Abstract
The activation of the interferon (IFN) system, which is triggered largely by the recognition of viral nucleic acids, is one of the most important host defense reactions against viral infections. Although influenza A and B viruses, which both have segmented negative-strand RNA genomes, share major structural similarities, they have evolutionarily diverged, with total genetic incompatibility. Here we compare antiviral-inducing mechanisms during infections with type A and B influenza viruses in human dendritic cells. We observed that IFN responses are induced significantly faster in cells infected with influenza B virus than in cells infected with type A influenza virus and that the early induction of antiviral gene expression is mediated by the activation of the transcription factor IFN regulatory factor 3 (IRF3). We further demonstrate that influenza A virus infection activates IFN responses only after viral RNA (vRNA) synthesis, whereas influenza B virus induces IFN responses even if its infectivity is destroyed by UV treatment. Thus, initial viral transcription, replication, and viral protein synthesis are dispensable for influenza B virus-induced antiviral responses. Moreover, vRNA molecules from both type A and B viruses are equally potent activators of IFN induction, but incoming influenza B virus structures are recognized directly in the cytosol, while influenza A virus is able to evade early recognition. Collectively, our data provide new evidence of a novel antiviral evasion strategy for influenza A virus without a contribution of the viral NS1 protein, and this opens up new insights into different influenza virus pathogenicities.
INTRODUCTION
Both influenza A and influenza B viruses are respiratory tract pathogens, which may cause widespread annual epidemics in humans. Type A viruses infect several host species from birds to mammals, and simultaneous infection with different strains can occur. This enables the exchange of gene material between different influenza A virus strains, giving rise to new reassortant viruses with pandemic potential. Instead, influenza B viruses infect mainly humans. Like influenza A viruses, type B viruses undergo significant antigenic drift, but the evolutionary rate is lower than that observed for type A viruses (27). Although influenza B virus infection can occasionally be severe and even fatal, the clinical picture of the disease is usually milder than that caused by type A viruses (26). Influenza A and B viruses do not naturally mix with each other, even though they share many similar properties (Fig. 1A). The genomes of influenza A and B viruses are composed of eight single-stranded RNA (ssRNA) gene segments that encode 13 or 11 different viral proteins, respectively. The viral structures are also similar: the ssRNA molecules are encapsidated by nucleoproteins (NPs), and the panhandle/corkscrew structure formed by the noncoding sequences at the ssRNA termini associates with the viral polymerase complexes (4, 28). The eight viral ribonucleoprotein (vRNP) complexes are packed underneath the viral matrix protein shell and the host-derived lipid envelope in which viral surface glycoproteins are embedded. Ten out of the 14 viral proteins show functional similarities between these two influenza virus types. Influenza B virus has an additional ion channel NB protein (2), and some type A virus strains express PB1-F2, PB1-N40, and the most novel PA-X protein, which all contribute to their virulence (3, 14, 42). Regardless of the similarities, these two types of influenza viruses clearly form distinct evolutionary branches, which leads us to presume that they may have evolved different strategies to combat host antiviral actions.
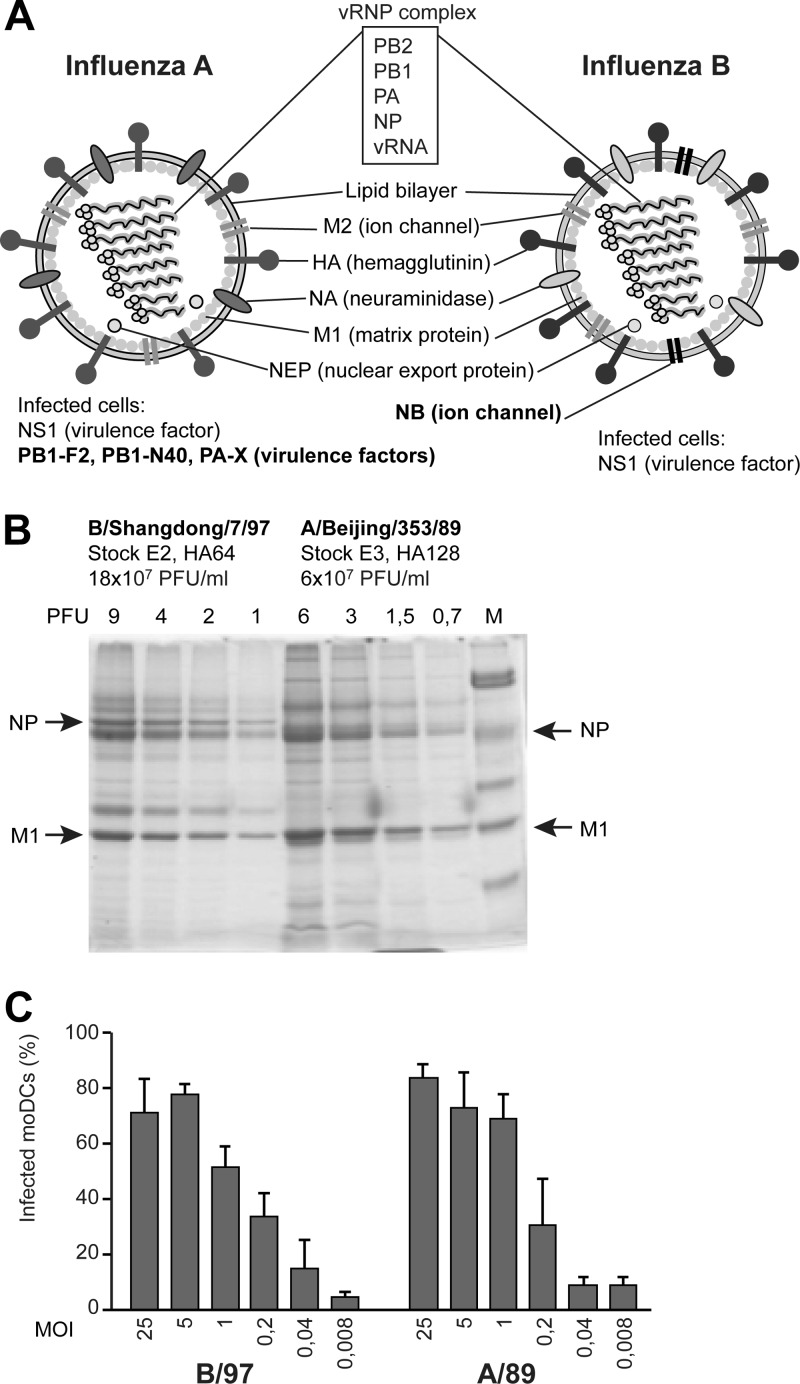
Fig 1.
Characterization of influenza A and B virus stocks. (A) Schematic representation of influenza A and B viruses and their proteins. (B) Influenza A/Beijing/353/89 and B/Shangdong/7/97 viruses were propagated in fertilized chicken eggs, and the infectivity of the virus stocks was determined by a plaque-forming assay. In addition, equal aliquots of virus stocks were purified through a sucrose cushion by ultracentrifugation, and viral proteins were then separated by electrophoresis and stained. (C) moDCs were infected with influenza B/Shangdong/7/97 and A/Beijing/353/89 viruses at MOIs of 25 to 0.008 for 18 h, and cells were collected for flow cytometry analysis. The proportion of infected cells was determined by staining with antibodies against influenza B or influenza A H3N2 virus glycoproteins. The results are from samples from four different blood donors analyzed separately and are presented as mean values with standard deviations.
