Abstract
The catalytic subunit of herpes simplex virus 1 DNA polymerase (HSV-1 Pol) has been extensively studied; however, its full complement of functional domains has yet to be characterized. A crystal structure has revealed a previously uncharacterized pre-NH2-terminal domain (residues 1 to 140) within HSV-1 Pol. Due to the conservation of the pre-NH2-terminal domain within the herpesvirus Pol family and its location in the crystal structure, we hypothesized that this domain provides an important function during viral replication in the infected cell distinct from 5′-3′ polymerase activity. We identified three pre-NH2-terminal Pol mutants that exhibited 5′-3′ polymerase activity indistinguishable from that of wild-type Pol in vitro: deletion mutants PolΔN43 and PolΔN52 that lack the extreme N-terminal 42 and 51 residues, respectively, and mutant PolA6, in which a conserved motif at residues 44 to 49 was replaced with alanines. We constructed the corresponding pol mutant viruses and found that the polΔN43 mutant displayed replication kinetics similar to those of wild-type virus, while polΔN52 and polA6 mutant virus infection resulted in an 8-fold defect in viral yield compared to that achieved with wild type and their respective rescued derivative viruses. Additionally, both polΔN52 and polA6 viruses exhibited defects in viral DNA synthesis that correlated with the observed reduction in viral yield. These results strongly indicate that the conserved motif within the pre-NH2-terminal domain is important for viral DNA synthesis and production of infectious virus and indicate a functional role for this domain.
INTRODUCTION
The efficiency of viral replication and spread of infectious progeny virus is dependent upon the expeditious and faithful replication of the parental genome. Herpes simplex virus 1 (HSV-1) encodes seven proteins that are essential for viral DNA synthesis: an origin binding protein (UL9), DNA polymerase holoenzyme (catalytic subunit UL30 [Pol] and processivity factor UL42), single-stranded DNA binding protein (SSB; ICP8), and the helicase-primase complex (UL52, UL5, and UL8) (2, 34). The exact mechanisms by which these proteins act in concert to initiate and efficiently replicate the HSV-1 genome in the infected cell are poorly understood. HSV-1 Pol is the central enzyme for synthesis of viral DNA and is a target for antiviral drugs; however, the efficacy of these treatments is limited, especially for immunocompromised patients with drug-resistant infections (8). Despite vigorous investigation into HSV-1 Pol function, all of the activities mediated by this enzyme have yet to be exhaustively characterized. Elucidation of conserved viral replication processes may identify factors that could serve as a target for new antiviral therapies.
HSV-1 Pol is a member of DNA polymerase family Pol B, which includes eukaryotic polymerases α, δ, and ε, in addition to viral and bacteriophage polymerases that are responsible for genomic DNA replication (15, 23, 35). The palm, fingers, thumb, and 3′-5′ exonuclease domains of HSV-1 Pol bear significant sequence and structural homology to corresponding domains in other Pol B enzymes and are the best-characterized regions of the protein (19, 25, 35, 46). Accordingly, HSV-1 Pol exhibits 5′-3′ polymerase and 3′-5′ exonuclease activities that are characteristic functions of the replicative Pol B family (20, 26, 35). Polymerase activity is essential for the production of infectious progeny virus (1, 13), while the 3′-5′ exonuclease activity is not required for viral replication but is important for replication fidelity during viral DNA synthesis (16, 17). The extreme C terminus of HSV-1 Pol is crucial for an interaction with processivity factor UL42 that is necessary for long-chain DNA synthesis and indispensable for viral replication (11). Most recently, HSV-1 Pol was found to possess apurinic/apyrimidinic and 5′-deoxyribose phosphate lyase activities consistent with base excision repair processes, which are typically functions of the repair polymerase family X (3, 35). Although the active site has yet to be mapped, DNA lyase activity has been localized to a 63-kDa C-terminal fragment of HSV-1 Pol (3). Unlike the C-terminal half of HSV-1 Pol, the N-terminal half has yet to be functionally characterized (25). Thus, further investigation of this region may elucidate novel activities and better characterize functions of HSV-1 Pol.
A crystal structure of HSV-1 Pol revealed a two-domain architecture of the N-terminal portion of the enzyme (25). One of the domains (NH2-terminal domain) contains three structural motifs that closely resemble NH2-terminal domain structures found in family B polymerases (25, 33, 39, 44, 45). The second domain, at the extreme N terminus of HSV-1 Pol, which was structurally distinct from the NH2-terminal domain, was dubbed the pre-NH2-terminal domain by Liu et al. (25). The pre-NH2-terminal domain is comprised of the first 140 residues of HSV-1 Pol, of which only residues 59 to 140 are visible in the published structure (25). Although the pre-NH2-terminal domain is well conserved among the members of the herpesvirus Pol family, a structural equivalent has yet to be identified in other published polymerase structures (12, 25, 30). The extreme N-terminal 42 residues, which are not present in the published crystal structure, are conserved in HSV-1 and -2 (25). Interestingly, a motif at HSV-1 Pol residues 44 to 49 is highly conserved among all human herpesviruses (Fig. 1A) (12, 25, 30). The pre-NH2-terminal domain is located at the periphery of the enzyme and outside the catalytic center for 5′-3′ polymerase activity (13, 19, 25), suggesting that it is unlikely to directly participate in polymerase activity. Due to its conservation among herpesviruses, we hypothesized that the pre-NH2-terminal domain provides an important function for viral DNA synthesis and production of infectious virus distinct from 5′-3′ polymerase activity. Accordingly, we generated mutants with pre-NH2-terminal mutations for analysis of in vitro 5′-3′ polymerase activity, viral DNA synthesis, and production of infectious virus.
Fig 1.
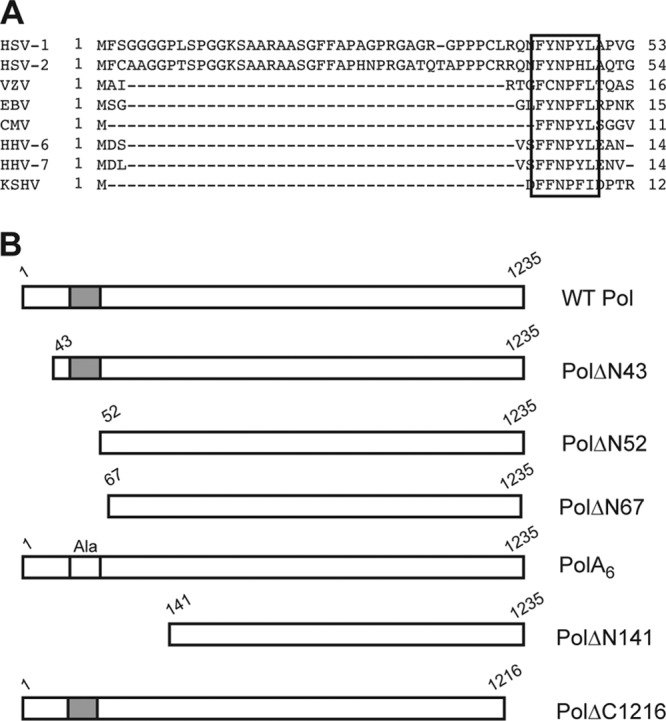
Sequence alignment and Pol mutant proteins. (A) Sequence alignment of the eight human herpesvirus Pol sequences (12, 30). A motif consisting of multiple hydrophobic and aromatic residues (FYNPYL) 44 to 49 of HSV-1 Pol is conserved in the human herpesvirus Pol family (box). Polymerase sequences of HSV-1 (14), HSV-2 (5), varicella-zoster virus (VZV) (9), Epstein-Barr virus (EBV) (10), cytomegalovirus (CMV) (7), human herpesvirus 6 (HHV-6) (18), HHV-7 (28), and Kaposi's sarcoma-associated herpesvirus (KSHV) (29) are shown. (B) Schematic diagram of selected Pol proteins analyzed in RRL assays. Amino-terminal deletions are numbered according to the first residue (downstream of the initiating AUG codon) of the WT protein present in the mutant, and carboxyl-terminal deletions are numbered according to the last amino acid present (upstream of the terminating UGA codon). The conserved motif is depicted by the gray box (not drawn to scale), which was replaced by alanines (Ala) in mutant PolA6.
