Abstract
To gain more insight into the phylogeny of Dabieshan virus (DBSV), carried by Niviventer confucianus and other Murinae-associated hantaviruses, genome sequences of novel variants of DBSV were recovered from Niviventer rats trapped in the mountainous areas of Wenzhou, China. Genetic analyses show that all known genetic variants of DBSV, including the ones identified in this study, are distinct from other Murinae-associated hantaviruses. DBSV variants show geographic clustering and high intraspecies diversity. The data suggest that DBSV is a distinct species in the genus Hantavirus. Interestingly, DBSV shows the highest sequence identity to Hantaan virus (HTNV), with a >7% difference in the sequences of the N, GPC, and L proteins, while N. confucianus is more closely related to Rattus norvegicus (the host of Seoul virus [SEOV]) than to Apodemus agrarius (the host of HTNV and Saaremaa virus [SAAV]). Further genetic analyses of all known Murinae-associated hantaviruses (both established and tentative species) show that many of them, including DBSV, may have originated from host switching. The estimation of evolutionary rates and divergence time supports the role of cross-species transmission in the evolution of Murinae-associated hantaviruses. The detection of positive selection suggests that genetic drift may contribute to the speciation of Murinae-associated hantaviruses and that adaptation has a role as well.
INTRODUCTION
New emerging viral pathogens, e.g., avian and swine influenza viruses (28, 41), severe acute respiratory syndrome (SARS) coronavirus (17), and human immunodeficiency virus (16, 51), cause epidemics (or pandemics) in humans by changing or expanding their host range. These pathogens are a considerable threat to human and/or wildlife health, agricultural production, and public security (5, 36). Almost all of the novel viruses have circulated in their reservoir hosts for a long time before emerging in humans or other animals (11, 36, 37). Zoonotic viral pathogens such as hantaviruses and rabies virus show high genetic diversity that depends on natural hosts or geographic origins (7, 22, 43). The role of cross-species transmission in the generation of a new virus species should be studied in greater detail (26), and better understanding of the evolutionary relationship between zoonotic pathogens and their hosts may help in the prevention and control of (re)emerging diseases.
The hantavirus genome consists of three RNA segments, i.e., small (S), medium (M), and large (L) segments; they encode, respectively, the nucleocapsid (N) protein (in some hantaviruses, the nonstructural NSs protein), the glycoprotein precursor (GPC) of the two envelope glycoproteins (Gn and Gc), and the viral RNA-dependent RNA polymerase (RdRP) (the L protein) (44). At least 23 established and 30 tentative hantavirus species have been identified worldwide in rodents and insectivores (42). Identification of insectivore-carried hantaviruses has increased especially rapidly during the last 5 years (22, 24). Each species of these known hantaviruses is specifically associated with one or several closely related rodent or insectivore hosts (42). As the phylogeny of hantaviruses may be congruent with their hosts, hantaviruses are considered to have coevolved (cospeciated) with their respective rodent or insectivore hosts (20, 21, 23, 25, 38, 39, 43, 44, 52). Recently, Ramsden et al. proposed that there was no codivergence between hantaviruses and their hosts and that the similarities between the phylogenies of hantaviruses and their hosts are the result of a more recent history of preferential host switching and local adaptation (46). Further studies are needed to determine if this is true.
The association between hantaviruses and their hosts is relatively specific, although host associations can include two or more animal species, such as Hantaan virus (HTNV) in mouse species Apodemus agrarius and Apodemus peninsulae (72) and Seoul virus (SEOV) in several rat species (Rattus norvegicus, Rattus flavipectus, Rattus losea, and Rattus nitidus) (61, 69, 70). Cross-species transmission (host switching or host jump) between more distantly related rodent hosts is suggested to occur during the evolution of several hantaviruses, e.g., Topografov virus (TOPV) (Lemmus/Microtus) (59) and Limestone Canyon virus (LSCV) (Peromyscus/Reithrodontomys) (47). Among the hantaviruses identified recently in insectivores, evidence for host switching between different families of insectivores within the order Soricomorpha was reported (4, 23). Recent studies show that hantaviruses carried by Murinae rodents are closely related to some Soricidae-carried viruses rather than to those associated with Cricetidae rodents (4, 23, 46), and Rockport virus (RKPV) in Scalopus aquaticus probably originated from rodents (24). In addition, host jumping of hantaviruses was observed frequently in rodents from the New World (10). Thus, cross-species transmission may be more common than previously shown and may be the important driving force in hantavirus evolution and speciation.
Dabieshan (or Da Bie Shan) virus (DBSV) was first isolated from Niviventer confucianus (Chinese white-bellied rat) captured in the Dabieshan mountainous area of Anhui Province, China (61). Genetically, the virus is more closely related to HTNV, carried by Apodemus mice (<15% amino acid difference), than to SEOV, carried by Rattus rats (>25% amino acid difference), where DBSV was initially recognized as a subtype of HTNV. As the Niviventer rat is more closely related to Rattus than to Apodemus, it was suggested that DBSV jumped to N. confucianus from Apodemus mice (61). Recently, a new lineage of DBSV was detected in N. confucianus captured in Yunnan Province, China (8). Interestingly, DBSV has not been found outside these two regions, whereas N. confucianus is widespread and abundant in 27 provinces of China (67). To further characterize the genetics of DBSV, we performed a survey in the mountainous areas in Wenzhou, Zhejiang Province, where hemorrhagic fever with renal syndrome (HFRS) is endemic (69) and performed (phylo)genetic analyses of the hantaviruses associated with N. confucianus. Additionally, to clarify the role of host switching and subsequent random and directional events in the speciation of Murinae-associated hantaviruses, we also analyzed the phylogenetic pattern of cross-species transmission from other Murinae-associated hantaviruses.
MATERIALS AND METHODS
Trapping of small animals and screening for hantaviruses.
During the spring and autumn of 2008, rodents were captured in the mountain areas in Wencheng (27°34′ to 27°59′N, 119°34′ to 120°15′E) and Yongjia (28°28′ to 28°33′N; 120°29′ to 120°36′E) counties in Wenzhou (Fig. 1), where more than 70% of the total area is mountainous. Small animals were trapped using a cage (20 cm by 20 cm by 56 cm) with a treadle release mechanism using deep-fried dough sticks as bait. The cages were set at 5-meter intervals according to the protocols described previously (34). Trapped animals were identified by morphological examination according to criteria reported by Chen (9) and further verified using sequence analysis of the cytochrome b (Cyt-b) gene (71). Lung and kidney tissue samples obtained from trapped animals were stored in liquid nitrogen. Hantavirus-specific antigen in lungs was detected using an indirect immunofluorescent assay (IFA) as described previously (68). In addition, hantaviral RNA was detected using reverse transcription-PCR (RT-PCR) as described by Klempa et al. (27).
Fig 1.
A map of China illustrating the locations of DBSV variants identified in Niviventer rats. The locations of trap sites where DBSV was detected in this study are shown as triangles (▲).
Preparation of viral RNA and rodent mtDNA.
TRIzol reagent (Invitrogen, Carlsbad, CA) was used to extract total RNA from the viral antigen-positive lung tissue samples according to the manufacturer's instructions. A genomic DNA extraction kit (SBS, Beijing, China) was used to extract mitochondrial DNA (mtDNA) from rat lung tissue samples according to the manufacturer's protocol.