The interferon (IFN) system is one of the major host defense mechanisms in the battle against viral infections. IFNs participate in the restriction of virus replication and the initiation of immune responses (36). The activation of the innate immune system during viral infection is triggered mainly by the recognition of viral nucleic acids. However, some viral protein structures have also been reported to induce an antiviral response, such as inflammasome activation by the influenza A virus M2 protein (13), the induction of the double-stranded RNA (dsRNA)-activated protein kinase (PKR) by influenza B virus vRNP (5), and IFN induction by measles virus nucleocapsids (39). Genomic viral RNAs (vRNAs), as well as RNA replication and transcription products, are potent activators of innate immune responses. The classical receptors responsible for vRNA recognition are RIG-I for the 5′-triphosphorylated RNA structures, Toll-like receptor 3 (TLR3) for the dsRNA, and TLR7/8 for the ssRNA molecules (43). Different types of vRNA molecules are produced during influenza virus infection, and total cellular RNA from virus-infected cells has been shown to be a potent inducer of IFN responses when transfected into cells (16, 32). Synthetic ssRNA molecules produced by using T7 polymerase can serve as efficient RIG-I agonists due to their 5′-triphosphate structure, and as we have recently shown, rather short (100-nucleotide [nt]) RNA molecules seem to be the most potent inducers of IFN expression (15). However, in influenza A virus infection, the 5′-triphosphorylated full-length vRNAs are the main ligands responsible for RIG-I activation (34). While RIG-I is considered to be the most important vRNA sensor, another RIG-I-like receptor, MDA5, has been reported to recognize longer and higher-order RNA structures (16, 33). Where the RIG-I pathway is dominant in influenza A virus infection, influenza B virus is also recognized by MDA5, suggesting that different RNA sensors play differential roles in the recognition of various types of viruses (17, 21). Indeed, there seems to be a great variation in the natural RNA structures produced in RNA virus-infected cells, which inevitably may lead to the activation of a wide range of antiviral signaling pathways.
Viruses have evolved highly sophisticated mechanisms to antagonize the activation of the host immune system. Influenza virus nonstructural protein 1 (NS1) interferes with multiple antiviral responses (11). The inhibition of RIG-I-mediated IFN induction is conserved among all influenza viruses. The RNA-binding properties of NS1 are essential for the inhibition of RIG-I-mediated IFN induction by type A viruses (38), whereas type B and C influenza virus NS1 proteins seem to be able to antagonize IFN induction independently of RNA-binding properties (6, 31). Unlike the influenza A virus NS1 protein, influenza B virus NS1 lacks the ability to interfere with the 3′-end processing and nuclear export of host cell pre-mRNAs, including IFN mRNAs (41). While the influenza A virus NS1 protein possesses functions associated with multiple cellular signaling pathways (11), less is known about the functions of the influenza B virus NS1 protein.
In the present study, we compare the mechanisms of IFN induction in influenza A and B virus-infected cells. By analyzing IFN gene expression and by monitoring the activation of transcription factors, we demonstrate that type B influenza virus induces antiviral activation clearly faster than type A virus. Furthermore, influenza B virus-induced interferon regulatory factor 3 (IRF3)-mediated antiviral gene expression takes place before viral primary transcription, replication, or protein synthesis, because even UV-irradiated influenza B viruses retained their IFN-inducing activity. Instead, infection with influenza A virus required viral RNA production for the activation of host innate immune responses, suggesting that incoming influenza A viruses are able to efficiently enter the cells without being recognized by the host.
MATERIALS AND METHODS
Cell cultures.
Monocyte-derived dendritic cells (moDCs) were differentiated from peripheral blood monocytes from healthy blood donors (Finnish Red Cross Blood Transfusion Service, Helsinki, Finland) according to a standard procedure (35). Peripheral blood mononuclear cells were fractionated by Ficoll-Paque (Pharmacia Biotech) gradient centrifugation followed by centrifugation on a Percoll gradient (Amersham Biosciences) and lymphocyte depletion with anti-CD3 and anti-CD19 magnetic beads (Dynal). moDCs were differentiated by culturing monocytes in RPMI medium (Sigma-Aldrich) supplemented with 0.6 μg/ml penicillin, 60 μg/ml streptomycin, 2 mM l-glutamine, and 20 mM HEPES in the presence of 10% fetal calf serum (FCS) (Integro), 10 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) (BioSource), and 20 ng/ml interleukin-4 (IL-4) (R&D Systems) for 1 week. In each experiment, cells from 3 to 4 donors were used separately for virus infection or RNA transfection experiments.
A549 human lung epithelial cells (ATCC CCL185), HEK293 human embryonic kidney cells (ATCC CLR1573), and Madin-Darby canine kidney (MDCK) cells (ATCC CCL34) were maintained with continuous growth in Eagle minimal essential medium (Eagle-MEM) (Sigma-Aldrich) supplemented with antibiotics, l-glutamine, HEPES, and 10% FCS. All cells were maintained at 37°C in 5% CO2.
Viruses and infections.
Human influenza A viruses A/Beijing/353/89 (H3N2) (A/89) and A/New Caledonia/20/99 (H1N1) (A/99), influenza B viruses B/Shangdong/7/97 (B/97) and B/Jiangsu/10/03 (B/03), and Sendai virus (strain Cantell) were grown from a 10−5 dilution of stock virus in allantoic cavities of 11-day-old embryonated chicken eggs at +36°C for 3 days. The recombinant influenza viruses A/Udorn/307/72 (H3N2) (obtained from R. Lamb) and B/Yamagata/16/88 (obtained from P. Palese) were propagated in MDCK cells with serum-free medium containing 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (TPCK-trypsin) (Thermo Scientific), as described elsewhere previously (10, 25), and virus stocks were produced in fertilized eggs. Virus titers were determined by a plaque assay on MDCK cells. Serially diluted viruses were absorbed onto confluent MDCK cells for 1 h at +37°C. After the cells were washed, they were covered with an agarose overlay (1% agarose and 1 μg/ml TPCK-trypsin in Eagle-MEM). Within 3 to 6 days, the plaques were counted to obtain the concentration of infective viruses as PFU/ml. The multiplicity of infection (MOI) is given according to the titers determined in MDCK cells.
Cells were infected with influenza viruses for different times, as indicated in the figure legends. UV irradiation of the stock viruses was performed with 600 mJ of UV light before the viruses were added to the cells. The efficiency of the UV irradiation was verified by the PFU titers of the UV-treated stocks, which were less than 102 PFU/ml. Viruses were fixed in the presence of 3% paraformaldehyde (PFA) (Sigma-Aldrich) with continuous rotation for 15 min, and the reaction was stopped by the addition of FCS to the mixture. Where indicated, protein synthesis was inhibited by the addition of 10 μg/ml cycloheximide (CHX) (Sigma-Aldrich) to cells 30 min before stimulation. As nonviral control stimuli, lipopolysaccharide (LPS) (Escherichia coli serotype O111:B4; Sigma-Aldrich) and the TLR7/8 ligand R848 (Alexis Biochemicals) were used at concentrations of 100 ng/ml and 10 μM, respectively.
Viral RNA preparations and RNA transfections.
Influenza B/Shangdong/7/97 and A/Beijing/353/89 viruses were purified and further concentrated by sedimentation though a 30% sucrose cushion by ultracentrifugation at 26,000 rpm in an SW32 rotor for 90 min (Beckman). vRNA was isolated from the virus pellet by use of the QIAmp viral RNA extraction kit (Qiagen). Synthetic vRNA was produced by amplifying the full-length gene segments from the plasmids of the recombinant virus systems by PCR. The reverse primer contained the T7 promoter sequence, allowing the subsequent production of influenza virus-specific ssRNAs by T7 RNA polymerase. The primer sequences will be provided upon request. The molecules were purified by high-performance liquid chromatography (HPLC), as previously described (15). Total cellular RNA from influenza virus-infected moDCs was isolated by use of the RNeasy minikit (Qiagen).
Where indicated, aliquots of vRNA were treated with FastAP thermosensitive alkaline phosphatase (Fermentas) to digest the 5′-triphosphate groups of RNA. The reactions were carried out under standard reaction conditions at +37°C for 10 min, followed by enzyme inactivation at +75°C for 10 min. After the treatment, an RNase inhibitor (Applied Biosystems) was added, and modified RNAs were purified by use of the QIAmp viral RNA extraction kit (Qiagen).
moDCs were transfected with total cellular (100 ng/ml) or viral (200 ng/ml) RNAs with Lipofectamine 2000 (Invitrogen). In RNA transfection mixtures, poly(I:C) (10 μg/ml; Invivogen) was used as a positive control, and poly(A), the carrier RNA in the QIAmp viral RNA, served as a negative control.