MATERIALS AND METHODS
Cells and viruses.
Vero (American Type Culture Collection) and polB3 cells, which inducibly express wild-type (WT) HSV-1 Pol upon infection and were kindly provided by Charles Hwang (17), were grown and maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% newborn calf serum, 1% penicillin and streptomycin, and 1% amphotericin B. HSV-1 was propagated and titrated on Vero or polB3 cells. Construction of mutant viruses from a bacterial artificial chromosome (BAC) clone of HSV-1 strain KOS is detailed below. Spodoptera frugiperda Sf9 cells (Invitrogen) were cultured under serum-free conditions with Sf-900II serum-free medium (Invitrogen) supplemented with 10 μg/ml gentamicin.
Plasmids and BACs.
All of the indicated pol constructs used for in vitro transcription/translation studies were cloned into the SpeI/HindIII (New England BioLabs) sites of pBluescript II KS+ (pBS; Stratagene). The HSV-1 WT pol gene was shuttled from plasmid HTC-Pol (kindly provided by Gloria Komazin-Meredith) to the pBS vector to generate pBS-Pol. Truncation mutants were amplified from pBS-Pol by employing KOD Hot Start DNA polymerase (EMD Biosciences) and sequence-specific primers that introduced a start codon (AUG) downstream of an SpeI site at the 5′ terminus and a stop codon (UGA) upstream of a HindIII site at the 3′ terminus. Plasmid pBS-PolA6 was generated via two sequential rounds of site-directed mutagenesis using the QuikChange method (Stratagene) that introduced alanine substitutions F44A, Y45A, N46A, P47A, Y48A, and L49A into pBS-Pol. Primer sequences are provided in Table S1, which can be found at http://coen.med.harvard.edu.
Escherichia coli strain GS1783 (41) (kindly provided by Greg Smith, Northwestern University) harboring a BAC clone of HSV-1 strain KOS (I. Jurak et al., unpublished data) was used to generate scarless pol mutants via the two-step Red recombination techniques outlined by Tischer et al. (42). Plasmids pEP-KanaS (42) and pBAD-I-SceI (42) were kindly provided by Nikolaus Osterrieder and B. Karsten Tischer (Cornell University). Manipulations to generate deletions, substitutions, and insertions within the pol locus in the BAC were performed as previously described (42). The presence of each mutation and lack of unintended mutations were verified by sequencing the pol gene in each mutant BAC clone. BAC-derived viruses were reconstituted via transfection of 2 μg of purified BAC DNA into 3 × 105 polB3 cells using Lipofectamine reagent (Invitrogen). At 5 days posttransfection, the viral supernatant was harvested and titrated on polB3 cells. A single plaque was isolated and subjected to an additional two rounds of plaque purification prior to amplification of pure viral stocks. Viral DNA was harvested from infected polB3 cells, and the pol locus was subjected to sequencing (data not shown). Restriction fragment length polymorphism analysis of purified BAC-derived viral DNA compared to DNA from WT HSV-1 strain KOS following digestion with BamHI (NEB) confirmed the overall integrity of the viral genome and the presence of each deletion at the 5′ end of the pol gene (see Fig. S1A, available at https://coen.med.harvard.edu). Rescued derivative viruses were generated by restoring the WT pol sequence in BAC clones polΔN52 and polA6 via two-step Red recombination prior to reconstitution in polB3 cells.
In vitro transcription/translation and DNA polymerase assays.
HSV-1 Pol proteins were expressed via rabbit reticulocyte lysate (RRL) using a TNT quick coupled transcription/translation systems kit (Promega). Six hundred nanograms of each plasmid DNA construct (or no DNA, as a negative control) and other kit components were added to 30 μl of RRL that had been supplemented with either [35S]methionine (Perkin Elmer) or cold methionine in parallel, and the mixture was incubated at 30°C for 90 min per the manufacturer's instructions. To assess the level of protein expression for each experiment, an aliquot from each radiolabeled reaction mixture was analyzed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. The unlabeled reaction mixtures were subjected to DNA polymerase assays similar to those that were previously described (13), with some modifications: individual RRL reaction mixtures were supplemented with a polymerase reaction mixture that brought the final concentration (in 100 μl) to 50 mM Tris-HCl, pH 7.5, 100 mM (NH4)2SO4, 50 μg/ml bovine serum albumin, 0.5 mM dithiothreitol (DTT), 7.5 MgCl2, 10 μg/ml activated calf thymus DNA, 5 μM (each) dCTP, dGTP, and dATP, and 2.5 μCi [α-32P]dTTP (Perkin Elmer). To assess stimulation of polymerase activity by UL42, reaction mixtures were supplemented with 2 pmol maltose binding protein (MBP)-UL42ΔC340 (21) (kindly provided by Gloria Komazin-Meredith) and incubated at 37°C for 30 min prior to addition of the reaction mixture. Aliquots were removed at the indicated time points, mixed with EDTA, and incubated on ice to halt further enzymatic activity. Samples were spotted onto DE81 anion-exchange filters (Whatman), washed twice with 5% (wt/vol) Na2HPO4 buffer (dibasic), and rinsed once with water and once with 100% ethanol. Filters were dried and subjected to liquid scintillation counting.
Recombinant baculovirus, protein expression, purification, and polymerase activity.
Bacmid donor plasmids were generated by shuttling polΔN52 and polA6 constructs from pBS-KS+ vectors to the pFastBac vector (Invitrogen) using SpeI and HindIII restriction sites. Sf9 cells were transfected with recombinant bacmid DNA to generate recombinant baculoviruses expressing N-terminal His6-tagged PolΔN52 and PolA6 proteins by using a Bac-to-Bac baculovirus expression system kit (Invitrogen). His6-WT Pol was expressed and purified from the corresponding recombinant baculovirus that was engineered in a similar fashion (kindly provided by Gloria Komazin-Meredith). Viral titers were determined using a BacPAK baculovirus rapid titer kit (Clontech). Mid-log-phase Sf9 cells were infected at a multiplicity of infection (MOI) of 2, and cells were harvested at 65 h postinfection (hpi). Cell pellets were washed with Dulbecco's phosphate-buffered saline (DPBS) and resuspended in buffer A (20 mM HEPES, pH 7, 1 mM DTT, 20% glycerol, 100 mM guanidine HCl, 200 mM NaCl, 20 mM imidazole, and two Roche complete protease inhibitor tablets/100 ml). His-tagged proteins were captured via batch purification with Ni2+-nitrilotriacetic acid resin (Qiagen) for 30 min with gentle agitation at 4°C. The absorbed resin was loaded onto a column and washed extensively with buffer A. Pol was eluted from the column with buffer A containing 500 mM imidazole. Fractions that contained His-tagged Pol were pooled and passed through a 1-ml HiTrap heparin HP column (GE Healthcare) that had been preequilibrated with buffer B (20 mM HEPES, pH 7, 2 mM DTT, 20% glycerol, 100 mM guanidine HCl, 200 mM NaCl). The column was washed with buffer B, and protein was eluted with a linear NaCl gradient of up to 1 M NaCl. Fractions containing Pol were pooled, concentrated with an Amicon Ultra-15 centrifugal unit (Millipore), and stored at −80°C.