Amplification of the viral genome and rodent cytochrome b (Cyt-b) gene and sequencing.
To amplify the L, M, and S segment sequences, primer P14 (49) was used to synthesize cDNA using avian myeloblastosis virus (AMV) reverse transcriptase (Promega, Beijing, China). Partial L segment sequences (nucleotides [nt] 3008 to 3325) were obtained using nested PCR with two primer pairs (27). The complete M and S segment sequences were amplified as described previously (74). The sequence of the rat Cyt-b gene was recovered using a standard PCR with primers CB1 and CB2 (31).
The obtained viral sequences and rat Cyt-b gene sequences were purified using agarose gel electrophoresis and sequenced using the ABI-PRISM dye termination sequencing kit and an ABI 373-A genetic analyzer.
Phylogenetic analyses.
The RDP, GENECONV, bootscan, maximum chi square, Chimera, SISCAN, and 3SEQ recombination detection methods used in RDP3 (33) were employed to detect potential recombinant viral sequences, identify likely parental viral sequences, and localize possible recombination breakpoints. The analyses were performed with default settings for the different test methods and a Bonferroni corrected P value cutoff of 0.05. When events were observed with two or more methods and with significant phylogenetic support, the viral sequences were considered recombinant and were excluded from this study.
The viral genome and rodent Cyt-b sequences were aligned using the Clustal W program (version 1.83). Their nucleotide and amino acid identities were calculated using the DNAStar program. The Metropolis-coupled Markov chain Monte Carlo (MCMC) method in MrBayes v3.1.2 was employed to estimate the phylogenetic trees. A general time-reversible (GTR) model with a gamma distribution of site rate heterogeneity and a proportion of invariable sites (GTR + Γ + I) was found to be the best model for the open reading frame (ORF) of the S segment and GTR + Γ to be best for the ORF of the M segment and the partial L segment; these were determined using jModelTest version 0.1 (45). Bayesian analysis consisted of 4 million MCMC generations sampled every 100 generations to ensure convergence across two runs of three hot chains and one cold chain. The performance was continued until the average standard deviation of split frequencies was less than 0.01 with a 25% burn-in. Convergence of parameters was assessed by calculating the effective sample size (ESS) using Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer) with an acceptable ESS threshold of over 200. The RAxML Blackbox web server was employed to construct maximum-likelihood (ML) trees (54).
The Dendroscope program (2.4) was used to visualize the tree files. Bayesian trees were readdressed to construct a tanglegram of rodent host and associated hantavirus using TreeMap software (2.0b) (21, 59). The Markov model in TreeMap was used to test significance by reconstructing 1,000 hantavirus trees with randomized branches and mapping these random trees onto the fixed host tree. When the level of congruence of the “real” virus tree was no more than that expected between randomly generated trees (P > 0.05 for both the number of codivergence events [CEs] and noncodivergence events [NCEs]), codivergence was not supported.
Estimating the rates of nucleotide substitution and the TMRCA of Murinae-associated hantaviruses.
The Bayesian MCMC approach available in the BEAST v1.6.0 software package (14) was used to estimate the rates of nucleotide substitution and divergence time (i.e., time to most recent common ancestor [TMRCA]) in Murinae-associated hantaviruses for the S and M segments, with uncertainty in all estimates reflected in the 95% high probability density (HPD) intervals. The ORF sequences of the S and M segments for the year of sampling that was available were used to generate the data sets with the recombinant sequences excluded. A total of 48 S and 40 M segment ORF sequences were compiled into data set 1 and data set 2, respectively (see Table S1 in the supplemental material).
The DAMBE program was used to determine the level of saturation at each codon position. When saturation was observed at the third position of the S segment and the first and third positions of the M segment, these positions were removed. BEAUTi v1.6.0 was used to generate BEAST XML input files with both the strict and uncorrelated log-normal distribution relaxed molecular clock model. When separate partitions of codon position sites were analyzed, we used GTR + Γ + I for the ORF sequence of the S segment and GTR + Γ for the M segment sequence determined by jModelTest version 0.1, and we used both the constant and the extended Bayesian Skyline trees before all analyses. Two independent runs were taken for each data set, with sampling every 1,000 generations. Each run was continued until the ESS of all parameters was larger than 200. Tracer v1.5 was employed to summarize, analyze, and visualize the resulting posterior sample. A maximum clade credibility (MCC) tree with a burn-in of 10% of the sampled trees was constructed to summarize the sample of trees produced by each BEAST run using the TreeAnnotator program (v1.6.0). The Bayes factor (BF) was estimated to determine the best clock and tree prior model with Tracer v1.5. An uncorrected log-normal distribution relaxed molecular clock model and extended Bayesian Skyline tree prior were used for both ORF sequences of the S and M segments according to the BF analysis data (see Table S2 in the supplemental material). The temporal signals in both data sets 1 and 2 were also evaluated. Under the best model, BEAST analyses were repeated for the data sets where sampling times were randomized running five times for the randomized data. When the mean rates and 95% HPDs from the real data set had major differences from those from resampled data, these samples were considered to contain a clear temporal structure.
We report posterior probabilities for the nodes in the MCC tree of >0.7 using FigTree v1.3.1 (http://tree.bio.ed.ac.uk). BEAST analyses were used to estimate rooted phylogenetic trees, where a time scale was incorporated according to rates of evolution estimated for each tree branch of the related viral sequences.
Determination of signature amino acid markers.
Based on the relationship of hantaviruses or their hosts, several groups were defined (Table 1; see Tables S3 and S4 in the supplemental material). Viral N and GPC proteins deduced from S and M segment sequences were aligned using MegAlign in the DNAStar program. If a specific amino acid exists in one species or one group but not in other species or group, this amino acid is considered a “signature amino acid” marker (or synapomorphy).
Table 1.
Comparison of amino acid signatures in the S and M segment amino acid sequences
| Species | Host(s) | No. of signature amino acids |
|
|---|---|---|---|
| S | M | ||
| SANGV | Hylomyscus simus | 18 | 70 |
| DOBV | Apodemus flavicllis | 1 | 10 |
| SAAV | A. agrarius | 2 | 14 |
| THAIV | Bandicota indica | 1 | 27 |
| SERV | R. rattus, R. tanezumi | 1 | 18 |
| SEOV | Rattus norvegicus | 3 | |
| GOUV | R. rattus | 4 | |
| DBSV | N. confucianus | 3 | 24 |
| HTNV | A. agrarius | 2 | 8 |
| ASV | A. peninsulae | 1 | 10 |
| SANGV + DOBV + SAAV | H. simus, Apodemus spp. | 14 | 28 |
| THAIV + SEOV + GOUV + DBSV + ASV + HTNV + SERV | B. indica, Rattus spp., Apodemus spp. | 8 | 19 |
| DOBV + SAAV | A. flavicllis, A. agrarius | 16 | 40 |
| SEOV + GOUV | R. norvegicus, R. rattus | 25 | 55 |
| SEOV + GOUV + SERV | R. norvegicus, R. rattus, R. tanezumi | 5 | |
| THAIV + SEOV + GOUV + SERV | R. norvegicus, R. rattus | 4 | 24 |
| ASV + HTNV | A. agrarius, A. peninsulae | 5 | 18 |
| DBSV + ASV + HTNV | N. confucianus, Apodemus spp. | 16 | 35 |
| ASV + HTNV + DOBV + SAAV | Apodemus spp. | 1 | 1 |
| SANGV + ASV + HTNV + DOBV + SAAV | Mice (H. simus, Apodemus spp.) | 1 | 3 |
| DBSV + SEOV + GOUV + THAIV + SERV | Rats (N. confucianus, Rattus spp., B. indica) | 2 | 2 |
| THAIV + SERV | B. indica, R. rattus, R. tanezumi | 19 | 36 |
Analysis of selection pressures.
