Abstract
Human endogenous retroviruses (HERVs), which are remnants of ancestral retroviruses integrated into the human genome, are defective in viral replication. Because activation of HERV-K and coexpression of this virus with HIV-1 have been observed during HIV-1 infection, it is conceivable that HERV-K could affect HIV-1 replication, either by competition or by cooperation, in cells expressing both viruses. In this study, we found that the release efficiency of HIV-1 Gag was 3-fold reduced upon overexpression of HERV-KCON Gag. In addition, we observed that in cells expressing Gag proteins of both viruses, HERV-KCON Gag colocalized with HIV-1 Gag at the plasma membrane. Furthermore, HERV-KCON Gag was found to coassemble with HIV-1 Gag, as demonstrated by (i) processing of HERV-KCON Gag by HIV-1 protease in virions, (ii) coimmunoprecipitation of virion-associated HERV-KCON Gag with HIV-1 Gag, and (iii) rescue of a late-domain-defective HERV-KCON Gag by wild-type (WT) HIV-1 Gag. Myristylation-deficient HERV-KCON Gag localized to nuclei, suggesting cryptic nuclear trafficking of HERV-K Gag. Notably, unlike WT HERV-KCON Gag, HIV-1 Gag failed to rescue myristylation-deficient HERV-KCON Gag to the plasma membrane. Efficient colocalization and coassembly of HIV-1 Gag and HERV-K Gag also required nucleocapsid (NC). These results provide evidence that HIV-1 Gag heteromultimerizes with HERV-K Gag at the plasma membrane, presumably through NC-RNA interaction. Intriguingly, HERV-K Gag overexpression reduced not only HIV-1 release efficiency but also HIV-1 infectivity in a myristylation- and NC-dependent manner. Altogether, these results indicate that Gag proteins of endogenous retroviruses can coassemble with HIV-1 Gag and modulate the late phase of HIV-1 replication.
INTRODUCTION
Human endogenous retrovirus (HERV) sequences comprise approximately 8% of human DNA (5, 35, 53). Although almost all HERV genomes appear to lack intact open reading frames (ORFs) and are therefore likely defective in replication, some of them, such as HERV-K113, have complete ORFs for all viral proteins (52). Nonetheless, HERV-K113 is poorly expressed and is not capable of replication (6, 9, 37). Recently, using bioinformatics approaches, two groups reconstructed infectious HERV-K sequences (18, 37). In one approach, a consensus sequence of HERV-K113 and closely related well-preserved HERV-Ks was determined and termed HERV-KCON (37). HERV-K molecular clones and their derivatives that encode these reconstructed sequences have become powerful tools to examine the biology of these ancient retroviruses.
While HERV-K is categorized as a betaretrovirus because of its sequence similarity to mouse mammary tumor virus (MMTV), in contrast to other betaretroviruses, which assemble particles in the cytoplasm (type B/D), HERV-K forms particles at the plasma membrane (PM) (type C), like human immunodeficiency virus type 1 (HIV-1) and murine leukemia virus (MLV) (8). Particle formation of retroviruses is driven by the precursor polyprotein Gag. HIV-1 Gag consists of four major domains, matrix (MA), capsid (CA), nucleocapsid (NC), and p6, as well as two spacer peptides, SP1 and SP2 (4). Using HERV-KCON (37) and another HERV-K113 derivative (termed oricoHERV-K113) in which 5 postinsertion mutations were reverted (21), recent studies unambiguously determined the domain organization of HERV-K Gag (21, 32). HERV-K Gag also has four domains, MA, CA, NC, p15, as well as three short peptide sequences, SP1, QP1, and QP2 (21, 32). In HIV-1, MA is required for Gag targeting and binding to the PM. CA and NC domains are essential for Gag multimerization. The p6 domain contains a late domain motif, Pro-Thr-Ala-Pro (PTAP), which recruits the cellular ESCRT complexes that facilitate release of virions. These domains give rise to individual mature Gag proteins upon proteolytic cleavage mediated by viral protease, which occurs during or immediately after virus particle release. The functions of these domains are less well understood for HERV-K Gag than in HIV-1 Gag, although some functional motifs are shared by both Gag proteins (21, 32). Notably, a single postinsertion mutation, which was corrected in both HERV-KCON and oricoHERV-K113, is responsible for the defect in assembly of HERV-K113 (26). Thus, these restored HERV-K113 sequences serve as good models for studying assembly of HERV-K Gag.
Since all human cells harbor HERV-K genomes and potentially express HERV-K proteins, HERV-K or some of its components might be coexpressed with HIV-1 in the same cell. In this regard, it is notable that a number of studies showed a correlation between increased expression of HERV-K and HIV-1 infection. Several groups have detected antibody responses to HERV-K in a majority of HIV-1-positive patients but not in uninfected donors (41, 54). Similarly, T cell responses to HERV-K were observed in HIV-1-infected patients but not in healthy donors (19, 49, 51). Furthermore, upregulation of HERV-K RNA was detected (14–17, 19, 40) in plasma samples of HIV-1-infected individuals, in HIV-1-infected T cells (17, 38), and in HIV-1 Tat-transfected T cells (23). In addition to RNA, HERV-K Gag proteins were also observed to increase upon HIV-1 infection (15, 17) and HIV-1 Tat transfection (23). These reports collectively suggest that HIV-1 infection enhances the HERV-K expression in T cells. Thus, it is possible that HERV-K Gag induced by HIV-1 infection coexists with HIV-1 Gag in the same host cells. However, the impact of such coexpression on HIV-1 replication is unknown.