Unless otherwise stated, polymerase assays using purified enzyme were conducted using 400 fmol Pol in the presence and absence of 1pmol MBP-UL42ΔC340, and samples were processed as described above. For concentration-dependent assays, basal polymerase activity was measured in individual reaction mixtures containing the indicated amount of purified enzyme following a 20-min incubation at 37°C. The amount of dTTP incorporation was calculated using a standard curve of known [32P]dTTP amounts.
Viral replication assays.
Vero cells (2.5 × 105) were infected in triplicate at an MOI of 10 or 20 PFU/cell, as indicated. After a 1-h adsorption period at 37°C, wells were washed with DPBS and replenished with 2 ml of DMEM containing 2% newborn calf serum. At each time point, whole-cell lysates were collected, frozen, and subsequently thawed and sonicated. Cellular debris was pelleted by centrifugation, and supernatants were titrated on polB3 cells in duplicate.
Indirect immunofluorescence.
Vero cells (1 × 105) were seeded on glass coverslips and infected with the indicated virus at an MOI of 20. At 6 hpi, cells were fixed with 3.8% formaldehyde for 15 min and permeabilized with 1% Triton X-100 for 10 min. Samples were incubated with blocking buffer (10% normal goat serum in phosphate-buffered saline [PBS]) at 4°C overnight. Cells were stained with 0.1 mg/ml anti-Pol antibody 1051c (38) (kindly provided by Robert Klemm and Charles Knopf, University of Heidelberg) for 1 h at room temperature. Samples were washed with PBS and reacted with Alexa Fluor 488-conjugated chicken anti-mouse antibody (Molecular Probes) at a 1:1,000 dilution for 1 h at room temperature. Coverslips were washed with PBS and mounted onto glass slides using ProLong gold antifade reagent (Invitrogen). Fluorescence microscopy was performed with a Yokogawa spinning disk confocal unit on a Nikon Ti inverted microscope using a 60× Plan Apo (numerical aperture, 1.4) objective lens. Sequential optical sections of 0.5-μm step size were collected with a Hamamatsu ORCA ER cooled charge-coupled-device camera. Images are presented as the median of the acquired z-series sections using MetaMorph (version 7) software.
Real-time PCR assay of viral DNA synthesis.
Vero cells (2.5 × 105) were infected with virus at an MOI of 20 in triplicate, and cell lysates were harvested at 12 and 16 hpi. DNA from mock- and HSV-infected Vero cells was isolated and processed as described previously for murine trigeminal ganglia (32). Viral DNA standards were generated by spiking 10-fold serial dilutions of purified HSV-1 DNA into mock-infected Vero cell lysate. Cellular DNA standards were prepared by making serial 3-fold dilutions of mock-infected lysate. Viral and cellular DNA standards were processed along with experimental samples as a control for the efficiency of DNA recovery from infected cell lysate. Real-time PCR assays for viral (32) and cellular (J. M. Pesola, unpublished results) DNA were conducted in 20-μl reaction mixtures with 2 μl of the experimental samples or standards (∼1/20 of entire sample), using 0.1 μM primers and SYBR green PCR master mix (Applied Biosystems). Each primer set selectively amplified the viral thymidine kinase gene or the cellular 1,3-alpha-galactosyltransferase gene, resulting in ≥96% PCR amplification efficiency in each assay (sequences of primers are provided in Table S2, available at http://coen.med.harvard.edu). Real-time reactions were performed on an Applied Biosystems StepOnePlus real-time PCR system per the manufacturer's instructions. Absolute and relative amounts of viral and cellular DNA, respectively, in each sample were interpolated from the generated standard curve (linear regression was performed on a plot of the threshold cycle versus log quantity DNA; all R2 values were ≥0.98; see Fig. S2, available at http://coen.med.harvard.edu). The amounts of cellular DNA detected in each sample were used to normalize viral DNA values, which are reported as viral DNA copy number. Statistical analyses were performed using GraphPad Prism software (GraphPad Software, San Diego, CA).
RESULTS
Analysis of HSV-1 Pol mutant protein activity in RRL.
We hypothesized that the pre-NH2-terminal domain is not directly involved in 5′-3′ polymerase activity due to its location outside the previously characterized active site (13, 19, 25). However, further examination of the crystal structure revealed an extensive hydrophobic network between the 3′-5′ exonuclease domain and antiparallel beta sheets within the pre-NH2-terminal domain located downstream of HSV-1 Pol residue 70 (25). Therefore, we anticipated that extensive deletions within these regions could lead to misfolding or destabilization of the Pol enzyme. A previous study had generated HSV-1 Pol mutant proteins via RRL for analysis of 5′-3′ polymerase activity (13). In order to avoid engineering viruses with mutations that could indirectly abrogate enzymatic activity and thereby result in viral lethality (1, 13), we utilized this expression system to test the 5′-3′ polymerase activity of Pol mutants in the presence and absence of viral processivity factor UL42.
Selected Pol mutants utilized in this study are depicted in Fig. 1B. Truncation mutant PolΔN43 was constructed to investigate the importance of the extreme N-terminal 42 residues. In an effort to explore the potential role of the conserved motif FYNPYL, we generated deletion mutant PolΔN52, which lacks the extreme N-terminal 51 residues, and substitution mutant PolA6, in which residues 44 to 49 were replaced by alanines. Additional mutants were tested in order to identify the most extensive truncation mutant that encoded an enzymatically active polymerase. One set of programmed reticulocyte lysates was supplemented with [35S]methionine in order to visualize the relative amounts of protein synthesized in each reaction via SDS-PAGE and autoradiography. Unlabeled lysates in which cold methionine was substituted for [35S]methionine were used to analyze the enzymatic activity of the HSV-1 Pol mutants. Basal polymerase assays were conducted by measuring the incorporation of [32P]dTTP into activated calf thymus DNA as previously described (13). In vitro reaction mixtures supplemented with purified MBP-UL42ΔC340, which is capable of stimulating HSV-1 Pol (11, 21), were used to assess processive polymerase activity. Two controls were included in each assay: a reaction mixture that lacked a pol construct in order to assess background activity of endogenous RRL protein and another that expressed Pol truncation mutant PolΔC1216, which cannot interact with UL42 and thereby is not stimulated by it (11).