The program Codeml in the PAML 4.4c software package was employed to detect positively selected sites in the N and GPC proteins (64). In each Murinae-associated hantavirus species, several sequences were selected for detection of positive selection. No sequence was identical to the others (57). Both data sets 1 and 2 were tested to determine if they were under positive selection. Three kinds of models (branch specific, site specific, and branch-site) were used to detect selective pressure among different branches and at different sites as described by Tang et al. (57). Comparing the models which do not allow for positive selection with the models in which positive selection is allowed, the likelihood ratio test (LRT) was used to find the presence of positively selected sites (3). It was assumed that all branches and sites in the phylogeny had the same ω ratio in the one-ratio model (M0) and that each branch in the phylogeny had an independent ω ratio in the free-ratio (FR) model. The difference in ω ratios could be determined by comparing M0 and FR to LRT. The discrete model (M8) was used to estimate ω for three classes of codons. The variability of selective pressure among sites was estimated by comparing M7 and M8. When positive selection (ω > 1) was found, posterior probabilities were estimated for site classes using the Bayes empirical Bayes (BEB) method (64).
The branch-site model, which assumes that the ω ratio varies both among sites and among branches (65, 66), was also used to find positively selected sites and was used when adaptive evolution occurred at a few time points and affected only a few amino acid residues. For branch-site model A, a given virus of interest (species or tentative species) was set as the foreground and the other viruses as the background. We assumed that selective constraint would change across sites in both the foreground and background, with a few sites that change only along foreground lineages. There were three ω ratios for the foreground (0 < ω0 < 1, ω1 = 1, and ω2 > 1) and two ω ratios for background (0 < ω0 < 1 and ω1 = 1) in branch-site model A. When positive selection (ω2 > 1) was found, posterior probabilities were estimated for site classes using the BEB method. The null model (model A′) was the same as model A except that ω2 = 1 was fixed. For the S and M segment sequences, we applied branch-site models to 14 groups on the trees for each. Thus, 0.0036 was used as the significance level for both S and M segment sequences.
RESULTS
Trapping of rodents and screening for hantaviruses.
From February to October 2008, a total of 149 small animals belonging to seven species were captured in the mountainous areas of Wenzhou, Zhejiang Province, China. Of these, 70 (41 N. confucianus, 15 A. agrarius, nine R. flavipectus, two R. losea, and three Suncus murinus) were trapped from Wencheng County and 79 (25 N. confucianus, 25 A. agrarius, two R. losea, one R. nitidus, 25 S. murinus, and one Callosciurus erythraeus) from Yongjia County. All small animals were screened for the presence of hantaviral antigens using IFA; hantaviral antigens were identified in the lung tissue samples from only three N. confucianus animals from Wencheng (samples Wencheng-Nc-427, Wencheng-Nc-469, and Wencheng-Nc-470) and four N. confucianus animals from Yongjia (samples Yongjia-Nc-15, Yongjia-Nc-38, Yongjia-Nc-58, and Yongjia-Nc-95). The results of the RT-PCR test were in full agreement with the IFA data.
Genetic analysis of viral sequences.
To characterize the N. confucianus-associated hantavirus found in Wenzhou, the complete hantaviral S and M sequences and also partial L segment sequences were recovered from all positive N. confucianus animals (see Table S1 in the supplemental material). The complete S segment has a total length of 1,725 nt, including 36 nt in the 5′ noncoding region (NCR), an ORF encoding the N protein of 429 amino acids, and the 399-nt-long 3′ NCR. Comparison of these complete S sequences showed they shared 95.3 to 99.9% nucleotide identity, which corresponded to 99.1 to 100% identity in the deduced amino acid sequences (see Table S5 in the supplemental material). Further comparison with other known hantaviruses showed that the novel strains were more closely related to DBSV strains Nc167 and AH09 identified in the Dabieshan mountain areas of Anhui Province (61) (88.7 to 89.7% nucleotide sequence identity and 98.4 to 98.8% amino acid sequence identity), followed by the DBSV strain YN509 identified in Yunnan Province (8) (83.4 to 84% nucleotide sequence identity and 97.9 to 98.4% amino acid sequence identity). In agreement with the previous studies (61), the DBSV variants carried by N. confucianus were more closely related to HTNV (78.0 to 79.1% nucleotide sequence identity and 92.1 to 93.5% amino acid sequence identity) than to SEOV (74.4 to 75.4% nucleotide sequence identity and 83.0 to 84.6% amino acid sequence identity) and other hantaviruses (34.6 to 79.9% nucleotide sequence identity and 45.9 to 93.2% amino acid sequence identity).
The M segment of the novel DBSV variants/strains has a total length of 3,623 nt (3,645 nt in Yongjia-Nc-38), including 46 nt of the 5′ NCR, an ORF for the GPC precursor of 1,133 aa, and 175 nt of the 3′ NCR (197 nt in Yongjia-Nc-38). Like the complete S segment sequences, the complete M and partial L segment sequences of these DBSV strains showed similar patterns of relatedness to other DBSV variants (from Dabieshan and Yunnan) and to other known hantaviruses (see Table S5 in the supplemental material).
Phylogenetic relationships of viral sequences.
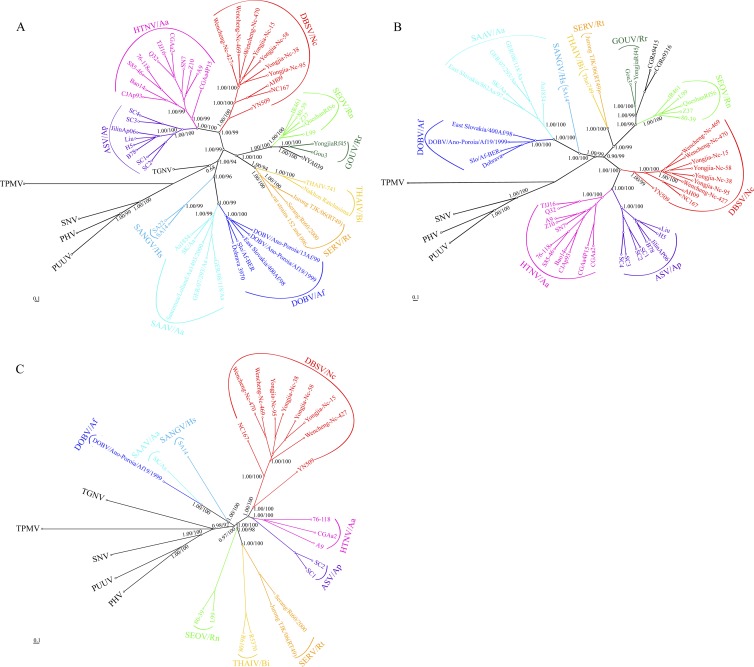
Phylogenetic analysis of the complete coding regions of the S and M segments and partial L segment sequences was performed using the Bayesian method implemented in the MrBayes v3.1.2 program package. The branching patterns of S, M, and L trees constructed using the classic ML method were similar to those based on the Bayesian method (>70% of bootstrap support values shown in Fig. 2). In the phylogenetic tree based on the S segment ORF, Wencheng and Yongjia variants from Zhejiang identified in this study clustered (Fig. 2A). They formed a well-supported group with the other DBSV strains (Nc167, AH09, and YN509) and showed three geographic lineages (Anhui, Zhejiang, and Yunnan Provinces). Notably, the strains identified in this study were more closely related to strains Nc167 and AH09 isolated in the Dabieshan mountain regions of Anhui Province, which shares a border with Zhejiang Province, than to strain YN509 detected in Yunnan Province, which is distant from Zhejiang Province (Fig. 1). In agreement with the previous studies (8, 61), DBSV shows a closer evolutionary relationship to HTNV and Amur/Soochong virus (ASV), carried by Apodemus mice, than to those carried by Rattus rats, even though Niviventer is more closely related to Rattus species than to Apodemus species (see Fig. 3). The topologies of the M and L trees were similar to that of the S tree (Fig. 2B and C). These results support the hypothesis of a cross-species transmission (host switching) of hantavirus between Niviventer rats and Apodemus mice (61), with a presumed direction of virus transmission not from Apodemus mice to Niviventer rats but in the opposite direction.