Coinfection and coexistence of two different lentiviruses in the same host have been observed naturally (2, 24, 31). In phylogenetic analysis, some of the primate lentiviruses were found to be recombinants of two distinct parental viruses, which must have arisen from coinfection of these two viruses in a single cell (3, 47). In cells coinfected with two such distinct parental viruses, two different Gag proteins would be expressed in the same cells, which raises a possibility that different but related Gag proteins coassemble into the same virions. Indeed, in cells coexpressing both HIV-1 and HIV-2, HIV-1 and HIV-2 Gag proteins colocalized at the PM and coassembled into the same virions (10). Gag proteins of more distantly related retroviruses also coassemble when modified by addition of a heterologous membrane binding motif (7) or exchange of CA (1, 36). However, coassembly between native Gag proteins of retroviruses of different genera, such as HIV-1 and HERV-K, has not been observed. Because diverse retroviruses rely on similar host factors, such as phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2] and ESCRT proteins, consequences that arise from coexpression of HIV-1 and HERV-K Gag proteins in the same cell might rather be competition for cellular cofactors and inhibition of assembly of one virus.
We hypothesized that if expression of HERV-K Gag is induced by HIV-1 infection, the HERV-K Gag might compete with HIV-1 Gag in the same host cell. In this study, we report that HERV-K Gag indeed inhibited the HIV-1 release efficiency and infectivity when Gag from HERV-KCON was coexpressed with HIV-1. Unexpectedly, HERV-K Gag colocalized with HIV-1 Gag at the PM and coassembled into the same virion. We found that both membrane binding and NC-mediated RNA binding of Gag are required for coassembly between HIV-1 and HERV-K Gag as well as for inhibition of progeny virus release and infectivity. To our knowledge, this is the first example in which a retroviral Gag coassembles with, and inhibits assembly of, another retrovirus Gag protein coexpressed in the same cells.
MATERIALS AND METHODS
Plasmids.
All HERV-K Gag constructs used in this study are based on HERV-KCON (37), unless otherwise specified. pCRVI/HERV-K/GagPro was a kind gift from P. Bieniasz. This plasmid encodes the HERV-KCON GagPro sequence following a cytomegalovirus (CMV) promoter and a sequence corresponding to the HIV-1 5′ untranslated region (nucleotides [nt] 428 to 785 in pNL4-3), along with ORFs encoding HIV-1 Rev, Tat, and Vpu. pCRVI/HERV-K/Gag-Flag, pCRVI/HERV-K/Gag-Venus, and pCRVI/HERV-K/Gag-mRFP were created from pCRVI/HERV-K GagPro by replacing the PR sequences with the Flag, Venus, and monomeric red fluorescent protein (mRFP) sequences, respectively. pCRVI/HERV-K/GagPro#09 is based on pCRVI/HERV-K/GagPro, and the sequence spanning from NC to Pro (nt 2753 to 3820) was replaced with a corresponding sequence of HERV-K-encoding cDNA derived from Jurkat cells. pCRVI/HERV-K/Gag/1GA-Flag, pCRVI/HERV-K/Gag/delNC-Flag, and pCRVI/HERV-K/Gag/PTAP(−)-Flag were generated by PCR mutagenesis. pCRVI/HERV-K/Gag/1GA-Flag encodes a Flag-tagged Gag derivative that contains a Gly-to-Ala substitution at the MA N terminus and therefore lacks myristoylation. pCRVI/HERV-K/Gag/delNC-Flag lacks a sequence corresponding to the NC zinc finger domain and basic amino acids (nt 2625 to 2804). pCRVI/HERV-K/Gag/PTAP(−)-Flag encodes Gag that contains a substitution at p15 amino acid residue 106 (Thr to Ala). pNL4-3/PR(−) was described previously (28). pNL4-3/delNC and pNL4-3/delNC/PR(−) were kind gifts from D. Ott (45). pCRVI/HIV-1/Gag-Flag, pCRVI/HIV-1/Gag-Venus, and pCRVI/HIV-1/Gag-mRFP were constructed using the pCRVI/HERV-K/Gag plasmids, and the Gag region was replaced with the HIV-1 Gag sequence derived from pNL4-3. pCRVI/HERV-K/Gag-Flag/psi(−), which lacks the HIV-1 packaging signal, and its derivatives with changes in Gag sequences were generated by removing a fragment between two SacI sites (nt 835 to 947). pCRVI/MLV/Gag-Flag and pCRVI/MLV/Gag-Venus were created from pCRVI/HERV-K Gag-Flag and pCRVI/HERV-K Gag-Venus by replacing the HERV-K Gag sequences with the MLV Gag sequences encoded by pNCA (12).