The results from the in vitro analysis of each pre-NH2-terminal mutant are summarized in Table 1. The two most conservative truncation mutants, PolΔN43 and PolΔN52, and substitution mutant PolA6 reproducibly exhibited time-dependent 5′-3′ polymerase activity similar to WT Pol in the presence and absence of UL42 (Fig. 2A and B and data not shown). The enhanced level of activity detected in reaction mixtures containing PolΔN43 is most likely due to increased protein expression in this experiment (Fig. 2C). PolΔN141, in which the entire pre-NH2-terminal domain was deleted, did not exhibit detectable enzymatic activity (Fig. 2A and B). Although a previous study had reported that a mutant lacking the extreme N-terminal 66 residues exhibited polymerase activity (13), we found that truncation mutant PolΔN67 reproducibly lacked polymerase activity above that for the negative controls in both the basal and processive polymerase assays (Table 1). Thus, removal of up to 51 residues at the extreme N terminus of HSV-1 Pol yielded an enzymatically active protein, while more extensive deletions negatively impacted 5′-3′ polymerase activity.
Table 1.
Summary of in vitro polymerase activity of selected Pol mutant proteins
| Polymerase mutant construct | Basal DNA polymerase activitya | Stimulated polymerase activity via UL42b |
|---|---|---|
| PolΔN43 | + | + |
| PolΔN52 | + | + |
| PolΔN67 | − | − |
| PolΔN141 | − | − |
| PolA6 | + | + |
| PolΔC1216 | + | − |
+, wild-type levels of activity; −, activity at or below that of background.
+ , wild-type levels of activity; − , activity at or below that of UL42-binding mutant protein PolΔC1216.
Fig 2.
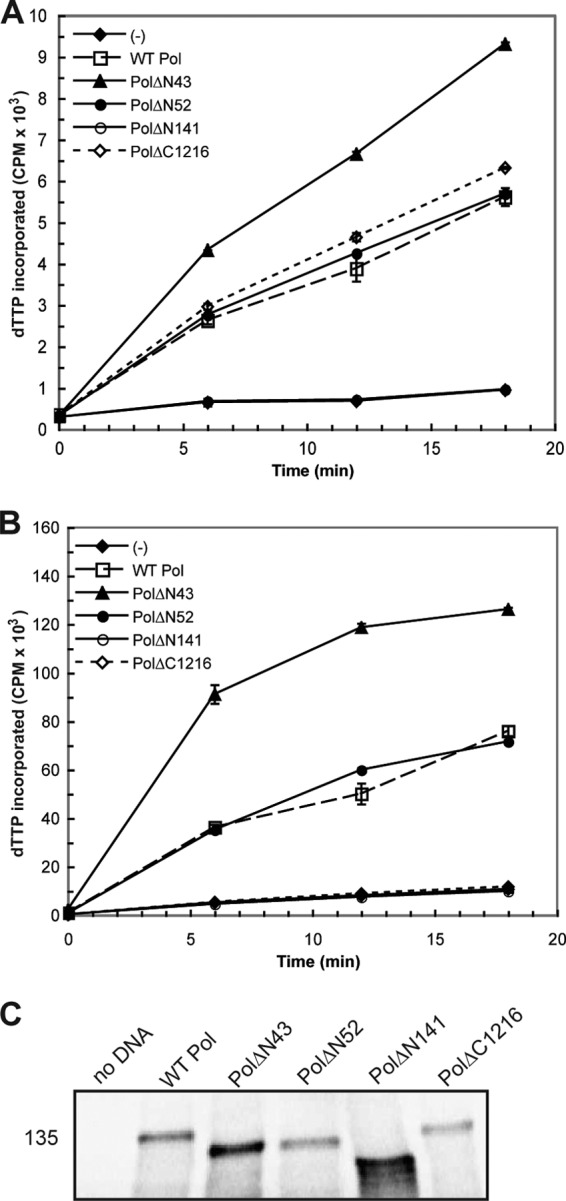
5′-3′ polymerase activity and Pol mutant protein expression in RRL. HSV-1 Pol constructs were engineered and expressed in RRL for evaluation of 5′-3′ polymerase activity. DNA was omitted from one in vitro reaction [indicated by (−)] in order to assess background levels of nucleotide incorporation. (A) Basal polymerase activity. Individual RRL reaction mixtures containing the constructs indicated in the key were supplemented with the polymerase reaction mixture and analyzed for enzymatic activity. The level of [α-32P]dTTP incorporation at each time point is reported as the number of counts per minute (CPM) and graphed as the mean of duplicate samples. (B) Processive polymerase activity. Parallel RRL reactions were supplemented with HSV-1 processivity factor UL42 prior to the addition of the polymerase reaction mixture in order to assess stimulated polymerase activity. Mutant PolΔC1216 cannot bind UL42 and thereby served as a negative control in this assay. (C) In vitro expression of Pol mutant proteins. Aliquots of RRL reaction mixtures supplemented with [35S]methionine were electrophoresed on a 5% SDS-polyacrylamide gel, dried, and exposed to a phosphorimager screen overnight. The position of a molecular mass marker (in kilodaltons) is included on the left side of the panel.
5′-3′ polymerase activity of purified HSV-1 Pol enzyme.
A previous study had reported that a purified HSV-1 Pol truncation mutant lacking the extreme N-terminal 42 residues retained 5′-3′ polymerase activity (25). In order to more quantitatively evaluate the activity of the remaining Pol mutants that maintained detectable activity in the RRL studies, recombinant baculoviruses were constructed for the generation of His-tagged fusion proteins PolΔN52 and PolA6. WT Pol, PolΔN52, or PolA6 proteins were purified to homogeneity from lysates of insect cells infected with the appropriate recombinant baculovirus. We assayed the ability of each enzyme to incorporate [32P]dTTP into activated calf thymus DNA in the presence or absence of purified MBP-UL42ΔC340.
In accordance with the results generated from the polymerase assays conducted in RRL (Table 1), we found that both purified mutant proteins displayed basal polymerase activity similar to that of WT Pol (Fig. 3A). The enhancement of polymerase activity in the presence of UL42 demonstrated a functional interaction with the viral processivity factor (11) (Fig. 3B). Lastly, we evaluated basal polymerase activity as a function of protein concentration for each enzyme (Fig. 3C). The average rates of dTTP incorporation for WT, PolΔN52, and PolA6 proteins were very similar: 620, 570, and 680 fmol/min/nmol, respectively. Therefore, the engineered mutations in HSV-1 Pol had little or no impact on 5′-3′ polymerase activity or the association with UL42.
Fig 3.