Fig 2.
Phylogenetic trees based on the entire coding regions of the genome sequences of Murinae-associated viruses, including the DBSV variants obtained in this study. The Bayesian/ML trees were based on the coding sequences of the S (A) and M (B) segments and the partial L (C) segment sequences. Numbers (>0.7/>70%) above or below the branches indicate posterior node probabilities or bootstrap values. Posterior node probabilities over 0.7 or a 70% bootstrap value was considered a node-supported value. All trees were rooted with Thottapalayam virus (TPMV). Scale bars represent the number of nucleotide substitutions per site. Rodent hosts abbreviations: Aa, Apodemus agrarius; Ap, Apodemus peninsulae; Af, Apodemus flavicollis; Nc, Niviventer confucianus; Rn, Rattus norvegicus; Rr, Rattus rattus; Rt, Rattus tanezumi; Bi, Bandicota indica; and Hs, Hylomyscus simus.
Fig 3.
Phylogenetic relationships between Niviventer rats captured in Wenzhou and other rodents with sequences found in GenBank. The Bayesian /ML trees were constructed using cytochrome b gene sequences. The sequences of Spalax ehrebergi were used as the outgroup. The sequences obtained in this study are shown in bold. Posterior node probabilities/bootstrap values (>0.7/>70%) are shown above or below the branches. The scale bar represents the number of nucleotide substitutions per site.
Phylogenetic relationships between Murinae-associated viruses and their rodent carriers.
Rodent migrations may have led to the current geographical distribution of hantaviruses (43). Phylogenetic analysis of the Cyt-b gene sequences of N. confucianus collected in this study may show the evolutionary relationships between DBSV and its host (N. confucianus). Overall, all available Cyt-b gene sequences of N. confucianus exhibited a high genetic diversity, up to 9.1%. The N. confucianus sequences obtained from Wenzhou showed 9.1% nucleotide divergence from the Yunnan sequences. As shown in Fig. 3, all Cyt-b gene sequences clustered and formed a well-supported N. confucianus clade that could be divided into two-well supported lineages (with posterior node probabilities of 0.98 and 1.00). Similar to the results of previous studies (55), N. confucianus appears to be more closely related to Rattus than to Apodemus, and all sequences were grouped into the two major groups corresponding to mice and rats (Fig. 3). The topologies of the trees constructed using the classical ML method and the Bayesian method were the same (Fig. 3 [only bootstrap values of >70% are shown]).
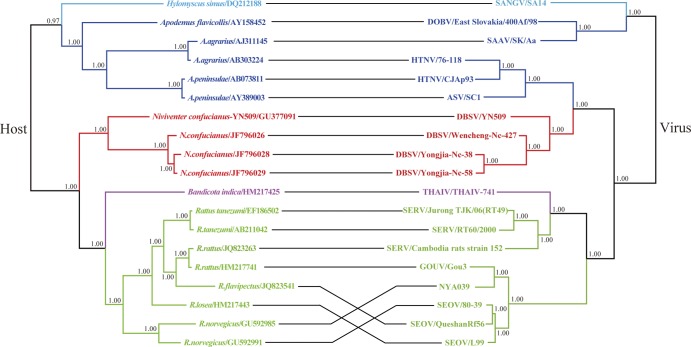
At present, the list of Murinae-associated hantaviruses defined by the International Committee on Taxonomy of Viruses includes five established species and five tentative species (42). To evaluate the evolutionary relationship between these species and their corresponding rodent hosts, TreeMap 2.0 was used to compare the viral S segment tree and the host Cyt-b gene tree. As shown in Fig. 4, the nodes of the viral phylogeny were similar in topology compared to the associated nodes of the Murinae host tree (P < 0.05) as measured by CE (P = 0.033 ± 0.004) and NCE (P = 0.019 ± 0.003) frequencies. All known Murinae-associated hantaviruses group together with some shrew-born hantaviruses, e.g., Tanganya virus (TGNV) from Africa (4, 23, 46). As the location of TGNV is nearest to the ancestral node separating the shrew-born hantaviruses from other species (Fig. 2) (46), the ancestor of the present known Murinae-associated rodent hantaviruses may have originated from the African Hylomyscus-like species. If this is true, at least nine cross-species transmission (host switching/host jump) events could be proposed to occur during evolution of the presently known Murinae-associated hantaviruses (Table 2). Six host switching events probably occurred between the different rodent genera: Hylomyscus mouse and Apodemus mouse, Hylomyscus (Apodemus) wood mouse and rat, Bandicota rat and Rattus rattus (R. tanezumi), Bandicota rat and R. norvegicus, Bandicota rat and Niviventer rat, and Niviventer rat and Apodemus mouse. Another three cross-species transmission events may have occurred between species within genus Apodemus or genus Rattus.
Fig 4.
Tanglegram constructed with the TreeMap2.0b program, illustrating the phylogenies of Murinae-associated hantaviruses and their rodent carriers. The host tree on the left was based on cytochrome b gene sequences, and the hantavirus tree on the right was based on the coding sequences of the S segment. The MrBayes v3.1.2 program package was used to construct the phylogenetic trees by the Bayesian method. Numbers (>0.7) above or below branches indicate posterior node probabilities. The reconciliation analysis showed a significant congruence between phylogenies of Murinae-associated hantaviruses and their hosts (P < 0.05).
Table 2.