Cells.
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Lonza) supplemented with 5% fetal bovine serum (FBS) (DMEM-5). The Molt4 T cell line was cultured in RPMI 1640 (Gibco) supplemented with 10% FBS (RPMI-10). CEM-GFP cells that harbor a green fluorescent protein (GFP) gene driven by the HIV-1 long terminal repeat (LTR) promoter were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Jacques Corbeil (22) and maintained in RPMI-10 containing 500 μg/ml Geneticin (Invitrogen).
RT assay.
HeLa cells were cotransfected with pNL4-3 and the indicated pCRVI plasmids using Lipofectamine 2000 according to the manufacturer's instructions. The ratio of pNL4-3 to pCRVI plasmids was 10:1 unless indicated otherwise. At 16 h posttransfection, the supernatants were filtered through 0.45-μm filters, and virions in the supernatants were pelleted by ultracentrifugation (35,000 rpm in a Thermo Scientific TH660 rotor, 4°C, 45 min). The pelleted viruses were resuspended in RPMI-10. The amount of virus was determined by a reverse transcriptase (RT) assay as described previously (55).
p24 ELISA.
Molt4 cells were cotransfected with pNL4-3 and the indicated pCRVI plasmids using an Amaxa system according to the manufacturer's instructions. The ratio of pNL4-3 to pCRVI plasmids was 10:1. At 2 days posttransfection, the supernatants were filtered through 0.45-μm filters, and virions in the supernatants were pelleted by ultracentrifugation (35,000 rpm, 4°C, 45 min). Gag proteins in the cell and virion lysates were quantified by p24 enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (ZeptoMetrix).
Virus release assay.
A virus release assay was performed as previously described (43). Briefly, HeLa cells were cotransfected with pNL4-3 and the indicated pCRVI plasmids. At 16 h posttransfection, virions in the supernatants were collected and pelleted as described above for the RT assay. Cells and virions were lysed with 0.5% Triton X lysis buffer (50 mM Tris-HCl [pH 7.5] containing 0.5% Triton X-100, 300 mM NaCl, and 10 mM iodoacetamide with protease inhibitor cocktail [Roche]). Gag proteins in the cell and virion lysates were detected by immunoblotting using HIV Ig (NIH AIDS Research and Reference Reagent Program), mouse monoclonal anti-Flag antibody (Sigma), rabbit polyclonal antihemagglutinin (anti-HA) antibody (Santa Cruz Biotech), or anti-HERV-K Gag antibody (HERM1831) (Austral) as the primary antibody. Alexa Fluor-488-conjugated anti-human IgG antibody (Invitrogen) and horseradish peroxidase (HRP)-conjugated anti-mouse Ig and anti-rabbit Ig antibodies (Amersham) were used as secondary antibodies. The fluorescence signal of the secondary antibody was quantified by using a Typhoon Trio imager (GE Healthcare). Detection using an HRP-conjugated secondary antibody was performed using the SuperSignal West Pico chemiluminescence detection kit (Thermo Scientific).
Fluorescence microscopy.
HeLa cells were plated in 8-well chamber slides (Nunc) at 1 day before transfection at 4.6 × 104 cells/well. At 16 h posttransfection, HeLa cells cotransfected with plasmids encoding yellow fluorescent protein(YFP)- and mRFP-tagged Gag proteins were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (PBS) for 30 min at room temperature, washed once with PBS, and stained with 4,6-diamidino-2-phenylindole (DAPI) (1 μg/ml) (Invitrogen) for 5 min at room temperature. The cells were then washed with PBS and mounted in Fluoromount-G (Dako). The images of 20 fields were recorded using a Zeiss LSM 700 laser-scanning confocal microscopy. Colocalization between YFP- and mRFP-tagged Gag was quantified using the ZEN software (Zeiss), with which we calculate the R strength of correlation. R = 1 represents perfect colocalization, and R = 0 represents random distributions of fluorescence intensities.
Virus particles prepared as described above were resuspended in PBS and plated on the poly-l-lysine-coated microscope slides (Polysciences). After 30 min, viruses on the slides were fixed and mounted as described above. The images of 20 fields were recorded using a Nikon TE2000 microscope and analyzed as described above.
Analysis of Gag processing in virions.
HeLa cells were cotransfected with pNL4-3/PR(−) and pCRVI plasmids encoding HERV-K Gag or GagPro. At 14 h posttransfection, the culture medium was changed to RPMI 1640 lacking both methionine (Met) and cysteine (Cys) and supplemented with 2% FBS [RPMI-2(−Met/−Cys)] and incubated for 30 min. Subsequently, these cells were metabolically labeled with [35S]Met/Cys (Perkin-Elmer) in fresh RPMI-2(−Met/−Cys) for 2 h. Virus lysates were prepared in 0.5% Triton X lysis buffer as described above for virus release assay. Viral proteins were detected by SDS-PAGE followed by autoradiography.
Coimmunoprecipitation assay.