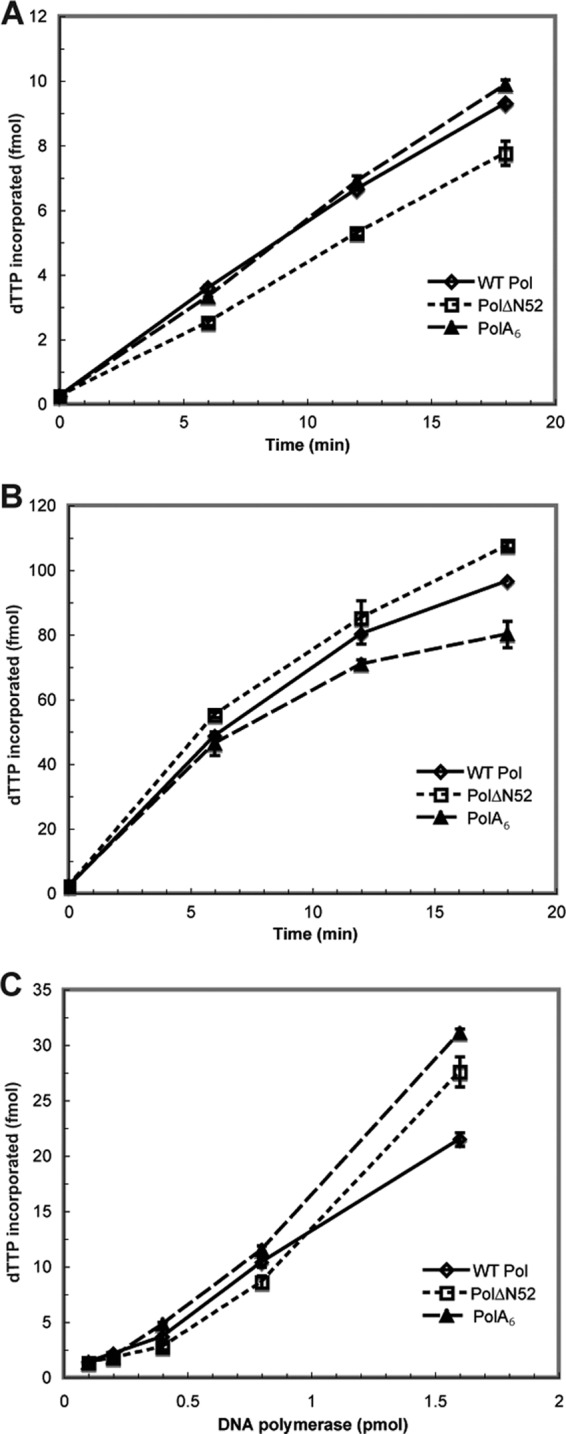
In vitro 5′-3′ polymerase activity of purified Pol protein. WT Pol and mutant Pol proteins were expressed and purified from insect cells infected with recombinant baculovirus. Purified protein was subjected to basal (A) and processive (B) polymerase assays as described in the legend to Fig. 2. Additionally, basal activity was measured and plotted as a function of enzyme concentration (C). The amount of dTTP incorporation (fmol) was calculated at each time point or enzyme concentration and graphed as the mean of duplicate samples.
Replication of BAC-derived HSV-1 pol mutant viruses.
In order to assess the biological significance of pol mutations within the context of infection, pol mutant viruses were engineered via manipulation of an infectious bacterial artificial chromosome (BAC) clone of HSV-1 strain KOS (I. Jurak et al., unpublished data). Using BAC recombineering techniques (42), pol coding sequences were deleted in order to produce viruses that expressed pre-NH2-terminal Pol mutants corresponding to the proteins included in our RRL analyses (Table 1). Mutant viruses polΔN43 and polΔN52 were analyzed to determine whether the extreme N-terminal 42 or 51 residues of HSV-1 Pol were essential for viral replication. Mutant virus polΔN141, in which the entire pre-NH2-terminal domain was deleted, was included to verify that a mutant lacking detectable polymerase activity in the in vitro RRL studies would be nonviable. Due to the potential for replication defects as a result of the introduced mutations, each BAC, including WT, was introduced into and propagated in polB3 cells, which inducibly express WT HSV-1 Pol upon infection (17). Each BAC clone and corresponding virus was sequenced at the pol locus in order to confirm the presence of the engineered mutation and verify the lack of adventitious mutations.
To assess the viability of reconstituted virus harvested from complementing polB3 cells, supernatant from cells transfected with BAC DNA was titrated on polB3 cells and noncomplementing Vero cells, and reconstituted virus was scored for the ability to form plaques on Vero cells relative to polB3 cells (plating efficiency) in comparison to the ability of WT virus (Table 2). As expected, WT BAC exhibited a plating efficiency of 100%, while mutant polΔN141 was unable to form plaques on Vero cells. Mutant virus polΔN43 exhibited a plating efficiency similar to that of WT (93%). Interestingly, mutant polΔN52 exhibited a lower plating efficiency (73%) and a small-plaque phenotype compared to WT and polΔN43 viruses (data not shown). This result suggested that the polΔN52 mutant could not replicate as well as WT in Vero cells. Analysis of single-cycle replication kinetics (see Fig. S1B, available at http://coen.med.harvard.edu) validated these initial observations: mutant polΔN141 failed to replicate, and mutant polΔN52 exhibited 5-fold and 7-fold decreases in viral yield at 12 and 16 hpi, respectively, while mutant polΔN43 replication kinetics were indistinguishable from those of WT. Thus, the extreme N-terminal 42 residues of HSV-1 Pol were dispensable for viral replication in cell culture, while a mutant in which the extreme N-terminal 51 residues were removed exhibited notably decreased viral replication.
Table 2.
Plating efficiencies of BAC-derived viruses
| Virusa | Titer (PFU/ml) on: |
Plating efficiency (%)b | |
|---|---|---|---|
| Vero cells | polB3 cells | ||
| WT | 2.7 × 107 | 2.7 × 107 | 100 |
| polΔN43 | 4.2 × 107 | 4.5 × 107 | 93 |
| polΔN52 | 4.9 × 106 | 6.7 × 106 | 73 |
| polΔN52R | 3.4 × 107 | 3.3 × 107 | 100 |
| polA6 | 5.2 × 106 | 6.5 × 106 | 80 |
| polA6R | 1.6 × 107 | 1.6 × 107 | 100 |
| polΔN141 | 0 | 7.5 × 106 | 0 |
The viral supernatant that was generated from transfection of polB3 cells with BAC DNA was harvested and subsequently titrated on the indicated cell line.
Calculated as the ratio of viral titers on Vero cells and polB3 cells.
Conserved motif FYNPYL is important for efficient production of infectious virus and viral DNA synthesis.
We hypothesized that the absence of the conserved motif accounted for the mutant polΔN52 replication defect. To test this possibility, we constructed mutant virus polA6 in which the six-residue motif was replaced by six alanines. Additionally, we wanted to test whether the observed replication defects were due to the engineered mutations. Accordingly, the polΔN52 and polA6 mutant BACs were used as the templates to restore the WT pol sequence and generate rescued derivative viruses polΔN52R and polA6R. WT, mutant, and rescued derivative viruses were tested for their ability to replicate in Vero cells. Interestingly, mutant polA6 mimicked mutant polΔN52 in forming small plaques (data not shown) and exhibiting a diminished plating efficiency (Table 2) and production of infectious virus (Fig. 4). We also found that WT-like plating efficiencies (Table 2) and replication kinetics in Vero cells (Fig. 4) were restored in both the polΔN52R and polA6R rescued derivatives. Each mutant virus exhibited a 6-fold and an 8-fold defect at 12 and 16 hpi, respectively, compared to its respective rescued derivative virus (Fig. 4). WT and rescued derivative viruses reached peak viral production at 24 hpi, and production remained up to 6-fold higher than that of the mutant viruses.
Fig 4.