Cross-species transmission events presumably occurring in the evolution of Murinae-associated hantaviruses
| Hosts |
Viruses |
Level | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species |
Genetic distance | Sister virusesa |
% Nucleotide/% amino acid difference between prospecies and neospecies |
|||||||
| Donor | Receptor | Prospecies | Neospecies | S | M | Gn | Gc | L | ||
| Hylomyscus simus | Apodemus agrarius | 0.192 | SANGV | SAAV | 22.1/11.4 | 27.1/19.5 | 28.9/22.9 | 24.6/15 | 27.9/14.3 | Genus |
| A. agrarius | A. flavicllis | 0.182 | SAAV | DOBV | 13.4/2.3 | 17.3/6.0 | 17.8/6.3 | 16.7/4.3 | 13.4/2.6 | Species |
| Bandicota indica | Rattus rattus | 0.144 | THAIV | SERV | 16.4/3.0 | 20.4/7.3 | 20.5/8.3 | 20.6/6.2 | 20.8/7.6 | Genus |
| B. indica | R. norvegicus | 0.155 | THAIV | SEOV | 22.6/13.5 | 25.9/18.5 | 27.0/21.8 | 24.6/14.4 | 25.8/15.2 | Genus |
| R. norvegicus | R. rattus | 0.126 | SEOV | GOUV | 12.0/1.4 | 15.4/3.4 | 15.6/3.7 | 15.4/3.1 | - | Species |
| B. indica | N. confucianus | 0.186 | THAIV | DBSV | 24.9/15.6 | 17.4/25.0 | 29.0/26.2 | 26.7/18.7 | 29.3/19.6 | Genus |
| N. confucianus | A. agrarius | 0.202 | DBSV | HTNV | 21.8/7.9 | 23.8/15.3 | 28.2/18.1 | 22.8/11.9 | 23.9/7.5 | Genus |
| A. agrarius | A. peninsulae | 0.169 | HTNV | ASV | 17.1/3.5 | 19.4/8.8 | 20.4/11 | 18.3/6.0 | 20.8/6.0 | Species |
Prospecies, presumable parental/sister species of new species; neospecies, presumable descendant species.
Rates of hantavirus evolution and diversification dates.
The date randomization test was used to determine if the structure and spread of the sequence ages were sufficient to estimate substitution rates and divergence times. The original estimate on data sets 1 and 2 was not recovered in the date-randomized data sets, suggesting that there was sufficient temporal structure in these data. Calculations based on data set 1 using the Bayesian MCMC analysis showed that the mean evolutionary rate in the Murinae-associated hantaviruses was 2.0 × 10−4 substitutions/site/year, with a 95% HPD from 1.1 × 10−4 to 2.9 × 10−4 substitutions/site/year (Table 3). As shown in Fig. 5A and Table 3, the estimated TMRCA were between 418 and 1,273 years before present (ybp) for all known DBSV variants based on the currently sampled genetic diversity, between 386 and 1,129 ybp for the HTNV variants, and between 78 and 199 ybp for the SEOV variants. Notably, the mean estimated divergence times were 1,590 ybp for DBSV and HTNV, 983 ybp for HTNV and ASV, 581 ybp for Dobrava-Belgrade virus (DOBV) and SAAV, and 515 ybp for Gou virus (GOUV) and SEOV.
Table 3.
Summary of evolution rates for the complete S segments
| Virus(es) | TMRCA (ybp) |
||||
|---|---|---|---|---|---|
| Mean | SE | Geometric mean | 95% HPD |
||
| Lower | Upper | ||||
| DBSV | 802 | 10.7 | 773 | 418 | 1,273 |
| HTNV | 754 | 9.8 | 732 | 386 | 1,129 |
| ASV | 536 | 6.7 | 518 | 280 | 836 |
| SEOV | 133 | 1.4 | 130 | 78 | 199 |
| GOUV | 206 | 2.3 | 199 | 107 | 318 |
| THAIV | 107 | 1.3 | 102 | 50 | 169 |
| SERV | 152 | 1.8 | 144 | 62 | 247 |
| DOBV | 149 | 1.8 | 145 | 80 | 233 |
| SAAV | 318 | 3.9 | 306 | 171 | 503 |
| SANGV | 63 | 0.8 | 60 | 29 | 107 |
| HTNV-ASV | 983 | 12.7 | 954 | 566 | 1,527 |
| HTNV-ASV-DBSV | 1,590 | 20.4 | 1,542 | 915 | 2,472 |
| DOBV-SAAV | 581 | 7.2 | 560 | 316 | 933 |
| DOBV-SAAV-SANGV | 1,614 | 21.6 | 1,549 | 824 | 2,601 |
| SEOV-GOUV | 515 | 3.6 | 506 | 400 | 738 |
| THAIV-SERV | 851 | 10.2 | 814 | 441 | 1,334 |
| SEOV-GOUV-THAIV-SERV | 1,896 | 24.8 | 1,836 | 1,058 | 2,932 |
| Mean rate | 2.0E−4 | 2.7E−6 | 1.9E−4 | 1.1E−4 | 2.9E−4 |
Fig 5.
Rooted phylogenetic trees with a molecular clock were reconstructed using the Bayesian MCMC method in BEAST based on the S segment sequences (A) and the M segment sequences (B). The divergence times, 95% high-probability density, and Bayesian posterior probabilities are given at the nodes leading to each major hantavirus-specific group.
The evolutionary rate estimated based on data set 2 was similar to that for the S segment (Fig. 5B; see Table S6 in the supplemental material). The evolutionary rates estimated in this study were in agreement with our recent results (29) and also those of Ramsden et al. (46); however, these rates appeared to be much higher than the previous estimates assuming a history of codivergence between hantaviruses and their hosts (∼10−6 to 10−7 substitutions/site/year) (20, 39, 52).
Genetic analysis of viruses originating from cross-species transmission.
For the viruses that may have originated via cross-species transmission, DBSV had >7% amino acid sequence difference in the complete N, GPC, and L protein sequences from a sister virus, HTNV (Table 2). An amino acid sequence difference of ≥7% in all three protein sequences was also observed between pairs of hantaviruses (Sangassou virus [SANGV] and SAAV, Thailand virus [THAIV] and SEOV, and THAIV and DBSV). However, a >7% amino acid sequence difference was found in GPC and L protein sequences between THAIV and Serang virus (SERV) and only in the GPC protein sequences between HTNV and ASV. Further, the difference was <7% in all three protein sequences in the pairs DOBV/SAAV and SEOV/GOUV.
Distinct hantaviruses, e.g., HTNV and Puumala virus (PUUV), possess specific “signature amino acids” in the N and GPC protein sequences, (52, 73). As shown in Table 1 and in Tables S3 and S4 in the supplemental material, eight of 10 species (all except SEOV and GOUV) have their own specific signature amino acid in the N protein sequence, and all 10 species have their own specific signature amino acid in the GPC protein sequence: SANGV (18/70), DOBV (1/10), SAAV (2/14), THAIV (1/27), SERV (1/18), SEOV (0/3), GOUV (0/4), DBSV (3/24), HTNV (2/8), and ASV (1/10). Remarkably, some signature amino acids are shared within the phylogenetic groups of hantavirus species, e.g., DBSV-HTNV-ASV (16/35), DOBV-SAAV (16/40), and SEOV-GOUV (25/55). Interestingly, some signature amino acids are also shared within the hantaviruses where their hosts are closely related, such as the mouse group (SANGV, ASV, HTNV, DOBV, and SAAV [1/3]) and the rat group (DBSV, SEOV, GOUV, THAIV, and SERV [2/2]), suggesting that these viruses may have had similar adaptive selection in mice or rats. In addition, the GPC protein may have faced a higher positive selective pressure than the N protein even if the nucleotide sequence divergences of the S and the M segments were similar (Tables 2 and 3; see Table S6 in the supplemental material).