Luciferase reporter assay.
HEK293 cells were transfected with an IFN-λ1 promoter-reporter construct (30) and expression plasmids for influenza virus NS1 with a Flag tag using TransIT-LT1 transfection reagent (Mirus Bio). After the transfection of the plasmids for 4 h, promoter activation was induced by Sendai virus. Luciferase activity was measured 16 h after virus infection by using a Dual Glo kit (Promega), and the results were normalized to the intensity of Renilla luciferase (Promega).
Quantitative PCR (qPCR).
The infected or transfected cells from different blood donors were harvested and pooled, and total cellular RNA was isolated by using the RNeasy minikit (Qiagen) including DNase digestion (RNase-free DNase kit; Qiagen). One microgram of total cellular RNA was transcribed to cDNA by using a TaqMan reverse transcriptase kit (Applied Biosystems) with random hexamers as primers or with oligo(dT) as the primer when distinguishing the viral mRNA levels. The cDNA was amplified by PCR using TaqMan Universal PCR Mastermix and gene expression assays (Applied Biosystems). The influenza A virus-specific NP primer-probe pair used was forward primer 5′-CCATAAGGACCAGGAGTGGA, reverse primer 5′-CCCTCCGTATTTCCAGTGAA, and probe Cy5-5′-CAGGCCAAATCAGTGTGCAACCTAC-black hole quencher (BHQ); the NS primer-probe pair used was forward primer 5′-TGAAAGCGAATTTCAGTGTGAT, reverse primer 5′-CTGGAAAAGAAGGCAATGGT, and probe Cy5-5′-CTAAGGGCTTTCACCGAAGAGGG-BHQ; the influenza B virus NP gene-specific oligonucleotides used were forward primer 5′-AACCAGTGGGACAACCAGAC, reverse primer 5′-TTCCGACATCAGCTTCACTG, and probe Cy5-5′-AATCATCAGACCAGCAACCCTTGC-BHQ; and the NS gene-specific oligonucleotides used were forward primer 5′-AAGCAGGAATTCTGGAGTGC, reverse primer 5′-CGGTCTTGACCAGGGTAGTC, and probe Cy5-5′-AAAGGCTTTCATGGCAAAGAGCCC. The data were normalized to 18S rRNA gene levels with a TaqMan endogenous control kit (Applied Biosystems) or to β-actin mRNA levels (gene expression assay; Applied Biosystems). Gene expression data are presented as the relative gene expression levels in relation to those of the unstimulated samples in order to calculate the fold change achieved by the stimulation.
ELISA.
The secreted levels of IFN-λ1 and IFN-λ2 from cell culture supernatants were analyzed by using a Duoset enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems). ELISAs to detect human IFN-β and IFN-α were supplied by PBL Biomedical Laboratories. The cytokine levels from cell culture supernatants of different blood donors were analyzed separately.
Immunoblotting.
For protein analyses, the cells from different blood donors were pooled, and whole-cell lysates were prepared in passive lysis buffer from the Dual Luciferase assay kit (Promega) containing 10 mM Na3PO4. Equal amounts of proteins were separated on SDS-PAGE gels and transferred onto Hybond-P polyvinylidene difluoride (PVDF) membranes (Amersham Biosciences). The membranes were blocked with 5% milk protein. The rabbit antibodies against influenza B virus NP and NS1 were prepared in a fashion similar to that described previously for IRF3 and influenza A virus NP and NS1 proteins (22, 29). Antibodies against the Flag tag were obtained from Sigma-Aldrich. Staining was done in blocking buffer at room temperature (RT) for 1 h. Antibodies for phosphorylated IRF3 (P-IRF3), P-p38, p38, phosphorylated Jun N-terminal protein kinase (JNK), and IκBα were obtained from Cell Signaling Technology, and staining was done in a buffer containing 5% bovine serum albumin (BSA) at +4°C overnight. Horseradish peroxidase (HRP)-conjugated antibodies (Dako) were used for secondary staining at RT for 1 h. Protein bands were visualized on HyperMax films by using an ECL Plus system (GE Healthcare).
Flow cytometry.
For infectivity analyses, infected moDCs were collected from each blood donor separately, cells were fixed with 3% PFA at RT for 20 min, and after washing, the cells were suspended in 1% BSA in phosphate-buffered saline (PBS). Cells were stained with rabbit antibodies specific for influenza B or influenza A H3N2 virus glycoproteins and with phycoerythrin (PE)-conjugated secondary antibodies (Invitrogen) at RT for 1 h (29). Flow cytometry was performed on a FACSCanto II cytometer and analyzed by using FACS Diva software (BD Biosciences).
RESULTS
Influenza A and B viruses induce IFN gene expression with different kinetics in human dendritic cells.
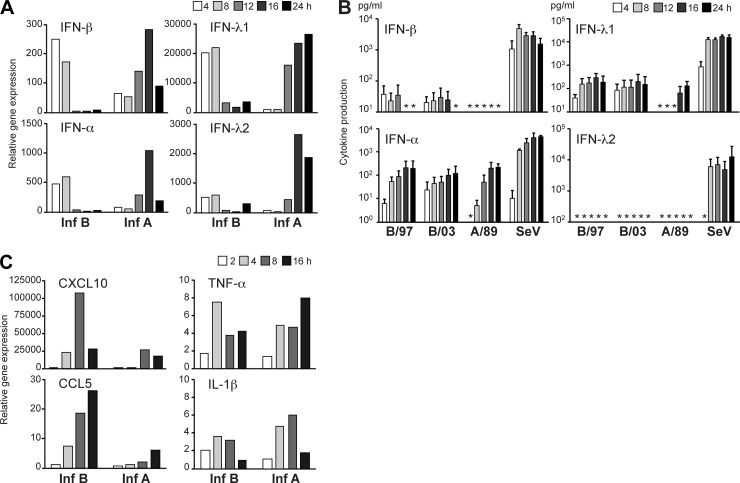
To compare the IFN-inducing abilities of influenza A and B viruses in human monocyte-derived dendritic cells (moDCs), we chose influenza virus strains A/Beijing/353/89 and B/Shangdong/7/97 as model viruses for this study. The virus stocks were propagated in fertilized eggs, and the infectivities of the viruses were measured to be at the same level, i.e., 6 × 107 PFU/ml for A/89 and 18 × 107 PFU/ml for B/97. The viral stock proteins were analyzed by electrophoresis to estimate the relative amounts of total viral proteins and their relation to infectivities, which were roughly equal between the viruses (Fig. 1B). Moreover, the infectivities of these viruses in moDCs were analyzed by detecting the proportion of cells infected by different virus amounts by flow cytometry, and a multiplicity of infection (MOI) of 5 showed a suboptimal infectivity rate for both viruses (Fig. 1C). For analyzing the cytokine responses, moDCs were infected with the model influenza A and B viruses at an MOI of 5, and the expression levels of IFN genes, i.e., IFN-α and IFN-β, representing type I IFNs, and IFN-λ1 and IFN-λ2, representing type III IFNs, were monitored over 24 h by qPCR. For influenza A virus infection, IFN induction was seen at late time points, while type B virus triggered IFN responses much faster, showing completely opposite IFN gene expression kinetics (Fig. 2A). At the protein level, the difference was not as clear but was still obvious (Fig. 2B). Influenza B viruses of both the Victoria (B/Shangdong/7/97) and Yamagata (B/Jiangsu/10/03) lineages were able to induce significant and rapid IFN production, while the responses detected in influenza A virus-infected cells were always weaker. Differences in CXCL10 and CCL5 chemokine and tumor necrosis factor alpha (TNF-α) and IL-1β proinflammatory cytokine gene expression profiles were not as obvious as those seen for IFN genes (Fig. 2C). The CXCL10 and CCL5 genes are regulated by IRF3, as are the IFN genes, whereas TNF-α and IL-1β are transcriptionally regulated by, e.g., mitogen-activated protein kinase (MAPK) pathways and NF-κB.