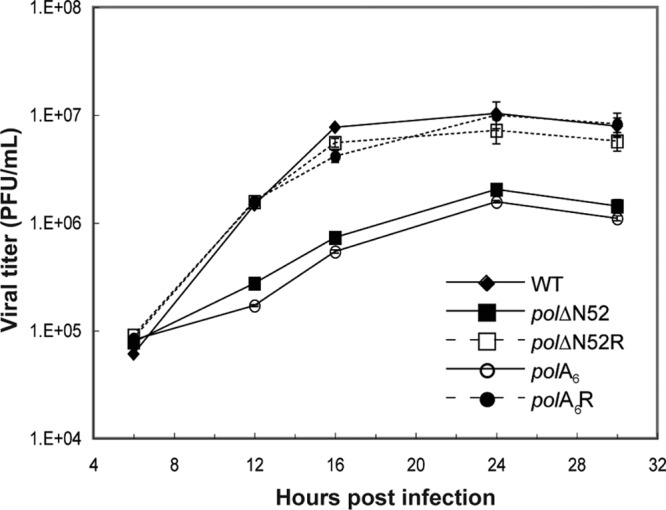
Single-cycle growth kinetics of pre-NH2-terminal mutant viruses. Vero cells were infected with BAC-derived virus at an MOI of 20. At the indicated time points, cells were harvested and virus was titrated on polB3 cells. Viral yield is reported as the number of PFU/ml, with each data point representing the mean ± SD of triplicate samples.
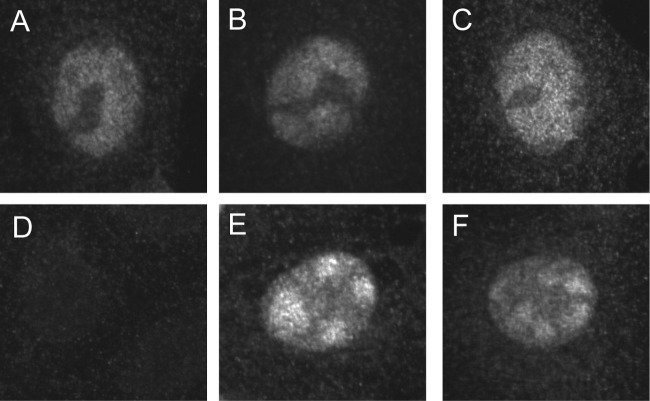
We sought to determine whether there were any effects of the engineered mutations on HSV-1 Pol localization during infection. During viral DNA synthesis in WT virus-infected cells, HSV-1 Pol localizes to replication compartments—large globular structures within the nucleus, which can be seen as early as 5.5 hpi (4, 24). Using indirect immunofluorescence, we observed that HSV-1 Pol staining was, as expected, predominantly found in large replication compartments that encompassed most of the nucleus in cells infected with WT, polΔN52R, and polA6R viruses (Fig. 5A to C). Pol staining was also mainly found in the nuclei of cells infected with the polΔN52 and polA6 mutants but was concentrated in smaller structures than in cells infected with the other viruses (Fig. 5E and F). No staining was observed in mock-infected cells (Fig. 5D). Localization of both PolΔ52 and PolA6 to replication compartments rather than punctate prereplicative sites indicated that viral DNA synthesis had not been drastically inhibited (4). Thus, the conserved motif is not required for nuclear localization of HSV-1 Pol. The presence of smaller replication compartments during polΔN52 and polA6 mutant infection suggested that the motif is required for WT-like levels of DNA synthesis.
Fig 5.
Localization of HSV-1 Pol in infected cells. Vero cells were either mock infected (D) or infected with BAC-derived virus at an MOI of 20 (A to C, E, F). Samples were fixed at 6 hpi and processed for indirect immunofluorescence with anti-Pol antibody. (A) WT virus; (B) polΔN52R virus; (C) polA6R virus; (D) mock-infected virus; (E) polΔN52 mutant; (F) polA6 mutant.
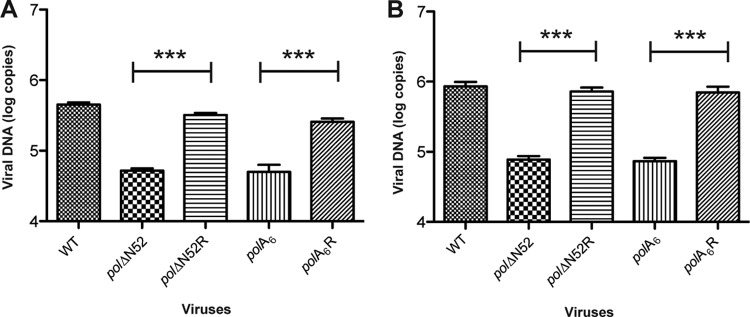
We then analyzed levels of viral DNA synthesis during the course of infection for each virus. DNA was isolated from mock- and HSV-infected cell lysates at 12 and 16 hpi. Viral and cellular DNA standards as well as experimental samples were subjected to real-time PCR with primers that targeted the viral and cellular genes thymidine kinase and 1,3-alpha-galactosyltransferase, respectively. Standard curves were generated in order to quantify the number of viral DNA copies, which was normalized to cellular DNA content (see Fig. S2, available at http://coen.med.harvard.edu). Both the polΔN52 and polA6 mutants exhibited a decrease in viral DNA production that corresponded to the observed defects in viral yield, with 6-fold and ∼10-fold defects at 12 and 16 hpi, respectively, compared to the appropriate rescued derivative (Fig. 6A and B). These differences were statistically significant, while the differences in viral DNA content between the polΔN52 and polA6 mutants were not, indicating that substitution of the conserved motif recapitulated the deletion mutant phenotype. There was no apparent instability of HSV-1 Pol mutant polypeptides PolΔN52 and PolA6, as indicated by Western blot analysis of infected cell lysates (S. L. Terrell and D. M. Coen, unpublished results). These data indicate that the loss of conserved motif FYNPYL is responsible for the observed defects in viral yield that correlate with decreased viral DNA production.
Fig 6.
Decreased viral DNA synthesis during pol mutant virus infection. Vero cells (2.5 × 105) were infected at an MOI of 20, and DNA was harvested from infected cell lysates at 12 (A) and 16 (B) hpi. The viral thymidine kinase gene was quantified via real-time PCR, the quantity was normalized to that of the cellular 1,3-alpha-galactosyltransferase gene as described in Materials and Methods, and values are reported as log viral DNA copy number. Means ± SDs of triplicate samples are plotted. ***, P < 0.0001 (by one-way analysis of variance with Bonferroni's multiple-comparison posttests).
DISCUSSION
We have shown in this report that three pre-NH2-terminal HSV-1 Pol mutants retained polymerase activity similar to that of WT Pol in vitro, while two of the corresponding mutant viruses exhibited decreased virus production. Our efforts identified a conserved motif at HSV-1 Pol residues 44 to 49 as necessary for the efficient production of viral DNA during infection. This decrease in viral DNA synthesis correlated with the reduced production of infectious viral progeny. Taken together, our data strongly suggest that the pre-NH2-terminal domain includes a function that is not important for 5′-3′ polymerase activity yet is crucial for efficient viral DNA synthesis during infection.
Enzymatic activity of pre-NH2-terminal Pol mutants in vitro.