At 14 h posttransfection, cells were labeled with [35S]Met/Cys as described above. Virus lysates were prepared as described above and subjected to immunoprecipitation with HIV-Ig.
Infectivity analysis of coassembled virions.
To measure the infectivity of coassembled virions, 5.6 × 105 HeLa cells were plated in 6-well plates and cotransfected the next day with 3.64 μg of pNL4-3 and 0.36 μg of pCRVI constructs that lack the HIV-1 Psi sequence present in the original pCRVI constructs. At 16 h posttransfection, virus-containing supernatants were filtered through a 0.45-μm filter and pelleted by ultracentrifugation (35,000 rpm, 4°C, 45 min). The pelleted viruses were resuspended in RPMI-10. Amounts of viruses were determined by an RT assay. For analysis of virus infectivity using CEM-GFP cells, 5 × 105 cells were inoculated with virus stocks normalized by RT activity (200,000 cpm of RT activity) for 2 h. To block the second-round infection, the CD4-blocking antibody Leu3a (0.25 μg/ml) (BD Biosciences) and the reverse transcriptase inhibitor zidovudine (AZT) (1 μM) were added to the medium at 12 h postinfection. At 2 days postinfection, cells were fixed in 4% paraformaldehyde in PBS and analyzed using a FACSCanto flow cytometer and FlowJo software version 8. 7. 1.
RESULTS
The release efficiency of HIV-1 is reduced by coexpression of HERV-K Gag.
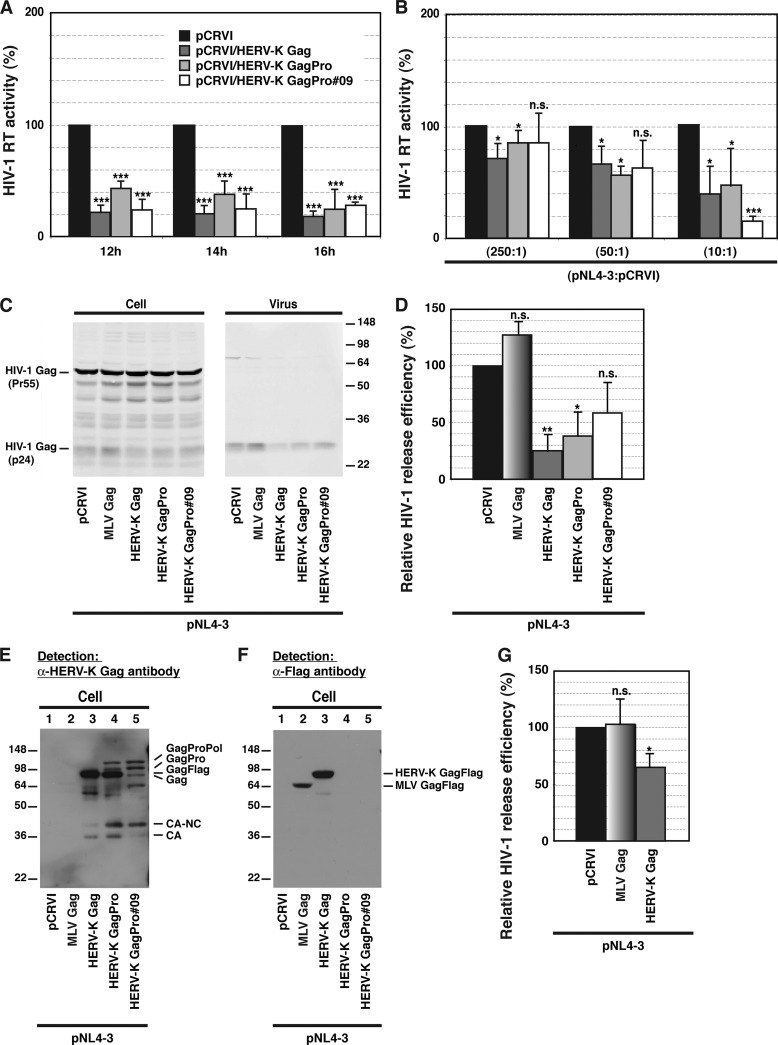
It has been observed that in T cells, HERV-K mRNA transcription and Gag expression increase upon HIV-1 infection (13–17, 19, 38, 40, 49, 51) and HIV-1 Tat transfection (23). These data raise an interesting possibility that HIV-1 Gag and HERV-K Gag would compete with each other for assembly cofactors such as PI(4,5)P2 or ESCRTs in HIV-1-infected cells. To investigate the impact of HERV-K Gag coexpression on HIV-1 release efficiency, we transfected HeLa cells with an HIV-1 molecular clone along with HERV-K Gag-encoding plasmids. The HIV-1 RT activities in supernatants collected 12, 14, and 16 h after transfection were 3- to 5-fold reduced upon coexpression of Flag-tagged Gag (Gag-Flag) of HERV-KCON (Fig. 1A). Transfection of plasmids encoding GagPro of HERV-KCON or HERV-K/GagPro#09, which is based on a HERV-K-encoding cDNA isolated from Jurkat cells, similarly reduced HIV-1 RT activities released in supernatants (Fig. 1A). The reduction of HIV-1 RT activity was less severe when the amount of the expression plasmids for HERV-K Gag-Flag or GagPro used for cotransfection was decreased, suggesting that the impact depends on the expression levels of HERV-K Gag-Flag and GagPro (Fig. 1B). The release efficiency of HIV-1 was consistently 3- to 5-fold reduced by overexpression of HERV-K Gag-Flag and GagPro but not MLV Gag-Flag (Fig. 1C and D). To confirm the expression of HERV-K and MLV Gag, cell lysates were examined by immunoblotting using anti-HERV-K Gag antibody or anti-Flag antibody. The band for transfected HERV-K Gag