Selection pressures in the hantavirus S and M segments.
Data sets 1 and 2 were used to analyze the adaptation of Murinae-associated hantaviruses to their respective hosts. The analyses of the branch-specific (FR) model showed that selective pressure varied along the branches for the S segment but not for the M segment (Table 4; see Tables S7 and S8 in the supplemental material). Further, the site-specific model (M8) did not identify any sites under positive selection, although the ω values were 3.00482 and 2.75162, respectively. For the S segment, no positive selection was found in all seven species as well when using the branch-site model. However, when closely related hantaviruses were grouped together, traces of positive selection were found in groups DOBV-SAAV, DOBV-SAAV-SANGV, GOUV-SEOV, and SERV-THAIV (Table 4; see Table S7 in the supplemental material). Both the LRT and BEB tests did not find these traces significant.
Table 4.
Detection of positively selected sites by maximum-likelihood estimation for 48 S segment and 40 M segment sequences of hantaviruses
| Model | Positively selected site(s) (position)a | |
|---|---|---|
| S segment | M segment | |
| Branch model (M0 vs FR) | Not allowed | Not allowed |
| Site model (M7 vs M8) | None | None |
| Branch-site model A (A′ vs A) with foreground group: | ||
| ASV | None | 144, 336 |
| DBSV | None | 285, 353, 364 |
| HTNV | None | 16, 51, 84 |
| SEOV | None | None |
| GOUV | None | 916 |
| DOBV | None | 932 |
| SAAV | None | 228, 932, 1076 |
| DOBV-SAAV | 256, 408 | 10, 56*, 207*, 309, 312, 336, 351*, 355, 364*, 430*, 653, 657, 1055*, 1071 |
| DOBV-SAAV-SANGV | 256, 408 | 14, 56, 101, 122, 218, 225, 232, 261, 309, 312, 322, 331, 332, 336, 351*, 364*, 583, 653, 657, 667*, 698, 703, 714, 908 , 932, 1055, 1089, 1114 |
| HTNV-ASV | None | 5, 101, 442, 885, 1025, 1060 |
| HTNV-ASV-DBSV | None | 55, 72*, 77, 223, 230, 232, 312, 333, 556, 699, 712, 892 |
| SEOV-GOUV | 234, 255, 259 | 52*, 214, 225, 228, 232 , 261, 326, 351, 502, 567, 569*, 667, 691, 703, 979, 1025, 1093 |
| THAIV-SERV | 74, 237, 253, 258, 263, 267, 272, 283, 286, 299, 412 | 43, 101, 261, 278, 351, 521, 703, 842, 911*, 1068 , 1071*, 1074 |
| THAIV-SERV-SEOV-GOUV | None | 90*, 100**, 108, 137, 191, 195, 198, 215**, 293**, 556, 650, 701, 703, 719*, 765, 793, 883**, 892, 956 |
* and **, positively selected sites identified with posterior probability P > 95% or P > 99%, respectively. The positive selection sites that are also the “signature amino acid” markers are shown in boldface.
For the M segment, three sites in the DBSV GPC protein may be subject to positive selection (ω = 40.35147 [P = 0.027]) when using the branch-site model. Presumable positive selection was observed in ASV, HTNV, and SAAV but not in SEOV, GOUV, and DOBV. The results, however, were not statistically significant. Similar to the observations made concerning the S segment, traces of positive selection were found in the following groups of closely related viruses: DOBV-SAAV, DOBV-SAAV-SANGV, ASV-HTNV, ASV-DBSV-HTNV, GOUV-SEOV, SERV-THAIV, and GOUV-SEOV-SERV-THAIV (Table 4; see Table S8 in the supplemental material). Finally, 68 of 139 (48.9%) positively selected sites in the N protein and GPC proteins appeared to be “signature amino acid” markers (Table 4).
DISCUSSION
The data here suggest that DBSV, identified in China, is a distinct species in the Hantavirus genus. Wang and colleagues first isolated the virus (strain Nc167) from Niviventer rats trapped in the Dabieshan mountain areas of Anhui Province in 2000 (61). They found that the virus was genetically related to but also distinct from HTNV and showed a 32-fold difference in titers from HTNV in a two-way cross-neutralization test. The virus is considered a lineage of HTNV generated by host switching from Apodemus mice to Niviventer rats (61). Recently, the virus was also found in Niviventer rats collected from Yunnan Province of China. This variant shared approximately 82% nucleotide sequence identity with strain Nc167 (8). At present, the virus carried by Niviventer rats is considered a tentative species in the Hantavirus genus and is designated DBSV (42). In this study, a new DBSV variant was found in the Niviventer rats captured in the mountainous areas of Wenzhou, Zhejiang Province, China. Analyses of the strains identified in this study and those found previously show that DBSV is carried by N. confucianus, in which no other hantaviruses have been found, and exhibits more than a 7% amino acid difference from any recognized hantaviruses in all three protein sequences. No reassortants have been found among the known variants of DBSV. Thus, DBSV meets all four criteria for species demarcation in the Hantavirus genus proposed by the International Committee on Taxonomy of Viruses (42). Further, the 15.4% amino acid difference in the M segment between DBSV and other recognized hantaviruses is also more than the 12% amino acid difference proposed by Maes et al. as the demarcation criterion of Hantavirus species (32). Thus, DBSV should be considered a distinct species of Hantavirus.
Generally, hantaviruses show close association with their respective rodent or insectivore host, and they are thought to present a good example of codivergence of a virus and a host (20, 21, 23, 25, 35, 38, 39, 43, 44, 52). In agreement with earlier studies (8, 61), our data show that DBSV is more closely related to HNTV and ASV than other Murinae-associated hantaviruses. For example, they share more signature amino acid than with any other Hantavirus species or group of species, suggesting that they share an ancestor. Moreover, our field trapping results also suggest that N. confucianus has more chances to contact A. agrarius than R. norvegicus, which was not found. Conversely, Niviventer rats have a closer evolutionary relationship with Rattus rats than with Apodemus mice (Fig. 3). Thus, the phylogeny of DBSV, HTNV, and SEOV is not consistent with their respective hosts (Fig. 4). One possible explanation for such a discrepancy would be that a host-switching event between rats and mice occurred. However, the data presented here did not support the direction from Apodemus mouse to Niviventer rat (61). With the discovery of novel Murinae-associated hantaviruses, the direction and time of the host switching will become clear.
Cross-species transmission among the hantaviruses was reported previously. Cross-species transmission of HTNV from A. agrarius to A. peninsulae, and even to Rattus, Niviventer, and Mus species, has been reported in China (61, 72, 73). Identification of SEOV in rat species other than R. norvegicus has also been reported (61, 69, 70). Other studies also report one hantavirus carried by several rodent species (39, 48, 50). Comparing the phylogenies of hantaviruses and their hosts (Fig. 2 and 4; Table 2), we conclude that most Murinae-associated viruses may originate via host switching. Recent work by Ramsden and colleagues suggests that there is no codivergence between hantaviruses and their hosts and that the congruence between the phylogenies of hantaviruses and their hosts is the result of a more recent history of preferential host switching and local adaptation (46). Here, our data show that cross-species transmission plays an important role in the speciation of the known Murinae-associated hantaviruses.