Fig 2.
Cytokine responses during influenza A and B virus infection. (A) Human primary monocyte-derived dendritic cells (moDCs) were infected with influenza virus strains A/Beijing/353/89 (Inf A) and B/Shangdong/7/97 (Inf B) at an MOI of 5 for the different time points indicated. Total cellular RNA was isolated, and IFN gene expression was measured by qPCR. The results depicted are presented as fold induction compared to the mock sample and are from a representative experiment out of three. (B) moDCs were infected with the viruses as described above and for comparison with another influenza B virus strain, B/Jiangsu/10/03, and Sendai virus (SeV) at an MOI of 5, and cytokine production was analyzed in the culture supernatants collected at various time points after infection. The results are presented as means with standard deviations from samples from four different blood donors. Asterisks indicate undetected values. (C) Influenza A and B virus-infected moDCs were analyzed for the expression of different cytokine/chemokine genes at the indicated time points by using qPCR. The data are representative of two independent experiments and indicate virus-induced relative expression over the mock sample.
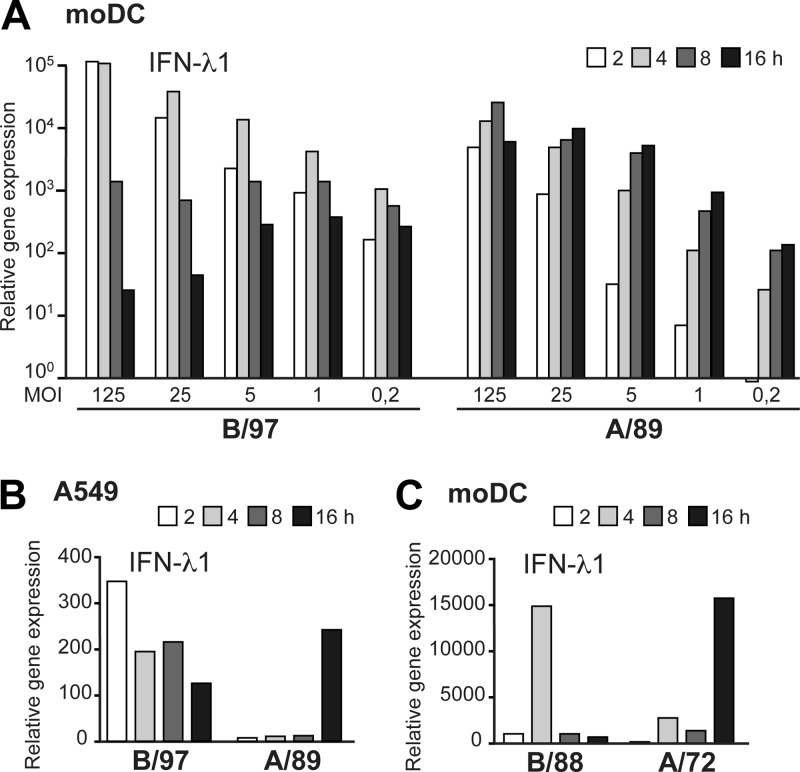
To further verify this phenomenon, we monitored IFN-λ1 gene expression in moDCs infected with the model influenza A and B virus strains at different MOI values during a 16-h follow-up period. The peak mRNA levels induced by influenza B virus were observed at 2 to 4 h postinfection, while in the case of influenza A virus, regardless of the virus dose, the highest IFN-λ1 mRNA levels were detected clearly later, at 8 to 16 h after infection (Fig. 3A). Moreover, the same temporal difference in the kinetics of IFN-λ1 gene expression between influenza A and B viruses was observed in human lung epithelial A549 cells (Fig. 3B) and in moDCs infected with other influenza virus strains, B/Yamagata/16/88 and A/Udorn/307/72 (Fig. 3C).
Fig 3.
Infection with influenza B virus induces faster IFN expression than infection with type A virus. (A) IFN-λ1 gene expression was analyzed in moDCs infected with influenza A/Beijing/353/89 and B/Shangdong/7/97 viruses at serial dilutions (MOIs of 125 to 0.2) for 16 h. (B) IFN-λ1 gene expression at different time points in A549 cells infected with the above-described viruses at an MOI of 2. (C) The same experiment was repeated with recombinant virus strains A/Udorn/307/72 and B/Yamagata/16/88 in moDCs at an MOI of 5, and IFN-λ1 gene expression was analyzed. All these experiments were repeated twice.
Early IFN gene expression in influenza virus infection correlates with phosphorylation of IRF3.
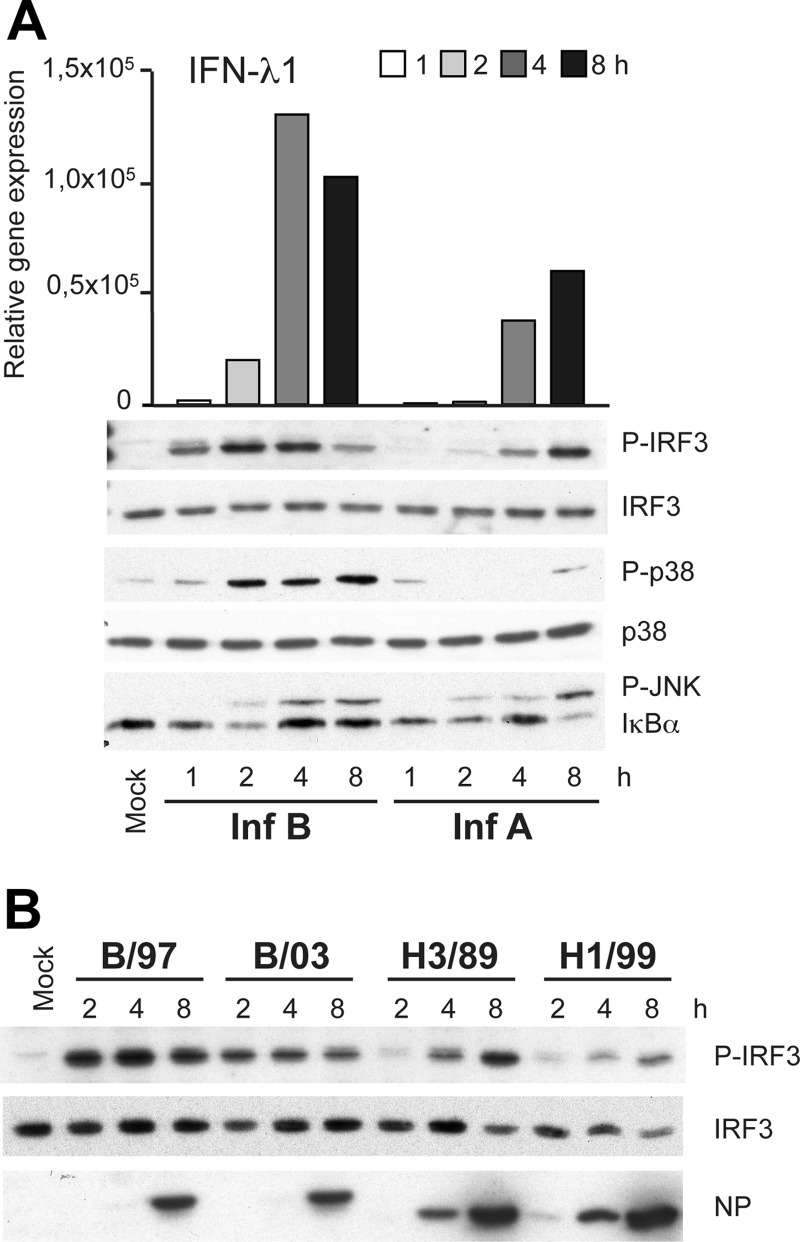
Antiviral IFN genes are induced mainly by the activation of IRFs, of which IRF3 plays the most crucial role (30). In addition to IRFs, NF-κB and different MAPK-regulated pathways are also important for the induction of IFNs and other innate immune genes (12). In order to examine the regulation involved in early influenza B virus-induced IFN gene expression, we analyzed the activation of different transcription factor pathways in moDCs during influenza A and B virus infections. Consistent with the kinetics of IFN-λ1 mRNA induction, IRF3 phosphorylation was observed at much earlier times in cells infected with influenza B virus than in those infected with influenza A virus (Fig. 4A). The phosphorylation of p38, indicating the activation of the MAPK-regulated pathway, was also strong and took place at relatively early time points in influenza B virus-infected cells. The activation of the NF-κB pathway was analyzed by monitoring the degradation of IκBα that was observed 1 to 2 h after infection; however, significant differences between the two viruses were not seen until the 8-h time point, when newly synthesized IκBα was degraded again by influenza A virus infection, correlating with the more sustained type A virus-induced IFN gene expression (Fig. 4A). In general, the transcriptional activation of the IFN response was observed at a later stage in influenza A virus infection than in influenza B virus infection, although the activated pathways were similar between these viruses. To study whether the temporal difference between influenza A and B virus-induced IRF3 phosphorylation is a general feature of these virus types, we analyzed Victoria (B/97) and Yamagata (B/03) lineage influenza B and H1N1 (A/99) and H3N2 (A/89) influenza A virus-stimulated IRF3 phosphorylation in human moDCs. Again, the difference between influenza A and B viruses was obvious, although some variation in the intensities of IRF3 phosphorylation was observed among the virus strains analyzed (Fig. 4B).