Analysis of 5′-3′ polymerase activity in RRL, as pioneered by Dorsky and Crumpacker (13), provided a rapid method for identification of catalytically active Pol mutants that warranted further analysis. The dynamic nature of the HSV-1 Pol protein made it difficult to predict how each mutation would affect protein function. In our studies, only mutants that contained deletions or substitutions within the extreme N-terminal 51 residues retained 5′-3′ polymerase activity similar to that of WT. Truncation mutant PolΔN67, in which the extreme N-terminal 66 residues were removed, was found to be catalytically inactive in our assays, which is in contrast to a previous report (13). This discrepancy may be due to the previously reported construct containing eight codons of nonnative sequence upstream of residue 67 (13). The pre-NH2-terminal domain engages in an extensive hydrophobic network with the adjacent 3′-5′ exonuclease domain (25), disruption of which may indirectly lead to a catalytically inactive protein. However, we cannot conclusively determine whether each inactive Pol mutant was misfolded or destabilized as a result of the mutation. Although our in vitro studies cannot exclude the possibility that any of the engineered mutations affected HSV-1 Pol activities, such as 3′-5′ exonuclease or lyase activity, we suggest that these possibilities are unlikely. The 3′-5′ exonuclease and lyase active sites have been mapped within the interior of the 3′-5′ exonuclease domain (17, 22) and a 63-kDa C-terminal fragment (3), respectively, and are separate from the sites of our engineered mutations within the extreme N-terminal 51 residues at the surface of the enzyme. Additionally, a previous study had reported that a mutant lacking detectable 3′-5′ exonuclease activity exhibited a 50-fold decrease in viral yield with only a 3-fold decrease in viral DNA production (40). The defect in viral yield observed in the previous study largely reflected an alteration in replication fidelity rather than a defect in viral DNA synthesis, as seen with our Pol mutants (40). Regardless, purification and assay of PolΔN52 and PolA6 proteins relative to WT Pol confirmed that the mutations did not induce any global effects on protein folding, as the mutant proteins exhibited robust 5′-3′ polymerase activity.
Importance of the pre-NH2-terminal domain during viral DNA synthesis.
Viral genetic analyses allowed us to evaluate the effect of each mutation within the context of infection. Despite 76% protein sequence identity in the extreme N-terminal 42 residues of Pol in HSV-1 and -2, our studies have shown that these were dispensable for viral replication in cell culture. There is a possibility that the extreme N-terminal 42 residues may be necessary for replication or pathogenesis in animal models of viral infection. We have demonstrated that the conserved motif FYNPYL contributes to the efficient synthesis of viral DNA and production of progeny virus. It seems reasonable, based on our results, that the defect in production of infectious virus is due to a defect in viral DNA synthesis.
Although the pre-NH2-terminal domain is conserved in the herpesvirus Pol family, a homologous domain is absent from the related bacteriophage RB69 Pol and other published family B polymerase structures (25, 33, 39, 44, 45). The conserved motif that we have identified as being important for viral DNA synthesis is a cluster of mostly aromatic and hydrophobic residues that are absent from the HSV-1 Pol crystal structure (25), suggesting that this motif is located within a disordered region of the protein. One could envision a scenario where this flexible segment near the extreme N terminus of HSV-1 Pol interacts with a factor that actively recruits the polymerase to the replication fork or otherwise positively impacts viral DNA production. UL8, a component of the viral helicase-primase complex, has been shown to interact with HSV-1 Pol in vitro (27). This interaction has been proposed to serve as a molecular tether for HSV-1 Pol at the leading strand of the replication fork (25). Another potential candidate is the viral SSB protein ICP8, which has been reported to stimulate HSV-1 Pol 5′-3′ polymerase activity (36). Protein complexes containing the HSV DNA polymerase holoenzyme, ICP8, and viral alkaline nuclease UL12 have been captured from infected cell lysate via immunoaffinity chromatography (43). Although a direct interaction has yet to be demonstrated, evidence supporting such an interaction includes that from a study that found that specific ICP8 mutants conferred altered sensitivity to viral DNA synthesis inhibitors (6). Additionally, HSV-1 Pol was unable to localize to prereplicative sites within the nucleus in the absence of a functional ICP8 protein (4). As yet, binding sites for ICP8 and UL8 have not been mapped on HSV-1 Pol, so the implications of disrupting such interactions during viral infection are unknown. Presumably, any other viral or cellular protein that participates in viral genome synthesis and maintenance is a potential candidate. For example, a recent study reported that cellular transcriptional regulator HCF-1 can interact simultaneously with HSV-1 Pol and histone chaperone Asf1b (31). Depletion of Asf1b prior to infection with HSV-1 resulted in 5- and 10-fold decreases in viral DNA and virus production, respectively, at 18 hpi, which suggested that HCF-1 and Asf1b are necessary for efficient viral DNA synthesis (31). These possibilities are under investigation. As yet, we have been unable to identify a binding partner whose association with Pol is disrupted as a result of the deletion or substitution mutation (unpublished results). More than likely, the presumed protein-protein interaction would represent a conserved replication mechanism exhibited by human herpesviruses.
The exact mechanism by which HSV-1 DNA replication is carried out within the infected cell has yet to be fully elucidated. Following DNA melting at the origin of replication, HSV-1 DNA synthesis is hypothesized to begin on a circular template via theta replication and is converted by an unknown mechanism to an exponential rolling-circle replication mechanism, which is responsible for the bulk of viral DNA synthesis (2, 34, 37). Interestingly, we observed that WT virus exhibited exponential viral growth at between 6 and 12 hpi, while mutant viruses polΔN52 and polA6 lacked this burst and maintained a diminished level of virus production throughout the course of infection. If viral yield were a direct function of viral DNA synthesis in this case, it could potentially represent a compromise in late-phase DNA replication. However, our studies do not clearly indicate whether the defect in viral DNA synthesis is due to a decrease in the rate of DNA production or a perturbation in the recruitment and retention of HSV-1 Pol at active replication forks. Identification of the mechanism responsible for the observed defect would lead to enhanced characterization of the functional HSV-1 replisome and provide valuable insight into processes that are essential for efficient HSV-1 replication in cell culture.
ACKNOWLEDGMENTS
We thank Charles Richardson, Sharmistha Ghosh, Dave Filman, Jennifer Baltz, Blair Strang, and Jeremy Kamil for helpful discussions. We are grateful to Gloria Komazin-Meredith for providing purified HSV-1 WT Pol (with corresponding recombinant baculovirus) and MBP-UL42ΔC340, as well as technical expertise and support. We thank Hyungsuk Oh, Mayuri Sharma, and Brian Bender for help with immunofluorescence protocols and the Nikon Imaging Center at Harvard Medical School for its assistance with microscopy. We thank Can Cui for generation of the infectious KOS BAC clone, Greg Smith for E. coli GS1783, Nikloaus Osterrieder and B. Karsten Tischer for providing plasmids pEP-KanaS and pBAD-I-SceI, Robert Klemm and Charles Knopf for providing anti-Pol antibody 1051c, Jean Pesola for designing the real-time PCR assay and performing statistical analyses, and Blair Strang and Jean Pesola for helpful comments on the manuscript.
This work was supported by NIH award F31-AI084490 (to S.L.T.) and NIH grant R01-AI019838 (to D.M.C.).
