed.
Fig 9.
PML.I influences ICP0 nuclear localization. (A) Locations of the regions responsible for PML.I interaction, ND10 localization, and the RING finger, nuclear localization sequence (nls), and USP7 binding domain within ICP0. (B) An ICP0 mutant lacking the C-terminal third of ICP0 (E52X) can be recruited to ND10 in a PML.I-dependent manner. Control HepaRG cells (shLuci), cells specifically depleted for PML.I (shPML.I), control cells expressing EYFP-PML.I (shLuci/EYFP-PML.I) and shPML cells expressing EYFP-PML.I, EYFP-PML.II, or EYFP-PML.IV were infected with HSV-1 mutant E52X at an MOI of 2 in the presence of MG132 and stained 4 h later for ICP0 and for endogenous PML when relevant.
PML.I depletion delays the degradation of other PML isoforms by ICP0.
To investigate whether PML.I specific targeting by ICP0 influences its effects on other PML isoforms, we constructed cells depleted specifically for PML.I using an shRNA targeting a sequence in PML.I exon 9 (shPML.I). This shRNA reduces the level of EYFP-PML.I expression in transduced cells (Fig. 11A), and the intensity of high-molecular-weight PML species when expressed in normal HepaRG cells (Fig. 11B). Moreover, the specificity of shPML.I targeting was confirmed, since none of the other PML isoforms tested was affected by its expression (data not shown). PML.I and PML.II are of similar size, and PML.I is more abundant than PML.II and the other isoforms (13). Therefore, the reduced intensity of the PML.I/II bands and their SUMO-modified species in shPML.I cells (Fig. 11B) is consistent with isoform-specific depletion of PML.I.
We reported previously that PML.I contributes to PML-mediated repression of ICP0-null mutant HSV-1 infection (15). Therefore, depletion of PML.I should increase the plaque formation efficiency of this mutant HSV-1. This hypothesis was tested using wt and ICP0-null mutant HSV-1 plaque assays in control, PML-depleted, and PML.I-depleted HepaRG cells (Fig. 11C). The plaque-forming efficiency of wt HSV-1 was similar in the three cell lines, whereas that of ICP0-null mutant HSV-1 was increased in both depleted cell lines (Fig. 11C). Although the effect of depleting PML.I alone was less than depletion of all PML isoforms, this would be expected because reconstitution of PML.I expression reversed only partially the improved plaque formation efficiency of the ICP0-null mutant in PML-depleted cells (15).
Depletion of PML.I did not affect the expression of ICP0, ICP4, or the typical early protein UL42 during wt HSV-1 infection (Fig. 11D and E), allowing direct comparison of the kinetics of PML isoform degradation by ICP0. At the MOIs used, all PML species in control cells were reduced by 3 h postinfection, and their levels continued to drop thereafter (Fig. 11D). In shPML.I cells, degradation of the remaining PML isoforms was less efficient than in the controls (Fig. 11E). Therefore, it appears that specific targeting of PML.I by ICP0 allows more efficient degradation of PML isoforms in general.
DISCUSSION
The core finding of the present study is that ICP0 interacts with PML.I, the most abundant of the PML isoforms, via isoform-specific sequences encoded by exon 9. This interaction enables SUMO-modification-independent targeting of PML.I by ICP0 and therefore represents a second mechanism, in addition to SUMO-dependent interactions (5), involved in ND10 disruption by ICP0. Given that the interaction occurs both in Co-IP and Y2H assays, is likely that the ICP0-PML.I interaction is direct.
ICP0 colocalization with PML.I is not dependent on the latter's SUMO modification sites, SIM or TRIM domain, although mutations in the coiled-coiled region of the TRIM reduce the degree of colocalization. The consistency of the colocalization, immunoprecipitation, Y2H, and degradation assays provides a solid case for the requirement for the PML.I isoform-specific sequences for the interaction. Little is known of the functional properties of this part of PML. It has been implicated in nucleolar targeting during stress, and there is a putative exonuclease III-like motif between residues 610 and 759, which overlaps exon 8a (12). The C-terminal region of PML.I also interacts with and recruits AML1b into ND10, and high-level expression of PML.I influences myeloid cell differentiation (45). Interestingly, it is the most highly conserved of the isoform-specific PML exons (13). Taken with our previous results on an antiviral role of PML.I (15), and the results using cells depleted of PML.I specifically (Fig. 11C), it appears that the ICP0-PML.I interaction reflects a countermeasure to PML-related antiviral restriction.