Fig 4.
The early influenza B virus-induced IFN response is mediated by IRF3 activation. (A) moDCs were infected with influenza A/Beijing/353/89 and B/Shangdong/7/97 viruses (MOI of 5), cells were collected at different time points after infection, and total cellular RNA or whole-cell lysates were prepared. Virus infection-induced IFN-λ1 gene expression (top) is shown in relation to the immunoblot analysis of phosphorylated forms of IRF3, p38, and JNK or IκBα. Total cellular IRF3 and p38 protein levels were analyzed to control for equal loading. The data are representative of three independent experiments. (B) Whole-cell lysates from moDCs infected with influenza B/Shandong/7/97, B/Jiangsu/10/03, A/Beijing/353/89 (H3N2), and A/New Caledonia/20/99 (H1N1) viruses at an MOI of 5 were collected at the 2-, 4-, and 8-h time points. Cellular proteins were separated on 10% SDS-PAGE gels, and immunoblot analysis of P-IRF3, IRF3, and influenza A and B virus nucleoproteins (NP) was carried out.
Early IFN gene expression is efficiently activated by influenza B virus rather than inhibited by influenza A virus.
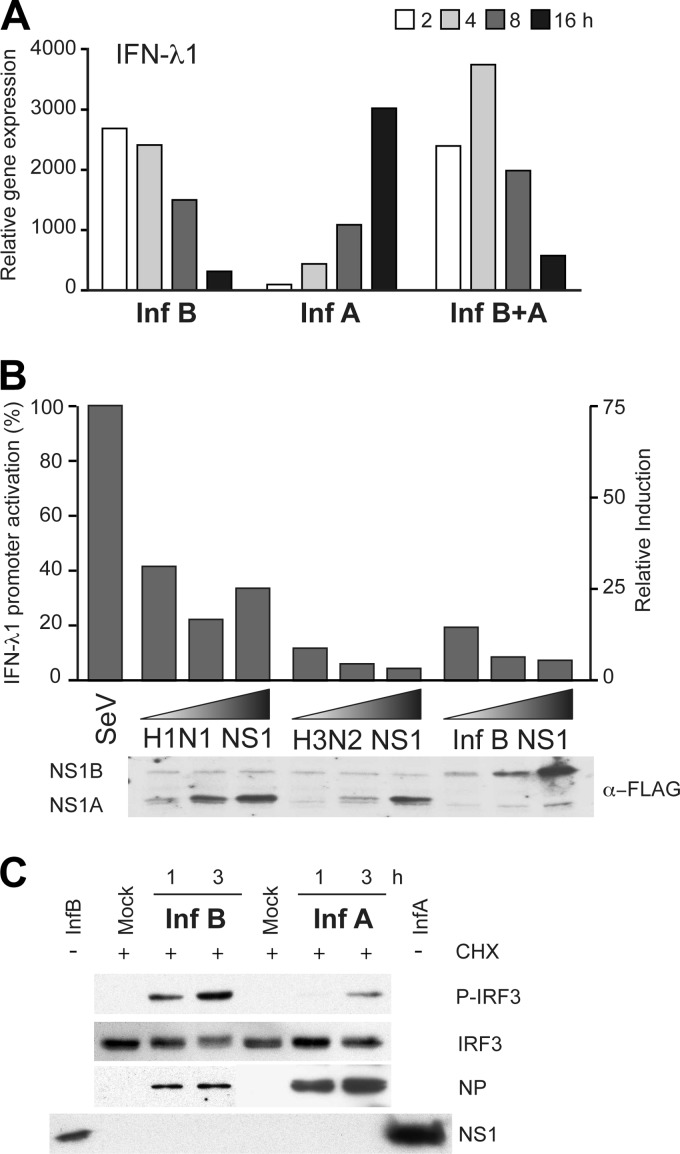
To study whether influenza B virus efficiently activates IFN gene expression, rather than influenza A virus inhibiting the response, we carried out a coinfection experiment with influenza A and B viruses. Since the kinetics of virus-induced IFN gene expression appeared to be independent of the virus dose (Fig. 3A), a moderate MOI value of 5 was selected for both viruses for this experiment. We observed that in coinfection, the kinetics and magnitude of IFN-λ1 gene expression followed the pattern for influenza B virus infection (Fig. 5A). This indicates that there is no influenza A virus protein or function that is able to block early influenza B virus-induced IFN-λ1 mRNA expression, but rather, influenza A virus appears to avoid the early activation of the IFN response. To confirm the IFN-antagonistic activity of influenza A and B virus NS1 proteins, a simplified luciferase reporter-based assay was set up. NS1 proteins of influenza A H1N1 and H3N2 and influenza B viruses were expressed together with an IFN-λ1 promoter-reporter construct in HEK293 cells, followed by infection with Sendai virus to obtain efficient IFN promoter activation. As shown in Fig. 5B, all tested influenza virus NS1 proteins were able to inhibit Sendai virus-induced IFN-λ1 promoter activation. Furthermore, to rule out the role of the NS1 protein as a factor controlling early IFN induction, we infected moDCs with influenza A and B viruses at high MOIs in the presence of the protein synthesis inhibitor cycloheximide (CHX). When NS1 protein expression was prevented by CHX treatment, we observed that influenza B virus infection still efficiently induced IRF3 phosphorylation, while induction by influenza A virus remained at a low level (Fig. 5C). These results show that influenza B virus-induced IRF3 phosphorylation is independent of the synthesis of any viral or host proteins.
Fig 5.
Role of the NS1 protein in influenza virus-induced IFN-λ1 gene expression and IRF3 phosphorylation. (A) For setting up a coinfection assay, moDCs were infected at an MOI of 5 with influenza B/Shangdong/7/97 or A/Beijing/535/89 virus or both viruses for 16 h. Cells were collected, total cellular RNA was isolated, and IFN-λ1 gene expression was measured by qPCR. Results are representative of two experiments. (B) HEK293 cells transfected with the IFN-λ1 promoter-luciferase reporter construct with increasing amounts of Flag-tagged NS1 expression plasmids for A/WSN/33 (H1N1), A/Udorn/72 (H3N2), and B/Shangdong/97 virus NS1 genes. Four hours after transfection, the cells were infected with Sendai virus (SeV) for 16 h. The luciferase activity in the whole-cell extracts was measured, and NS1 protein expression was visualized by using anti-Flag antibodies. Luciferase results are presented as the percentage of activation in relation to Sendai virus-induced promoter activation (100%) and as relative induction. The experiment was repeated three times. (C) After moDCs were treated with cycloheximide (CHX) at 10 μg/ml for 30 min, the cells were infected with influenza A (A/89) and B (B/97) viruses at a high MOI of 30. After 1 and 3 h postinfection, the cells were collected, and immunoblot analysis for P-IRF3, IRF3, and influenza A and B virus NP and NS1 proteins was carried out. Cell lysates from influenza A or B virus-infected samples (16 h) without CHX treatment were included in the analysis to control for NS1 staining.