precursor was detected at a position corresponding to 80 kDa. Compared to that in cells expressing HERV-K GagPro, the ratio of HERV-K GagPro- and Gag-sized bands in cells expressing HERV-K/GagPro#09 was altered due to one nucleotide insertion that causes a frameshift near the end of the gag ORF (Fig. 1E, compare lane 5 with lane 4). This construct was still efficient in reducing the virus release measured by RT activity (Fig. 1A and B), but its impact on the virus release efficiency was somewhat decreased (Fig. 1C and D). Although expression of Flag-tagged MLV Gag was lower than that of HERV-K Gag-Flag in these experiments (Fig. 1F), even when HERV-K Gag expression was lowered to the level of MLV Gag expression by reducing the amount of transfected plasmid, it still reduced the release efficiency of HIV-1 Gag (data not shown). In T cells, the release of HIV-1 was moderately but significantly reduced by overexpression of HERV-K Gag but not by that of MLV Gag (Fig. 1G). Altogether, these results indicate that expression of HERV-K Gag but not MLV Gag inhibits the release of HIV-1.
Fig 1.
Expression of HERV-K Gag reduces HIV-1 release efficiency. (A) HeLa cells were cotransfected with HIV-1 molecular clone pNL4-3 and the indicated pCRVI plasmids at a 10:1 ratio. HIV-1 RT activity of virus pellets from transfected cells was measured at 12, 14, and 16 h posttransfection. An empty vector, pCRVI, was used as a control. pCRVI/HERV-K Gag-Flag expresses Flag-tagged HERV-K Gag. pCRVI/HERV-K GagPro and pCRVI/HERV-K GagPro#09 express both HERV-K Gag and GagPro precursors, leading to the presence of precursor HERV-K Gag and processed HERV-K Gag. The sequence of HERV-K GagPro#09 is derived from cDNA isolated from Jurkat cells. (B) HeLa cells were cotransfected with pNL4-3 and the indicated plasmids at different ratios. HIV-1 RT activity was measured at 14 h posttransfection. (C) Cell and viral lysates from cotransfected cells were subjected to SDS-PAGE and analyzed by immunoblotting with HIV Ig. (D) Virus release efficiency was calculated by dividing the amount of p24CA in the viral lysate by the total amount of Gag in the cell and viral lysates. (E and F) Cotransfected cell lysates were subjected to SDS-PAGE and analyzed by immunoblotting using anti-HERV-K Gag antibody (E) and anti-Flag antibody (F). Note that only pCRVI/HERV-K Gag and pCRVI/MLV Gag encode Flag-tagged Gag proteins. (G) HIV-1 p24CA levels in cell and virus lysates from cotransfected T cells were measured by ELISA at 2 days posttransfection. Virus release efficiency was calculated by dividing the amount of Gag/p24CA in the viral lysate by the total Gag/p24CA in the cell and viral lysates. Data from three independent experiments are shown as means ± standard deviations. P values were determined using Student's t test. *, P < 0.01; **, P < 0.001; ***, P < 0.0001; n.s., not significant.
HERV-K Gag colocalizes with HIV-1 Gag at the plasma membrane.