Definition of the ICP0 sequences that are involved in the interaction with PML.I is not straightforward. Y2H assays indicate that the ICP0 region responsible lies in the N-terminal half of the protein. It is likely that the motif involved is present in both HSV-1 and HSV-2, as EYFP-PML.I.C-term is recruited into ICP0 foci during infection by both viruses (data not shown). The most conserved motifs in this region of the protein are the RING finger (ICP0 HSV-1, positions 116 to 156), the SUMO interaction motif related sequence SLS-4 (ICP0 HSV-1, IVISDS, positions 364 to 369) (5), the phosphorylated regions Phos 1 (ICP0 HSV-1, S224, T226, T231, and T232) and Phos 2 (ICP0 HSV-1, S367, S369 and S373) (16), and the recently reported FHA domain interaction motif centered on residue T67 (ICP0 HSV-1, positions 58 to 70) (10). The RING finger itself can be excluded (Fig. 6G and 8C) and also the N-terminal 241 residues in the context of the Y2H assay (Fig. 8C). Of the remaining motifs, mutation of SLS-4 did not affect the interaction with PML.I in the Y2H assay, and the major phosphorylation motifs were not required for colocalization of ICP0 with PML.I.C-term (data not shown). Therefore, the precise motif of ICP0 required for its interaction with PML.I remains to be determined.
Our results demonstrate that PML can be targeted by ICP0 by two distinct mechanisms: (i) degradation of all SUMO-modified PML isoforms via a SUMO-dependent mechanism and (ii) through an interaction with PML.I that is likely to be direct. This poses the question of which, if either, is the dominant mechanism during the early stages of HSV-1 infection. It is possible that ICP0 interaction with PML.I, the most abundant isoform, confers sensitivity to all other SUMO-modified PML isoforms because of their heterodimerization with PML.I. This explanation is supported by the fact that the degradation of the other SUMO-modified PML isoforms is less efficient in the absence of PML.I (Fig. 11E). In addition, we observed that SUMO-modified Sp100 is degraded more slowly in the absence of PML.I (data not shown), suggesting further consequences of the ICP0 and PML.I interaction early after infection. On the other hand, the N-terminal two-thirds of ICP0 is able to interact with PML.I and to induce its degradation very efficiently and yet localizes to ND10 less efficiently, especially during a normal infection. Among the many motifs within the C-terminal third of ICP0 are three SIM-like sequences that contribute to the efficiency of ICP0-mediated degradation of SUMO-modified PML (5). Given that the SUMO-modified forms of PML are targeted by ICP0 more efficiently than the bulk of SUMO-modified proteins, we have suggested that substrate specific motifs within selected SUMO-modified proteins might confer increased sensitivity to ICP0 (5). The present study strongly suggests that PML.I provides just such an example, and therefore it becomes moot whether the PML.I isoform-specific or the SUMO-dependent mechanism is the more dominant; in practice they may well be acting in concert.
Our data demonstrate not only that ICP0 accumulates at existing foci containing PML.I but also that the location of PML.I is actively influenced by ICP0 (in the presence of MG132). For example, PML.I mutants with a diffuse nuclear expression pattern, the EYFP-PML.I-C-term fusion protein, PML.I.ΔCC, and PML.I.ΔRING, all relocalize into ICP0 foci (Fig. 3B and 6B). This property is conferred by the PML.I isoform-specific sequences since it did not occur with an EYFP fusion protein with the isoform-specific C-terminal portion of PML.II (EYFP-exon 7b [data not shown]). It is attractive to suggest that the portion of PML.I that is diffuse in the nucleus can be recruited to ICP0 foci whether or not they are at preexisting ND10. The notion that PML.I is able to respond to a stimulus by changing nuclear localization and then recruiting the other PML isoforms is supported by the observation that all PML isoforms accumulate in the nucleolus in response to genotoxic stress, but this behavior is PML.I dependent (12).