Influenza B virus-induced early IFN gene expression occurs independently of viral RNA replication.
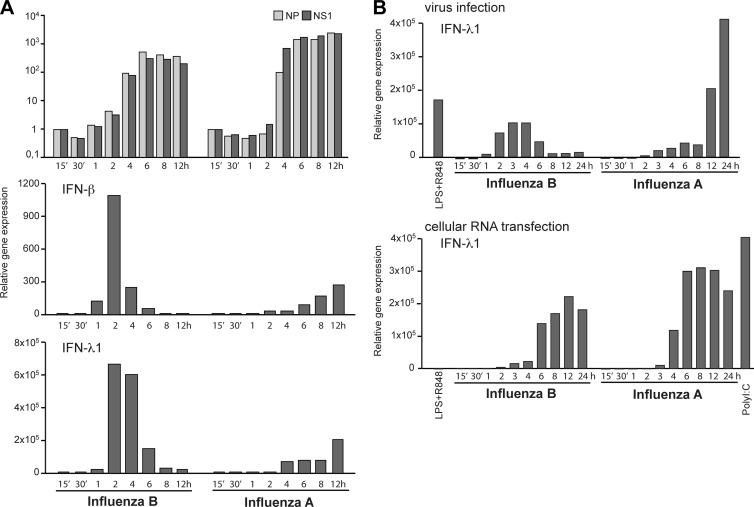
Given that viral genomic RNA is the most potent ligand for RIG-I during RNA virus infection (34), we evaluated the replication kinetics of influenza A and B viruses in moDCs in relation to their ability to induce IFN gene expression. For total cellular RNA isolation, virus-infected cells were collected at different time points after infection, and virus replication was analyzed by measuring the relative amounts of viral NP and NS RNAs (both vRNA and mRNA) by qPCR. Although the overall replication curves for influenza A and B viruses were almost identical, IFN-β and IFN-λ1 mRNA expression levels displayed a clear temporal difference (Fig. 6A). Moreover, equal amounts of total cellular RNAs isolated at various time points after infection were transfected into a fresh set of moDCs in order to analyze the IFN-inducing activity of vRNA products during infection (Fig. 6B). Surprisingly, IFN-λ1 gene expression kinetics were similar with both viruses, and the early IFN responses induced by type B virus infection were lost (Fig. 6B), indicating that the early IFN response induced by influenza B virus infection was triggered by early events other than the replicating vRNA. Instead, influenza A virus seems to induce IFN gene expression in relation to its replication and expression of vRNA molecules.
Fig 6.
Viral replication is not required for the early influenza B virus-induced IFN response. (A) moDCs were infected with influenza A/89 and B/97 viruses at an MOI of 1, and the RNA samples were isolated from cells collected at different time points from 15 min to 12 h after infection. The expression levels of viral NP- and NS1-specific RNAs were analyzed by qPCR, and the expression level is shown in relation to that of the 15-min sample, representing increases in the levels of viral RNAs during the time course of infection. IFN-β and IFN-λ1 gene expression levels determined by qPCR analysis are presented as fold induction over the values for the mock samples. The results are representative of three individual experiments. (B) Influenza virus strains A/89 and B/97 were used to infect moDCs at an MOI of 5 for 24 h, cells were collected at different time points postinfection, and total cellular RNA was isolated. The gene expression of IFN-λ1 was analyzed by qPCR, and the gene expression level is shown in relation to values for unstimulated samples. Costimulation with LPS and R848 was used as a positive control for IFN induction. The total cellular RNA from this experiment was transfected into a fresh set of moDCs. After 4 h of transfection, the cells were harvested for the isolation of total cellular RNA and further analysis of IFN-λ1 gene expression by qPCR. The experiment was repeated twice.
Both genomic and synthetic viral RNAs of influenza A and B viruses induce IFN responses equally well.
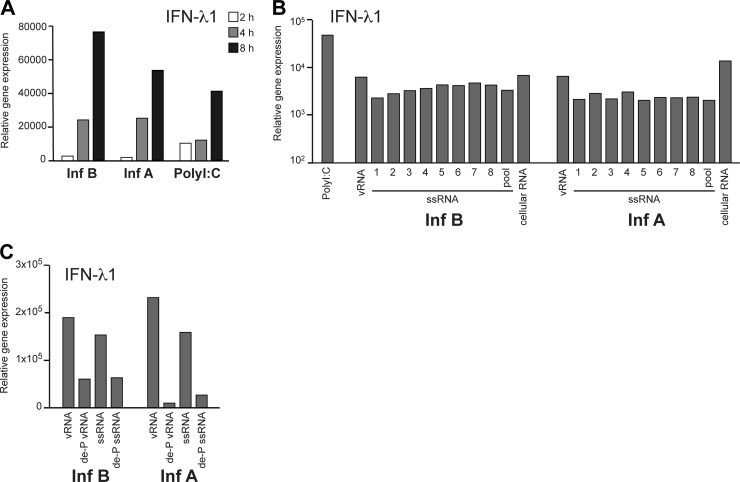
As it seems that influenza B virus induces IFN responses independently of viral replication or viral protein synthesis, we postulated that the influenza A and B virus genomic vRNAs might be different in their abilities to induce IFN gene expression. To test this experimentally, we isolated vRNA directly from purified virus stocks. By transfecting equal amounts of vRNA from influenza A and B viruses into moDCs, we observed that IFN-λ1 gene expression was induced with comparable kinetics that were similar to those induced by poly(I:C) (Fig. 7A). Next, we synthesized influenza virus-specific ssRNAs separately for each viral gene segment in vitro by using T7 RNA polymerase. Using these RNA molecules, we observed that there were no sequence-specific differences in the IFN induction abilities of influenza A and B virus-derived RNAs (Fig. 7B). To rule out the possibility that the eight gene segments as a mixture could form some secondary structures that would be more prone to triggering antiviral signaling, we also compared the synthetic ssRNA segments in combinations of all eight segments. However, no differences between the homologous or heterologous ssRNA mixtures were seen.
Fig 7.
Purified influenza virus RNA-triggered IFN induction is similar between influenza types. (A) Viral RNA (vRNA) was isolated from purified A/89 and B/97 viruses and transfected into moDCs. Poly(I:C) was used as a control. At 2, 4, and 8 h after transfection, cells were collected, cellular RNA was isolated, and IFN-λ1 gene expression was analyzed by qPCR. (B) Synthetic ssRNAs specific for each of the eight full-length viral gene segments of influenza A (A/Udorn/307/72) and B (B/Yamagata/16/88) viruses were produced in vitro by the use of T7 RNA polymerase. In vitro-produced single ssRNA molecules, their 8 segment pools, virus-isolated vRNAs (as described above for panel A), and total cellular RNAs from infected cells were transfected into moDCs, and IFN induction was analyzed after 4 h by qPCR. Poly(I:C) served as a positive control. (C) Virus particle-derived genomic vRNAs (as described above for panel A) and synthetic ssRNA pools (as described above for panel B) were enzymatically dephosphorylated (de-P). Parallel reactions with and without the enzyme were performed, and the RNAs were transfected into moDCs, followed by the isolation of cells at 4 h after transfection and analysis of IFN-λ1 gene expression by qPCR. All experiments were repeated twice.