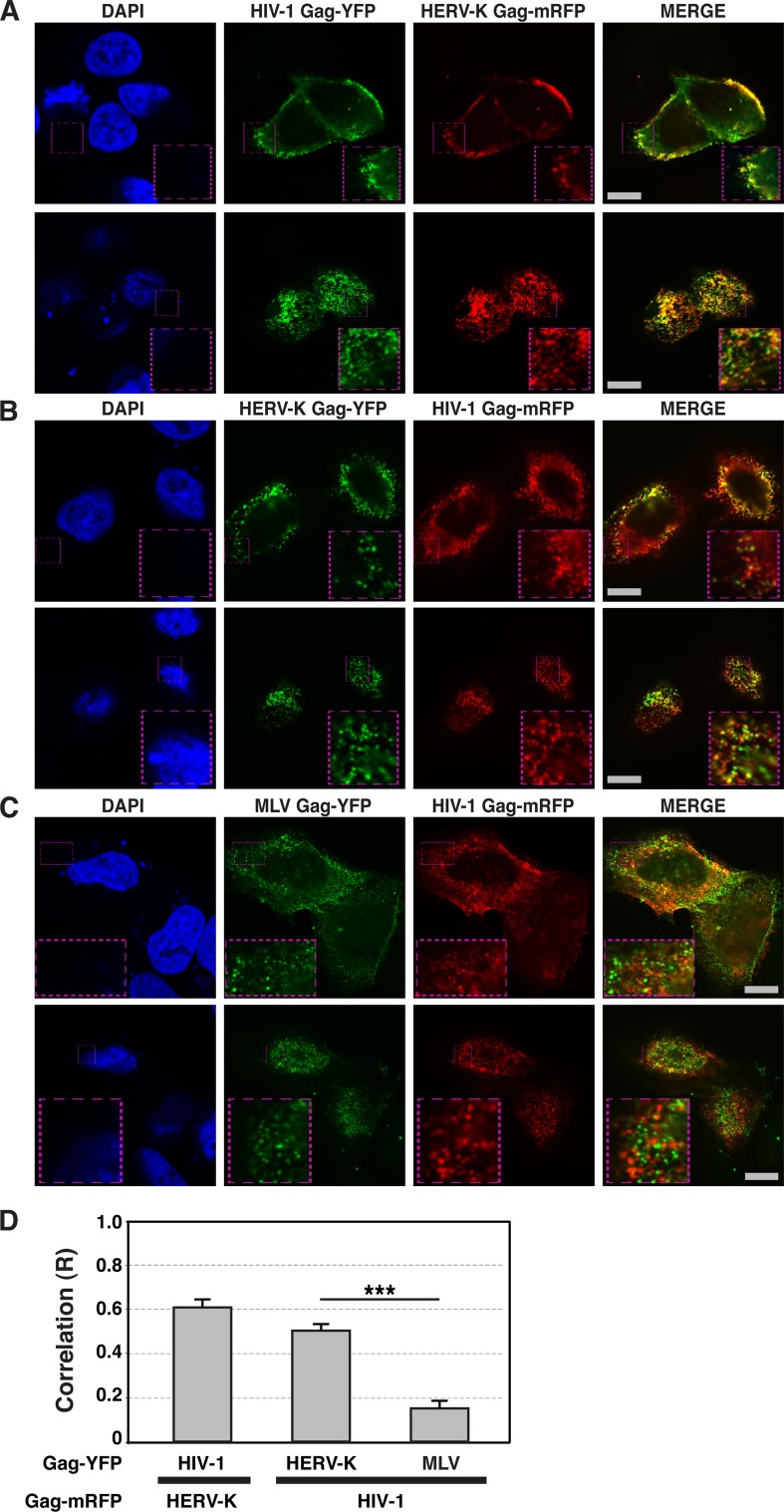
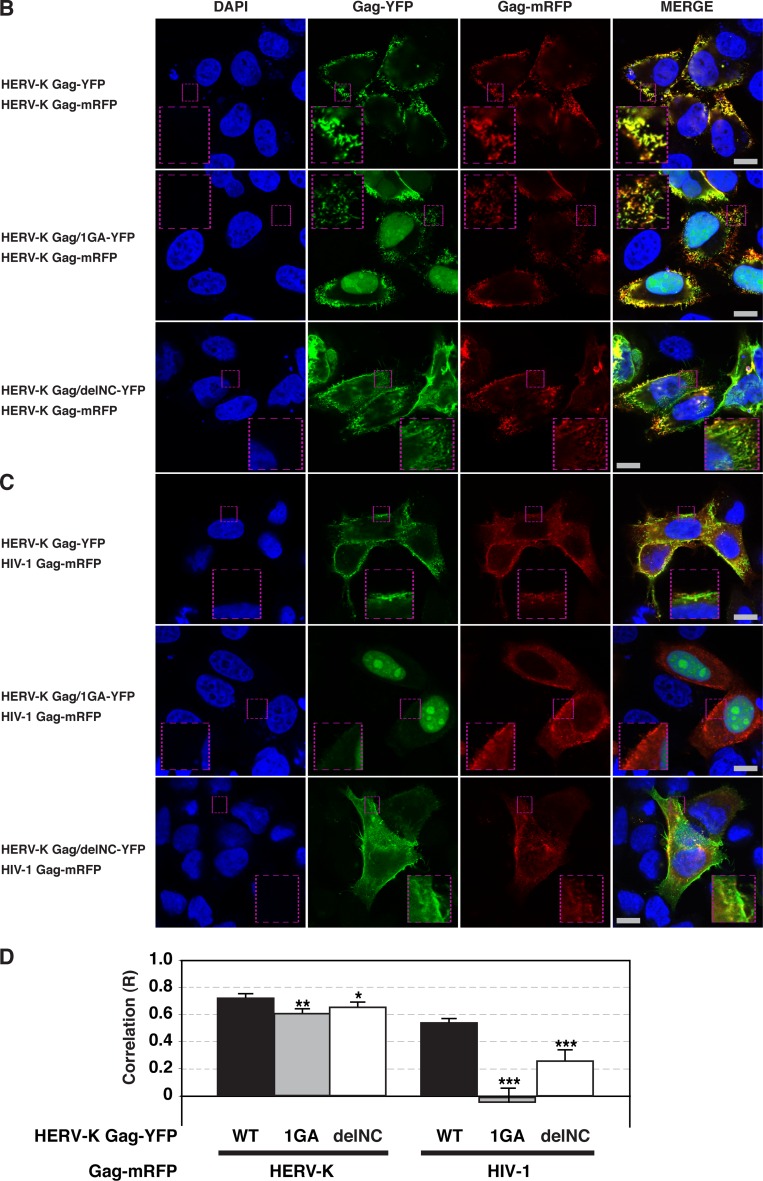
Since HIV-1 release efficiency is reduced by HERV-K Gag coexpression, it seemed possible that HERV-K Gag localizes at the PM similarly to HIV-1 Gag, which in turn could inhibit the binding of HIV-1 Gag to the PM via competition for host factors. To determine whether HERV-K Gag and HIV-1 Gag localize at the PM in the same cells, we examined localization of these Gag proteins on the cell surface using fluorescence microscopy. HeLa cells were cotransfected with plasmids encoding HERV-K Gag and HIV-1 Gag, each with different fluorescent protein tags, and observed after fixation at 16 h posttransfection. As observed previously (37), HERV-K Gag showed punctate localization at the PM. Unexpectedly, we found a substantial colocalization between HIV-1 Gag and HERV-K Gag puncta at the PM in coexpressing cells (Fig. 2A, B, and D). In contrast, consistent with a report by others (1), colocalization between HIV-1 Gag and MLV Gag was significantly lower even though both Gag proteins were at the PM (Fig. 2C and D). These results indicate that HERV-K Gag specifically colocalizes with HIV-1 Gag and suggests the possibility that HERV-K Gag might coassemble with HIV-1 Gag into the same virions.
Fig 2.
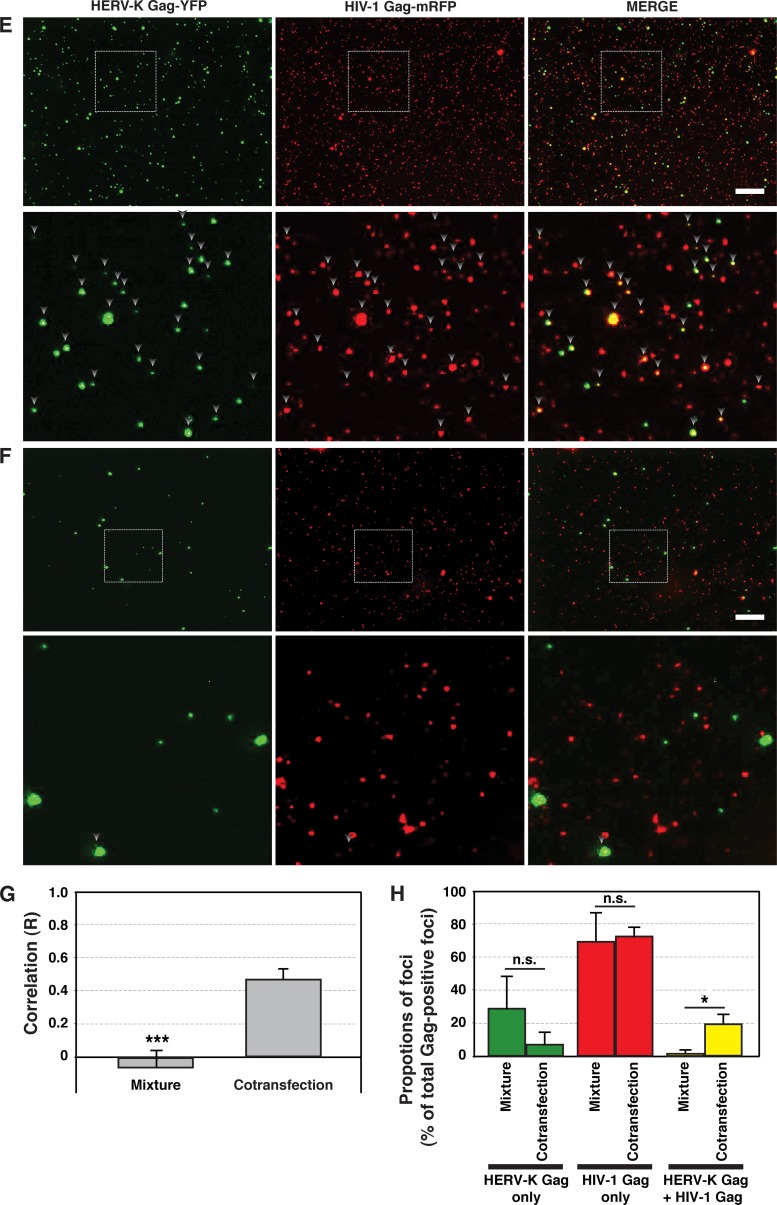
HIV-1 Gag colocalizes with HERV-K Gag, but not MLV Gag, at the PM. HeLa cells coexpressing YFP-tagged (green) and mRFP-tagged (red) Gag proteins of different retroviruses were fixed, stained with DAPI, and examined using fluorescence microscopy at 16 h after cotransfection (A to C). Top panels, Images