Footnotes
Published ahead of print 8 August 2012
REFERENCES
- 1. Aron GM, Purifoy DJ, Schaffer PA. 1975. DNA synthesis and DNA polymerase activity of herpes simplex virus type 1 temperature-sensitive mutants. J. Virol. 16:498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boehmer PE, Lehman IR. 1997. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66:347–384 [DOI] [PubMed] [Google Scholar]
- 3. Bogani F, Boehmer PE. 2008. The replicative DNA polymerase of herpes simplex virus 1 exhibits apurinic/apyrimidinic and 5′-deoxyribose phosphate lyase activities. Proc. Natl. Acad. Sci. U. S. A. 105:11709–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bush M, et al. 1991. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J. Virol. 65:1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chibo D, Mijch A, Doherty R, Birch C. 2002. Novel mutations in the thymidine kinase and DNA polymerase genes of acyclovir and foscarnet resistant herpes simplex viruses infecting an immunocompromised patient. J. Clin. Virol. 25:165–170 [DOI] [PubMed] [Google Scholar]
- 6. Chiou HC, Weller SK, Coen DM. 1985. Mutations in the herpes simplex virus major DNA-binding protein gene leading to altered sensitivity to DNA polymerase inhibitors. Virology 145:213–226 [DOI] [PubMed] [Google Scholar]
- 7. Chou S, et al. 1999. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Adult AIDS Clinical Trials Group CMV Laboratories. Antimicrob. Agents Chemother. 43:1500–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coen DM, Schaffer PA. 2003. Antiherpesvirus drugs: a promising spectrum of new drugs and drug targets. Nat. Rev. Drug Discov. 2:278–288 [DOI] [PubMed] [Google Scholar]
- 9. Davison AJ, Scott JE. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759–1816 [DOI] [PubMed] [Google Scholar]
- 10. de Jesus O, et al. 2003. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 84:1443–1450 [DOI] [PubMed] [Google Scholar]
- 11. Digard P, Chow CS, Pirrit L, Coen DM. 1993. Functional analysis of the herpes simplex virus UL42 protein. J. Virol. 67:1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di Tommaso P, et al. 2011. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39:W13–W17 doi:10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dorsky DI, Crumpacker CS. 1988. Expression of herpes simplex virus type 1 DNA polymerase gene by in vitro translation and effects of gene deletions on activity. J. Virol. 62:3224–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibbs JS, et al. 1985. Sequence and mapping analyses of the herpes simplex virus DNA polymerase gene predict a C-terminal substrate binding domain. Proc. Natl. Acad. Sci. U. S. A. 82:7969–7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hubscher U, Maga G, Spadari S. 2002. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71:133–163 [DOI] [PubMed] [Google Scholar]
- 16. Hwang YT, Hwang CB. 2003. Exonuclease-deficient polymerase mutant of herpes simplex virus type 1 induces altered spectra of mutations. J. Virol. 77:2946–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hwang YT, Liu BY, Coen DM, Hwang CB. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 71:7791–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isegawa Y, et al. 2009. Human herpesvirus 6 ganciclovir-resistant strain with amino acid substitutions associated with the death of an allogeneic stem cell transplant recipient. J. Clin. Virol. 44:15–19 [DOI] [PubMed] [Google Scholar]
- 19. Knopf CW, Weisshart K. 1988. The herpes simplex virus DNA polymerase: analysis of the functional domains. Biochim. Biophys. Acta 951:298–314 [DOI] [PubMed] [Google Scholar]
- 20. Knopf KW. 1979. Properties of herpes simplex virus DNA polymerase and characterization of its associated exonuclease activity. Eur. J. Biochem. 98:231–244 [DOI] [PubMed] [Google Scholar]
- 21. Komazin-Meredith G, et al. 2008. The positively charged surface of herpes simplex virus UL42 mediates DNA binding. J. Biol. Chem. 283:6154–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuhn FJ, Knopf CW. 1996. Herpes simplex virus type 1 DNA polymerase. Mutational analysis of the 3′-5′-exonuclease domain. J. Biol. Chem. 271:29245–29254 [DOI] [PubMed] [Google Scholar]
- 23. Lehman IR, Kaguni LS. 1989. DNA polymerase alpha. J. Biol. Chem. 264:4265–4268 [PubMed] [Google Scholar]
- 24. Liptak LM, Uprichard SL, Knipe DM. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu S, et al. 2006. Crystal structure of the herpes simplex virus 1 DNA polymerase. J. Biol. Chem. 281:18193–18200 [DOI] [PubMed] [Google Scholar]
- 26. Marcy AI, Olivo PD, Challberg MD, Coen DM. 1990. Enzymatic activities of overexpressed herpes simplex virus DNA polymerase purified from recombinant baculovirus-infected insect cells. Nucleic Acids Res. 18:1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marsden HS, et al. 1997. The catalytic subunit of the DNA polymerase of herpes simplex virus type 1 interacts specifically with the C terminus of the UL8 component of the viral helicase-primase complex. J. Virol. 71:6390–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Megaw AG, Rapaport D, Avidor B, Frenkel N, Davison AJ. 1998. The DNA sequence of the RK strain of human herpesvirus 7. Virology 244:119–132 [DOI] [PubMed] [Google Scholar]
- 29. Neipel F, Albrecht JC, Fleckenstein B. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217 [DOI] [PubMed] [Google Scholar]
- 31. Peng H, Nogueira ML, Vogel JL, Kristie TM. 2010. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc. Natl. Acad. Sci. U. S. A. 107:2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pesola JM, Zhu J, Knipe DM, Coen DM. 2005. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J. Virol. 79:14516–14525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodriguez AC, Park HW, Mao C, Beese LS. 2000. Crystal structure of a pol alpha family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9 degrees N-7. J. Mol. Biol. 299:447–462 [DOI] [PubMed] [Google Scholar]
- 34. Roizman B, Knipe DM. 2001. Herpes simplex viruses and their replication, p 2399–2459 In Knipe DM, et al. (ed), Fields virology, 4th ed Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 35. Rothwell PJ, Waksman G. 2005. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 71:401–440 [DOI] [PubMed] [Google Scholar]
- 36. Ruyechan WT, Weir AC. 1984. Interaction with nucleic acids and stimulation of the viral DNA polymerase by the herpes simplex virus type 1 major DNA-binding protein. J. Virol. 52:727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strang BL, Stow ND. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79:12487–12494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strick R, Hansen J, Bracht R, Komitowski D, Knopf CW. 1997. Epitope mapping and functional characterization of monoclonal antibodies specific for herpes simplex virus type I DNA polymerase. Intervirology 40:41–49 [DOI] [PubMed] [Google Scholar]
- 39. Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. 2009. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 16:979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tian W, Hwang YT, Lu Q, Hwang CB. 2009. Finger domain mutation affects enzyme activity, DNA replication efficiency, and fidelity of an exonuclease-deficient DNA polymerase of herpes simplex virus type 1. J. Virol. 83:7194–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless Red recombination system. Methods Mol. Biol. 634:421–430 [DOI] [PubMed] [Google Scholar]
- 42. Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 43. Vaughan PJ, Banks LM, Purifoy DJ, Powell KL. 1984. Interactions between herpes simplex virus DNA-binding proteins. J. Gen. Virol. 65:2033–2041 [DOI] [PubMed] [Google Scholar]
- 44. Wang J, et al. 1997. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 89:1087–1099 [DOI] [PubMed] [Google Scholar]
- 45. Wang J, Yu P, Lin TC, Konigsberg WH, Steitz TA. 1996. Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions. Biochemistry 35:8110–8119 [DOI] [PubMed] [Google Scholar]
- 46. Wang TS, Wong SW, Korn D. 1989. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 3:14–21 [DOI] [PubMed] [Google Scholar]