The 5′-triphosphate of ssRNA is an essential structural element that regulates the activation of the RIG-I pathway and subsequent IRF3 phosphorylation (15, 34). To elucidate whether influenza A and B virus genomic vRNAs have additional IFN-inducing elements apart from 5′-triphosphate groups, we transfected dephosphorylated vRNAs, isolated from the viral stocks or in vitro-synthesized ssRNA pools, into moDCs and analyzed their abilities to induce IFN gene expression. We observed that both influenza A and B virus vRNAs were sensitive to dephosphorylation in their ability to induce IFN gene expression (Fig. 7C).
Early influenza B virus-induced IFN responses are independent of vRNA transcription.
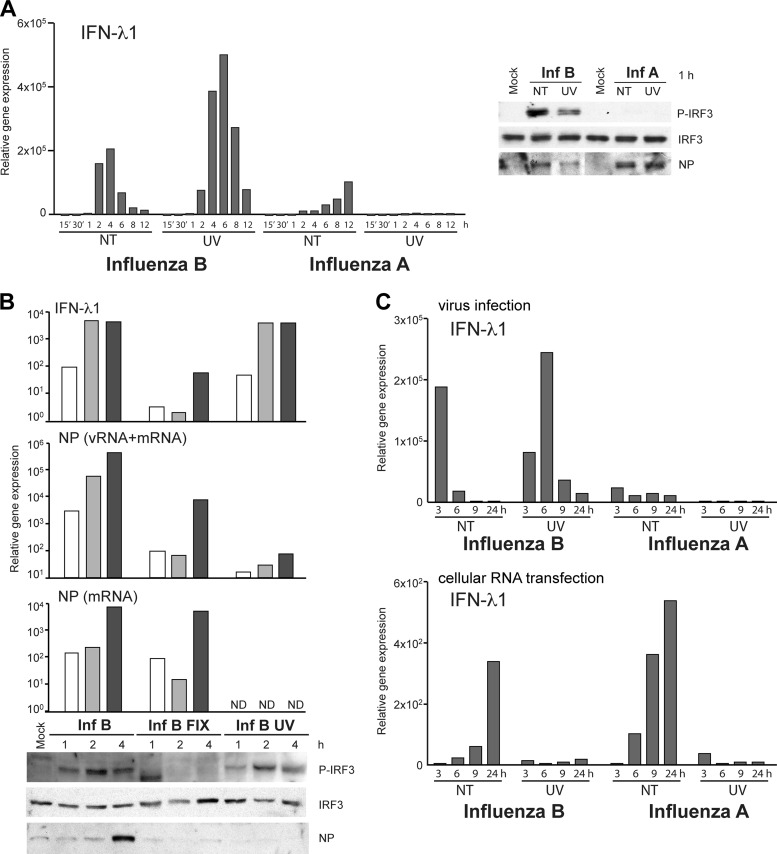
Incoming influenza virus particles contain vRNPs in which genomic vRNAs are covered by NPs and viral polymerase complexes. vRNPs need to be transported into the nucleus, where primary transcription by vRNP-associated viral polymerases takes place. The initiation of viral protein synthesis and vRNA replication can subsequently occur. To explore the role of the early steps of virus entry in relation to influenza B virus-induced IFN gene expression, we cross-linked vRNP complexes in viral particles by UV irradiation. moDCs infected with UV-irradiated influenza A or B viruses were analyzed for the activation of IFN-λ1 gene expression. Interestingly, UV-irradiated influenza B viruses were still able to induce significant IFN gene activation, which correlated with the phosphorylation of IRF3 (Fig. 8B), whereas type A virus-infected cells lacked early IRF3 phosphorylation regardless of UV treatment (Fig. 8A). As shown by UV treatment, influenza B virus-induced IFN gene expression took place regardless of viral transcriptional activation (Fig. 8B). However, when influenza B virus particles were fixed with paraformaldehyde (PFA), which prevents the endosomal uncoating and cytosolic release of the vRNPs, the IFN-λ1 gene expression level was dramatically decreased (Fig. 8B). Still, separately expressed influenza B virus proteins from the plasmids of the recombinant virus system did not induce the phosphorylation of IRF3 in transfected HEK293 cells (data not shown). To study the effect of UV treatment on influenza virus RNA-induced IFN-λ1 gene expression, we infected moDCs with intact and UV-irradiated stock viruses, isolated total cellular RNA from the cells, and used this RNA to further stimulate noninfected moDCs. Even though UV-irradiated influenza B virus was able to efficiently induce IFN-λ1 expression, the total cellular RNA isolated from these cells was incapable of inducing an IFN response in RNA-transfected moDCs (Fig. 8C).
Fig 8.
Viral transcription is dispensable for influenza B virus-triggered IFN induction. (A) UV-irradiated or nontreated (NT) infectious influenza A/89 and B/97 viruses were used to infect moDCs at an MOI of 5. Infected cells were collected at different times, total cellular RNA was isolated, and IFN-λ1 gene expression was measured by qPCR. Data are representative of four independent experiments. For the analysis of IRF3 phosphorylation, cells were infected with high doses of UV-treated or live viruses (MOI of 30). Cells were collected, and cellular proteins were separated on 10% SDS-PAGE gels and immunoblotted with anti-P-IRF3, anti-IRF3, and influenza A and B virus NP-specific antibodies. (B) Influenza B/97 viruses were fixed with 3% paraformaldehyde and irradiated by UV light or were left untreated prior to the infection of moDCs with a dilution that corresponds to an MOI of 5. The gene expression levels of IFN-λ1 and viral NP vRNA or mRNA were measured from total cellular RNA and are shown as fold increases over the value for the mock sample. Phosphorylated and total IRF3 and viral NP levels in whole-cell lysates were visualized by immunoblotting. ND, not detected. (C) Cellular RNA from a virus infection experiment similar to that described above for panel A was analyzed for IFN-λ1 gene expression by qPCR (top) and further transfected into a fresh set of moDCs, and RNA-induced IFN-λ1 gene expression at 4 h posttransfection was analyzed by qPCR (bottom).
DISCUSSION
In virus infection, the interaction between the virus and the host determines the outcome of the disease. Both influenza A and B viruses cause large epidemics and contribute to a significant economical burden. However, influenza B virus epidemics seem to be somewhat milder, with fewer patients needing hospital care. In order to understand the mechanisms of pathogenesis of these different influenza virus types, we analyzed the activation of the antiviral responses of human dendritic cells to influenza A or B virus infection. Interestingly, influenza B virus activated IRF3-dependent IFN responses much faster than type A virus did. This could contribute to a faster and more effective restriction of influenza B virus replication and, eventually, a faster clearance of the infection.
The current impression is that influenza A virus evades host defense systems due to the functions of the NS1 protein. Via its dsRNA-binding abilities, NS1 interferes with the activation of antiviral signaling by RIG-I and inhibits the functions of the antiviral proteins PKR and the 2′-5′ oligoadenylate synthetase (OAS) (23, 24, 38). In addition, by direct interactions, NS1 inhibits the functions of PKR and TRIM25, which is needed for the activation of RIG-I (9). Importantly, unlike type B virus NS1, type A virus NS1 is able to block host antiviral gene expression by interfering with the processing of host pre-mRNA molecules (41). Consistent with our present and previous results (29), Kim and coworkers suggested previously that the initial antiviral response was suppressed in influenza A virus infection by the NS1-mediated inhibition of mRNA production (18). Moreover, it was shown that both type A and type B viruses were able to induce IRF3-mediated gene activation and that this transcriptional activation was independent of viral or host protein synthesis (18). In the present study, we have analyzed these mechanisms in more detail and have taken a clear step forward in an understanding of how the two viruses differ in their abilities to induce the host antiviral response. Indeed, at very early time points of infection, before any viral replication, type B virus induced the activation of transcription factors responsible for initial antiviral gene expression significantly earlier than did type A influenza virus, indicating an additional, NS1-independent counteraction of the antiviral defense by type A virus.