acquired at the midsection of the cells. Bottom panels, Images acquired at the top of the cells. Bars, 10 μm. (D) The R strength of correlation between fluorescence intensities of pairs of the indicated Gag-fluorescent protein chimeras was calculated for cells coexpressing these Gag proteins. Data from 21 to 27 cells are shown as means ± standard errors of the means (SEM). P values were determined using Student's t test. ***, P < 0.0001. (E) HeLa cells were cotransfected with plasmids encoding HERV-K Gag-YFP and HIV-1 Gag-mRFP. The supernatants of cotransfected HeLa cells were concentrated by ultracentrifugation at 16 h posttransfection. Particulate materials positive for fluorescently tagged Gag in the supernatants were examined with a fluorescence microscope as described in Materials and Methods. (F) HeLa cells were separately transfected with plasmids encoding HERV-K Gag-YFP and HIV-1 Gag-mRFP. The supernatants of transfected HeLa cells were mixed and concentrated by ultracentrifugation at 16 h posttransfection. Particulate materials positive for fluorescently tagged Gag in the supernatants were examined with a fluorescence microscope as described in Materials and Methods. Magnified views of boxed areas in the top-row images are shown in the bottom rows in panels E and F. Arrowheads indicate foci showing both green and red fluorescence signals. Bars, 10 μm. (G) The R strength of correlation between fluorescence intensities of HERV-K Gag-YFP and HIV-1 Gag-mRFP was calculated. Data from 15 images are shown as means ± SEM. P values were determined using Student's t test. ***, P < 0.0001. (H) The