We provide here another example of the diverse means by which ICP0 substrate selection is mediated. E3 ubiquitin ligases have modular structures, with one part (in this case the RING [58]) required for catalytic activity and interaction with E2 ubiquitin-conjugating enzymes, while other domains interact either directly or via adaptor proteins with specific substrates. An emerging picture is that the many functions of ICP0 are mediated by the varied motifs throughout the protein that engage with other proteins. For example, ICP0 includes multiple SIM-like sequences that are required for its ability to induce the widespread proteasome-dependent degradation of SUMO-conjugated proteins, including SUMO-conjugated PML (5, 14). This provides a general mechanism for the targeting of several proteins, although preferential substrate selectivity occurs within this group of proteins, since SUMO-modified PML is degraded more rapidly than SUMO-modified proteins in general (5). Distinct SUMO-independent substrate-specific interaction domains within ICP0 have now been identified for USP7 (9), RNF8 (10, 37), SIAH-1 (43), and now PML.I. Whether the targeting of other proteins that are degraded in cells expressing ICP0, such as CENP-C (20) and DNA-PK (46), is dependent on substrate-specific binding motifs that are yet to be defined, or indirectly through interactions with adaptor proteins, or as a secondary consequence of ICP0 activity on other substrates remains to be determined.
ACKNOWLEDGMENTS
This study was supported by the Medical Research Council. M.G. was supported by an EC FP6 Marie Curie Fellowship (PIEF-GA-2009-251948).
We thank Keith Leppard for cDNAs encoding PML isoforms, Philippe Gripon for HepaRG cells, Didier Trono for pCMV.DR8.91, and Tom Gilbey (Beatson Institute for Cancer Research, Glasgow, United Kingdom) for FACS enrichment of EYFP-positive cells. We are particularly grateful to Steven McFarlane for Y2H technical advice. Chris Boutell and members of the R.D.E. laboratory were very helpful in discussions throughout the course of this work.
Footnotes
Published ahead of print 8 August 2012
REFERENCES
- 1. Bernardi R, Pandolfi PP. 2003. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 22:9048–9057 [DOI] [PubMed] [Google Scholar]
- 2. Bernardi R, Pandolfi PP. 2007. Structure, dynamics, and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell. Biol. 8:1006–1016 [DOI] [PubMed] [Google Scholar]
- 3. Bischof O, et al. 2002. Deconstructing PML-induced premature senescence. EMBO J. 21:3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971–982 [PubMed] [Google Scholar]
- 5. Boutell C, et al. 2011. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 7:e1002245 doi:10.1371/journal.ppat.1002245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boutell C, Orr A, Everett RD. 2003. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 77:8686–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boutell C, Sadis S, Everett RD. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brand P, Lenser T, Hemmerich P. 2001. Assembly dynamics of PML nuclear bodies in living cells. PMC Biophys. 3:3 doi:10.1186/1757-5036-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Canning M, Boutell C, Parkinson J, Everett RD. 2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 279:38160–38168 [DOI] [PubMed] [Google Scholar]
- 10. Chaurushiya MS, et al. 2012. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: viral mimicry of a cellular phosphorylation mark targets the RNF8 FHA domain. Mol. Cell 46:79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chelbi-Alix MK, de The H. 1999. Herpesvirus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941 [DOI] [PubMed] [Google Scholar]
- 12. Condemine W, Takahashi Y, Le Bras M, de The H. 2007. A nucleolar targeting signal in PML-I addresses PML to nucleolar caps in stressed or senescent cells. J. Cell Sci. 120:3219–3227 [DOI] [PubMed] [Google Scholar]
- 13. Condemine W, et al. 2006. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 66:6192–6198 [DOI] [PubMed] [Google Scholar]
- 14. Cuchet-Lourenco D, et al. 2011. SUMO pathway-dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 7:e1002123 doi:10.1371/journal.ppat.1002123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuchet D, et al. 2011. PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J. Cell Sci. 124:280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davido DJ, von Zagorski WF, Lane WS, Schaffer PA. 2005. Phosphorylation site mutations affect herpes simplex virus type 1 ICP0 function. J. Virol. 79:1232–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dellaire G, Bazett-Jones DP. 2004. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26:963–977 [DOI] [PubMed] [Google Scholar]
- 18. Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24:1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Everett RD, Chelbi-Alix MK. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830 [DOI] [PubMed] [Google Scholar]
- 20. Everett RD, Earnshaw WC, Findlay J, Lomonte P. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Everett RD, et al. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Everett RD, Lomonte P, Sternsdorf T, van Driel R, Orr A. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112:4581–4588 [DOI] [PubMed] [Google Scholar]