Host cells recognize influenza viruses via their RNA molecules, and the RIG-I-mediated cascade, activated by 5′-triphosphate-bearing RNAs, is considered to be the most crucial antiviral signaling pathway involved in the clearance of influenza virus infection. Recently reported evidence suggests that influenza virus infection, viral genomic RNA, viral replication RNA intermediates, reconstituted vRNPs, and in vitro-produced 5′-phosphorylated RNA molecules can efficiently activate innate immunity via RIG-I (1, 15, 16, 32, 34). In order to reveal the mechanisms behind the differential induction of IFN responses between influenza A and B virus infections, we demonstrate here that vRNAs isolated from type A or B viruses as well as total cellular RNA isolated from influenza A or B virus-infected cells were equally effective in activating IFN gene expression. This finding indicates that potentially different kinds of secondary structures between influenza A and B virus RNA molecules or different types of replication intermediates show no differences in their innate immune activation potentials. However, by cross-linking the vRNP complexes inside the viral particles that destroy vRNA synthesis, we were still able to see early IFN responses in influenza B virus infection, which is consistent with data described previously (18). In influenza A virus infection, instead, early IFN induction was missing, and IRF3 phosphorylation and subsequent IFN gene expression took place only after efficient virus replication. Interestingly, transcriptionally inactive vRNP complexes from vesicular stomatitis virus (VSV) are able to activate IRF3-mediated IFN responses (40). Mechanistically, the VSV data are comparable to those found in the present work, clearly indicating that early influenza B virus-induced antiviral responses are triggered by the incoming viral complexes rather than by any newly produced vRNA molecules or viral protein synthesis.
Influenza virus uses membrane fusion to penetrate the host cell, and clathrin-mediated entry has been identified as one of the major routes of entry (20). Dynamin-independent micropinocytosis was recently observed to be an alternative route for influenza A virus entry (7). After internalization, influenza A viruses are transported into late endosomes, where viral hemagglutinin (HA) is subjected to conformational changes due to the low-pH environment (pH ∼5), which triggers viral and endosomal membrane fusion, leading to the release of vRNPs into the cell cytosol. However, some other viruses, like Semliki forest virus and VSV, introduce their genomes into the cytoplasm in early endosomes at higher luminal pH values of ∼6 (20). By using immunofluorescence methods, it was visualized previously that influenza A viruses are already localized in early endosomes at 10 min postinfection and then reach late endosomes within 40 min after infection (37). Moreover, it was shown that the trafficking of these virus-containing endocytic vesicles was microtubule dependent and that late endosomes ended up in the perinuclear region (19). However, there are no detailed reports on how the entry of influenza B viruses takes place. VSV, which penetrates though early endosomes, has been demonstrated to activate IRF3-dependent antiviral activation directly by its vRNP complexes (40). Consequently, one could assume that, similarly to VSV, influenza B virus could also use the early-endosomal entry route and release its vRNPs into the pericytoplasmic region and thus be more potent for early recognition by the host. Furthermore, the pathogenicity of influenza A H5N1 virus strains was recently shown to correlate with the optimal pH-dependent activation of the viral HA protein for membrane fusion (8). These distinct cellular control points suggest possible therapeutic applications that are not restricted to antiviral control points in general but are more strain specific between influenza viruses.
We propose that some of the early virus-host cell interactions are quite different between influenza A and B viruses. While influenza B virus activates IRF3-mediated antiviral responses by the incoming virus particles, the early steps of influenza A virus infection appear to remain undetected by the host cell. This indicates that even if influenza A and B viruses are structurally and functionally similar, there are still fundamental differences in the ways in which the host recognizes these pathogens.
ACKNOWLEDGMENTS
We thank Robert Lamb and Peter Palese for kindly providing the recombinant influenza virus systems. We are grateful to Hanna Valtonen, Sari Maljanen, Riitta Tarkiainen, and Marjukka Soininen for their skillful technical assistance.
The Academy of Finland supported this study with grants 128080 (to P.Ö.) and 115279, 252252, and 256159 (to I.J.) from the Research Council for Health and grants 250113 and 256069 (to M.M.P.) from the Research Council for Biosciences and Environment. This study was also funded by the Sigrid Jusélius Foundation (to M.M.P.).
Footnotes
Published ahead of print 1 August 2012
REFERENCES
- 1. Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Betakova T, Nermut MV, Hay AJ. 1996. The NB protein is an integral component of the membrane of influenza B virus. J. Gen. Virol. 77(Pt 11):2689–2694 [DOI] [PubMed] [Google Scholar]
- 3. Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3:1414–1421 doi:10.1371/journal.ppat.0030141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crow M, Deng T, Addley M, Brownlee GG. 2004. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 78:6263–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dauber B, et al. 2009. Influenza B virus ribonucleoprotein is a potent activator of the antiviral kinase PKR. PLoS Pathog. 5:e1000473 doi:10.1371/journal.ppat.1000473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dauber B, Schneider J, Wolff T. 2006. Double-stranded RNA binding of influenza B virus nonstructural NS1 protein inhibits protein kinase R but is not essential to antagonize production of alpha/beta interferon. J. Virol. 80:11667–11677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Vries E, et al. 2011. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 7:e1001329 doi:10.1371/journal.ppat.1001329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DuBois RM, et al. 2011. Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity. PLoS Pathog. 7:e1002398 doi:10.1371/journal.ppat.1002398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gack MU, et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hai R, et al. 2008. Influenza B virus NS1-truncated mutants: live-attenuated vaccine approach. J. Virol. 82:10580–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 12. Hiscott J. 2007. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18:483–490 [DOI] [PubMed] [Google Scholar]
- 13. Ichinohe T, Pang IK, Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 11:404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jagger BW, et al. 28 June 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science doi:10.1126/science.1222213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang M, et al. 2011. Innate immune responses in human monocyte-derived dendritic cells are highly dependent on the size and the 5′ phosphorylation of RNA molecules. J. Immunol. 187:1713–1721 [DOI] [PubMed] [Google Scholar]
- 16. Kato H, et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kato H, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 18. Kim MJ, Latham AG, Krug RM. 2002. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc. Natl. Acad. Sci. U. S. A. 99:10096–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 100:9280–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lakadamyali M, Rust MJ, Zhuang X. 2004. Endocytosis of influenza viruses. Microbes Infect. 6:929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loo YM, et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melen K, et al. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 81:5995–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Min JY, Krug RM. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 103:7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Min JY, Li S, Sen GC, Krug RM. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236–243 [DOI] [PubMed] [Google Scholar]
- 25. Neumann G, et al. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nicholson KG, Wood JM, Zambon M. 2003. Influenza. Lancet 362:1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nobusawa E, Sato K. 2006. Comparison of the mutation rates of human influenza A and B viruses. J. Virol. 80:3675–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noda T, Kawaoka Y. 2010. Structure of influenza virus ribonucleoprotein complexes and their packaging into virions. Rev. Med. Virol. 20:380–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Osterlund P, et al. 2005. Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J. Virol. 79:9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. 2007. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J. Immunol. 179:3434–3442 [DOI] [PubMed] [Google Scholar]
- 31. Pachler K, Vlasak R. 2011. Influenza C virus NS1 protein counteracts RIG-I-mediated IFN signalling. Virol. J. 8:48 doi:10.1186/1743-422X-8-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pichlmair A, et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001 [DOI] [PubMed] [Google Scholar]
- 33. Pichlmair A, et al. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rehwinkel J, et al. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408 [DOI] [PubMed] [Google Scholar]
- 35. Sallusto F, Lanzavecchia A. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179:1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Samuel CE. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sieczkarski SB, Whittaker GR. 2003. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4:333–343 [DOI] [PubMed] [Google Scholar]
- 38. Talon J, et al. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989–7996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. tenOever BR, Servant MJ, Grandvaux N, Lin R, Hiscott J. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. tenOever BR, et al. 2004. Activation of TBK1 and IKKvarepsilon kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 78:10636–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang W, Krug RM. 1996. The RNA-binding and effector domains of the viral NS1 protein are conserved to different extents among influenza A and B viruses. Virology 223:41–50 [DOI] [PubMed] [Google Scholar]
- 42. Wise HM, et al. 2009. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 83:8021–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoneyama M, Fujita T. 2010. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 20:4–22 [DOI] [PubMed] [Google Scholar]