Both stochastic events (e.g., genetic drift or bottleneck) and deterministic processes (e.g., selection or adaptation) are occurring in a population. In a large population, even weak selection on a mutant may play an important role in its evolution (1). However, viruses can be particularly susceptible to the effects of genetic drift because interhost transmission frequently involves population size “bottlenecks” that occur independently of viral fitness (12). Earlier studies found genetic drift effects during both intrahost and interhost infection of viruses (2, 6, 15). All hantaviruses except SAAV and GOUV, which presumably originate from cross-species transmission, show >7% sequence differences from their presumable parental/sister species in at least one of the encoded proteins (Table 2). Further, each species of Murinae-associated hantavirus has its own specific signature amino acid markers (Table 1). However, our analyses show that only a few amino acid sites may have been under weak positive selection in the M segment proteins for ASV, DBSV, DOBV, GOUV, HTNV, and SAAV (Table 4; see Table S8 in the supplemental material). Hantavirus may infect only a small number of individuals when it first jumps into a new rodent population, where genetic drift effects may play a major role in the fixation of virus mutations in a new rodent host.
Although ASV, GOUV, SAAV, and SERV probably all originate from cross-species transmission, the number and type of amino acid differences between these viruses and their sister viruses varied greatly (Table 2). The age of the virus in the new host after cross-species transmission may be responsible for the variation, which may be caused by the accumulation of fixation of random mutations in viruses in the new hosts.
Usually, the accumulation of adaptive changes can facilitate the successful colonization in a new host species. HIV (60), influenza virus (13), and SARS coronavirus (57) provide good examples. The closer the donor and recipient host species are in phylogenetic space, the fewer changes are likely to be required for adaptation to the new host (19, 26). Different characteristics of hantaviruses have emerged as adaptations to the distinct genetic environments of their rodent hosts (43). Each hantavirus species or genetic lineages of the same species can possess specific amino acid “signatures,” such as those in the N protein sequences for Puumala virus (52) or the GPC protein sequences for HTNV (73). Similar to previous studies (20, 62), we did not find convincing evidence for positive selection in Murinae-associated hantaviruses. However, each species of Murinae-associated hantavirus has its own specific signature amino acid markers (Table 1; see Tables S3 and S4 in the supplemental material). More nonsynonymous substitutions have been found in the GPC protein (Table 2), which is known to mediate cell attachment and fusion and to be the major element involved in induction of neutralizing antibodies during hantavirus infection (25). Further, presumably adaptive evolution was detected in the GPC protein, especially when the closely related sister viruses were considered a group. Therefore, adaptation to host species (codivergence) may have occurred and facilitated the speciation and the further genetic diversity of hantaviruses as well.
Despite advances in understanding the patterns and processes of microevolution in RNA viruses, little is known about the determinants of viral diversification at the macroevolutionary scale, particularly the processes by which viral lineages diversify into different “species” (26). Several studies show that within host populations both pathogen and host are able to adapt in response to the interactions, resulting in coevolution (18, 56, 58, 63). However, there is also a debate on how the microevolutionary changes can influence the patterns of speciation of the interacting species at the macroevolutionary levels (26). For hantaviruses, there is an ongoing dispute concerning coevolution/coadaptation of viruses and their hosts (43, 46). The data presented here indicate that cross-species transmission, the subsequent genetic drift effect, and adaptation in the new host population may substantially contribute to the speciation of the Murinae-associated hantaviruses.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Chinese Ministry of Science and Technology (grants 2002DIB40095 and 2003BA712A08-02) and by the State Key Laboratory for Infectious Disease Prevention and Control (2011SKLID101).
Footnotes
Published ahead of print 1 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ali A, et al. 2006. Analysis of genetic bottlenecks during horizontal transmission of Cucumber mosaic virus. J. Virol. 80:8345–8350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alizon S, Luciani F, Regoes RR. 2011. Epidemiological and clinical consequences of within-host evolution. Trends Microbiol. 19:24–32 [DOI] [PubMed] [Google Scholar]
- 3. Anisimova M, Bielawski JP, Yang Z. 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18:1585–1592 [DOI] [PubMed] [Google Scholar]
- 4. Arai S, et al. 2008. Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides). Proc. Natl. Acad. Sci. U. S. A. 105:16296–16301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bell DM, et al. 2009. Pandemic influenza as 21st century urban public health crisis Emerg. Infect. Dis. 5:1963–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Betancourt M, Fereres A, Fraile A, GarcíA-Arenal F. 2008. Estimation of the effective number of founders that initiate an infection after aphid transmission of a multipartite plant virus. J. Virol. 82:12416–12421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bourhy H, et al. 1999. Ecology and evolution of rabies virus in Europe. J. Gen. Virol. 80:2545–2557 [DOI] [PubMed] [Google Scholar]
- 8. Cao ZW, et al. 2010. Genetic analysis of a hantavirus strain carried by Niviventer confucianus in Yunnan province, China. Virus Res. 153:157–160 [DOI] [PubMed] [Google Scholar]
- 9. Chen HX. 1987. Classification and identification of medical animals. The Institute of Epidemiology and Microbiology, Chinese Academy of Preventive Medicine; Beijing, China [Google Scholar]
- 10. Chu YK, Owen RD, Jonsson CB. 2011. Phylogenetic exploration of hantaviruses in Paraguay reveals reassortment and host switching in South America. Virol. J. 8:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cleaveland S, Haydon DT, Taylor L. 2007. Overviews of pathogen emergence: which pathogens emerge, when and why? Curr. Top. Microbiol. Immunol. 315:85–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeFilippis VR, Villarreal LP. 2001. Virus evolution, p 353–370 In Knipe DM, Howley P. (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 13. Dos RM, Tamuri AU, Hay AJ, Goldstein RA. 2011. Charting the host adaptation of influenza viruses. Mol. Biol. Evol. 28:1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214 doi:10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Edwards CT, et al. 2006. Population genetic estimation of the loss of genetic diversity during horizontal transmission of HIV-1. BMC Evol. Biol. 6:28 doi:10.1186/1471-2148-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao F, et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- 17. Guan Y, et al. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278 [DOI] [PubMed] [Google Scholar]
- 18. Herniou EA, Olszewski JA, O'Reilly DR, Cory JS. 2004. Ancient coevolution of baculoviruses and their insect hosts. J. Virol. 78:3244–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holmes EC. 2009. The evolution and emergence of RNA viruses. Oxford Univ Press, Oxford, United Kingdom [Google Scholar]
- 20. Hughes AL, Friedman R. 2000. Evolutionary diversification of protein-coding genes of hantaviruses. Mol. Biol. Evol. 17:1558–1568 [DOI] [PubMed] [Google Scholar]
- 21. Jackson AP, Charleston MA. 2004. A cophylogenetic perspective of RNA-virus evolution. Mol. Biol. Evol. 21:45–57 [DOI] [PubMed] [Google Scholar]
- 22. Jonsson CB, Figueiredo LT, Vapalahti O. 2010. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 23:412–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kang HJ, et al. 2009. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii). Virology 388:8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang HJ, Bennett SN, Hope AG, Cook JA, Yanagihara R. 2011. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J. Virol. 85:7496–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khaiboullina SF, Morzunov SP, St Jeor SC. 2005. Hantaviruses: molecular biology, evolution and pathogenesis. Curr. Mol. Med. 5:773–790 [DOI] [PubMed] [Google Scholar]
- 26. Kitchen A, Shackelton LA, Holmes EC. 2011. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 108:238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klempa B, et al. 2007. Novel hantavirus sequences in Shrew, Guinea. Emerg. Infect. Dis. 13:520–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuiken T, et al. 2006. Host species barriers to influenza virus infections. Science 312:394–397 [DOI] [PubMed] [Google Scholar]
- 29. Lin XD, et al. 2012. Migration of rats resulted in the worldwide distribution today: evidence for China as a radiation center of the present Seoul virus. J. Virol. 86:972–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reference deleted.