- 23. Everett RD, Meredith M, Orr A. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73:417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Everett RD, et al. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Everett RD, Murray J, Orr A, Preston CM. 2007. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J. Virol. 81:10991–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Everett RD, Orr A, Elliott M. 1991. High-level expression and purification of herpes simplex virus type 1 immediate-early polypeptide Vmw110. Nucleic Acids Res. 19:6155–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Everett RD, Orr A, Preston CM. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Everett RD, et al. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu L, et al. 2005. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol. Biol. Cell 16:4905–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gripon P, et al. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. U. S. A. 99:15655–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishov AM, et al. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jamieson DR, Robinson LH, Daksis JI, Nicholl MJ, Preston CM. 1995. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus type 1 Vmw65 mutants. J. Gen. Virol. 76:1417–1431 [DOI] [PubMed] [Google Scholar]
- 34. Jensen K, Shiels C, Freemont PS. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223–7233 [DOI] [PubMed] [Google Scholar]
- 35. Lafarga M, et al. 2002. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell 13:2771–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lallemand-Breitenbach V, et al. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lilley CE, et al. 2010. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 29:943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maul GG, Everett RD. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223–1233 [DOI] [PubMed] [Google Scholar]
- 39. Maul GG, Yu E, Ishov AM, Epstein AL. 1995. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell Biochem. 59:498–513 [DOI] [PubMed] [Google Scholar]
- 40. Meredith M, Orr A, Elliott M, Everett R. 1995. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology 209:174–187 [DOI] [PubMed] [Google Scholar]
- 41. Muller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nabetani A, Yokoyama O, Ishikawa F. 2004. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J. Biol. Chem. 279:25849–25857 [DOI] [PubMed] [Google Scholar]
- 43. Nagel CH, et al. Herpes simplex virus immediate-early protein ICP0 is targeted by SIAH-1 for proteasomal degradation. J. Virol. 85:7644–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Negorev D, Maul GG. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234–7242 [DOI] [PubMed] [Google Scholar]
- 45. Nguyen LA, et al. 2005. Physical and functional link of the leukemia-associated factors AML1 and PML. Blood 105:292–300 [DOI] [PubMed] [Google Scholar]
- 46. Parkinson J, Lees-Miller SP, Everett RD. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reineke EL, Kao HY. 2009. PML: an emerging tumor suppressor and a target with therapeutic potential. Cancer Ther. 7:219–226 [PMC free article] [PubMed] [Google Scholar]
- 48. Salomoni P, Pandolfi PP. 2002. The role of PML in tumor suppression. Cell 108:165–170 [DOI] [PubMed] [Google Scholar]
- 49. Schenk P, Ludwig H. 1988. The 65 K DNA binding protein appears early in HSV-1 replication. Arch. Virol. 102:119–123 [DOI] [PubMed] [Google Scholar]
- 50. Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. 2006. The mechanisms of PML-nuclear body formation. Mol. Cell 24:331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Showalter SD, Zweig M, Hampar B. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sternsdorf T, Guldner HH, Szostecki C, Grotzinger T, Will H. 1995. Two nuclear dot-associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand. J. Immunol. 42:257–268 [DOI] [PubMed] [Google Scholar]
- 53. Stow ND, Stow EC. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J. Gen. Virol. 67:2571–2585 [DOI] [PubMed] [Google Scholar]
- 54. Stuurman N, et al. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773–784 [DOI] [PubMed] [Google Scholar]
- 55. Takahashi Y, Lallemand-Breitenbach V, Zhu J, de The H. 2004. PML nuclear bodies and apoptosis. Oncogene 23:2819–2824 [DOI] [PubMed] [Google Scholar]
- 56. Tavalai N, Stamminger T. 2009. Interplay between herpesvirus infection and host defense by PML nuclear bodies. Viruses 1:1240–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tavalai N, Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 1783:2207–2221 [DOI] [PubMed] [Google Scholar]
- 58. Vanni E, Gatherer D, Tong L, Everett RD, Boutell C. 2012. Functional characterization of residues required for the herpes simplex virus type-1 E3 ubiquitin ligase ICP0 to interact with the cellular E2 ubiquitin-conjugating enzyme UBE2D1 (UbcH5a). J. Virol. 86:6323–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yao F, Schaffer PA. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhong S, Salomoni P, Pandolfi PP. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85–E90 [DOI] [PubMed] [Google Scholar]