- 31. Luo J, et al. 2004. Molecular phylogeny and biogeography of Oriental voles: genus Eothenomys (Muridae, Mammalia). Mol. Phylogenet. Evol. 33:349–362 [DOI] [PubMed] [Google Scholar]
- 32. Maes P, et al. 2009. A proposal for new criteria for the classification of hantaviruses, based on S and M segment protein sequences. Infect. Genet. Evol. 9:813–820 [DOI] [PubMed] [Google Scholar]
- 33. Martin DP, et al. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mills JN, Childs JE, Ksiazek TG, Peters CJ, Velleca WM. 1995. Methods for trapping and sampling small mammals for virologic testing. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 35. Monroe MC, et al. 1999. Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerg. Infect. Dis. 5:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morens DM, Folkers GK, Fauci AS. 2004. The challenge of emerging and re-emerging infectious diseases. Nature 430:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morse SS. 1995. Factors in the emergence of infectious diseases. Emerg. Infect. Dis. 1:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morzunov SP, et al. 1998. Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J. Virol. 72:57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nemirov K, Henttonen H, Vaheri A, Plyusnin A. 2002. Phylogenetic evidence for host switching in the evolution of hantaviruses carried by Apodemus mice. Virus Res. 90:207–215 [DOI] [PubMed] [Google Scholar]
- 40. Reference deleted.
- 41. Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Plyusnin A, et al. 2011. Bunyaviridae, p 693–709 In King AMQ, Lefkowitz EJ, Adams MJ, Carstens EB. (ed), Virus taxonomy: 9th report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA [Google Scholar]
- 43. Plyusnin A, Morzunov SP. 2001. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 256:47–75 [DOI] [PubMed] [Google Scholar]
- 44. Plyusnin A, Vapalahti O, Vaheri A. 1996. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 77:2677–2687 [DOI] [PubMed] [Google Scholar]
- 45. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 46. Ramsden C, Holmes EC, Charleston MA. 2009. Hantavirus evolution in relation to its rodent and insectivore hosts: no evidence for codivergence. Mol. Biol. Evol. 26:143–153 [DOI] [PubMed] [Google Scholar]
- 47. Sanchez AJ, Abbott KD, Nichol ST. 2001. Genetic identification and characterization of limestone canyon virus, a unique Peromyscus-borne hantavirus. Virology 286:345–353 [DOI] [PubMed] [Google Scholar]
- 48. Schlegel M, et al. 2009. Dobrava-belgrade virus spillover infections, Germany. Emerg. Infect. Dis. 15:2017–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmaljohn CS, Jennings GB, Hay J, Dalrymple JM. 1986. Coding strategy of the S genome segment of Hantaan virus. Virology 155:633–643 [DOI] [PubMed] [Google Scholar]
- 50. Schmidt-Chanasit J, et al. 2010. Extensive host sharing of central European Tula virus. J. Virol. 84:459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sharp PM, et al. 2001. The origins of acquired immune deficiency syndrome viruses: where and when? Philos.Trans. R. Soc. B Biol. Sci. 356:867–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sironen T, Vaheri A, Plyusnin A. 2001. Molecular evolution of Puumala hantavirus. J. Virol. 75:11803–11810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reference deleted.
- 54. Stamatakis AP, Hoover J, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758–771 [DOI] [PubMed] [Google Scholar]
- 55. Steppan SJ, Adkins RM, Spinks PQ, Hale C. 2005. Multigene phylogeny of the Old World mice, Murinae, reveals distinct geographic lineages and the declining utility of mitochondrial genes compared to nuclear genes. Mol. Phylogenet. Evol. 37:370–388 [DOI] [PubMed] [Google Scholar]
- 56. Switzer WM, et al. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376–380 [DOI] [PubMed] [Google Scholar]
- 57. Tang X, et al. 2009. Differential stepwise evolution of SARS coronavirus functional proteins in different host species. BMC Evol. Biol. 9:52 doi:10.1186/1471-2148-9-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Timms R, Read AF. 1999. What makes a specialist special? Trends. Ecol. Evol. 14:333–334 [DOI] [PubMed] [Google Scholar]
- 59. Vapalahti O, et al. 1999. Isolation and characterization of a hantavirus from Lemmus sibiricus: evidence for host switch during hantavirus evolution. J. Virol. 73:5586–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wain LV, et al. 2007. Adaptation of HIV-1 to its human host. Mol. Biol. Evol. 24:1853–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang H, et al. 2000. Genetic diversity of hantaviruses isolated in China and characterization of novel hantaviruses isolated from Niviventer confucianus and Rattus rattus. Virology 278:332–345 [DOI] [PubMed] [Google Scholar]
- 62. Woelk CH, Holmes EC. 2002. Reduced positive selection in vector-borne RNA viruses. Mol. Biol. Evol. 19:2333–2336 [DOI] [PubMed] [Google Scholar]
- 63. Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin R. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32:569–577 [DOI] [PubMed] [Google Scholar]
- 64. Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555–556 [DOI] [PubMed] [Google Scholar]
- 65. Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19:908–917 [DOI] [PubMed] [Google Scholar]
- 66. Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22:2472–2479 [DOI] [PubMed] [Google Scholar]
- 67. Zhang RZ, et al. 1997. Distribution of mammalian species in China, p 185–211 China Forestry Publishing House, Beijing, China [Google Scholar]
- 68. Zhang YZ, et al. 2009. Seoul virus and hantavirus disease, Shenyang, People's Republic of China. Emerg. Infect. Dis. 15:200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang YZ, et al. 2010. Hantaviruses in small mammals and humans in the coastal region of Zhejiang Province, China. J. Med. Virol. 82:987–995 [DOI] [PubMed] [Google Scholar]
- 70. Zhang YZ, Zou Y, Fu ZF, Pluusnin A. 2010. Hantavirus infections in humans and animals, China. Emerg. Infect. Dis. 16:1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang YZ, et al. 2007. Detection of phylogenetically distinct Puumala-like viruses from red-grey vole Clethrionomys rufocanus in China. J. Med. Virol. 79:1208–1218 [DOI] [PubMed] [Google Scholar]
- 72. Zhang YZ, et al. 2007. Isolation and characterization of hantavirus carried by Apodemus peninsulae in Jilin, China. J. Gen. Virol. 88:1295–1301 [DOI] [PubMed] [Google Scholar]
- 73. Zou Y, et al. 2008. Molecular diversity of hantaviruses in Guizhou, China: evidence for origin of Hantaan virus from Guizhou. J. Gen. Virol. 89:1987–1997 [DOI] [PubMed] [Google Scholar]
- 74. Zou Y, et al. 2008. Genetic characterization of hantaviruses isolated from Guizhou, China: evidence for spillover and reassortment in nature. J. Med. Virol. 80:1033–1041 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.