foci containing YFP only, mRFP only, or both YFP and mRFP were counted, and proportions relative to total Gag-positive foci were calculated. Data from three independent images are shown as means ± standard deviations. P values were determined using Student's t test. *, P < 0.01; n.s., not significant.
HERV-K Gag coassembles with HIV-1 Gag into virions.
To investigate whether HIV-1 Gag and HERV-K Gag coassemble into single virions, we examined virus-like particles (VLPs) using fluorescence microscopy. VLP suspensions were prepared from supernatants of HeLa cells cotransfected with plasmids encoding HERV-K Gag-YFP and HIV-1 Gag-mRFP, and fluorescent foci that represent VLPs and other Gag-positive particulate materials (e.g., membrane fragments) were examined using a fluorescence microscope. The R strength of correlation between HERV-K Gag-YFP and HIV-1 Gag-mRFP signals was about 0.5 (Fig. 2E and G). Approximately 20% of fluorescent foci contained both HERV-K Gag-YFP and HIV-1 Gag-mRFP (Fig. 2E and H). These results support the possible coassembly between HERV-K Gag and HIV-1 Gag. Aggregation of VLPs during pelleting is unlikely to fully explain these results, because virus particles in a mixture of culture supernatants of HeLa cells singly expressing HERV-K Gag-YFP or HIV-1 Gag-mRFP showed predominantly single-color fluorescence (Fig. 2F, G, and H). However, it is still possible that HERV-K and HIV-1 particles assembled at the same PM areas may be tethered to each other by molecules such as tetherin, thereby forming two-color spots in microscopy analysis.
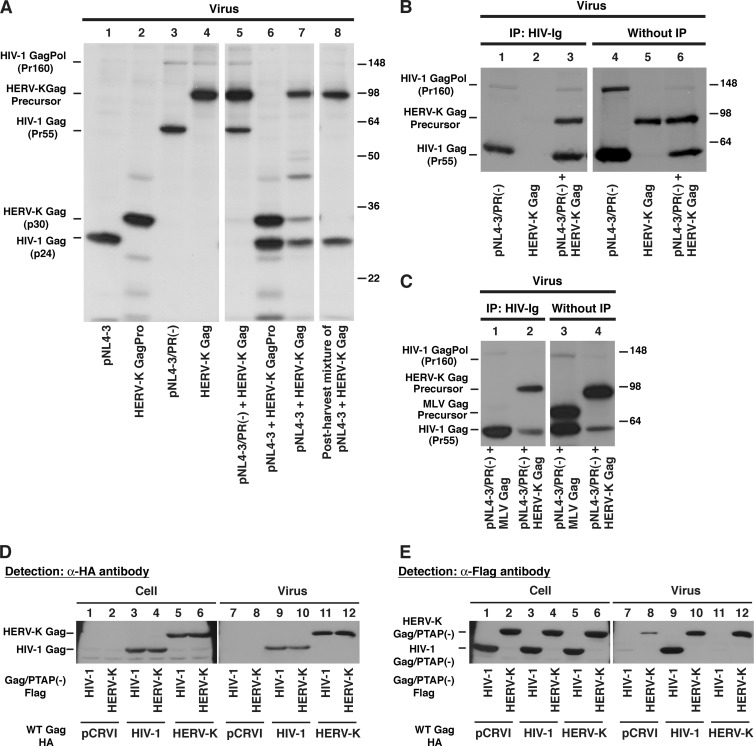
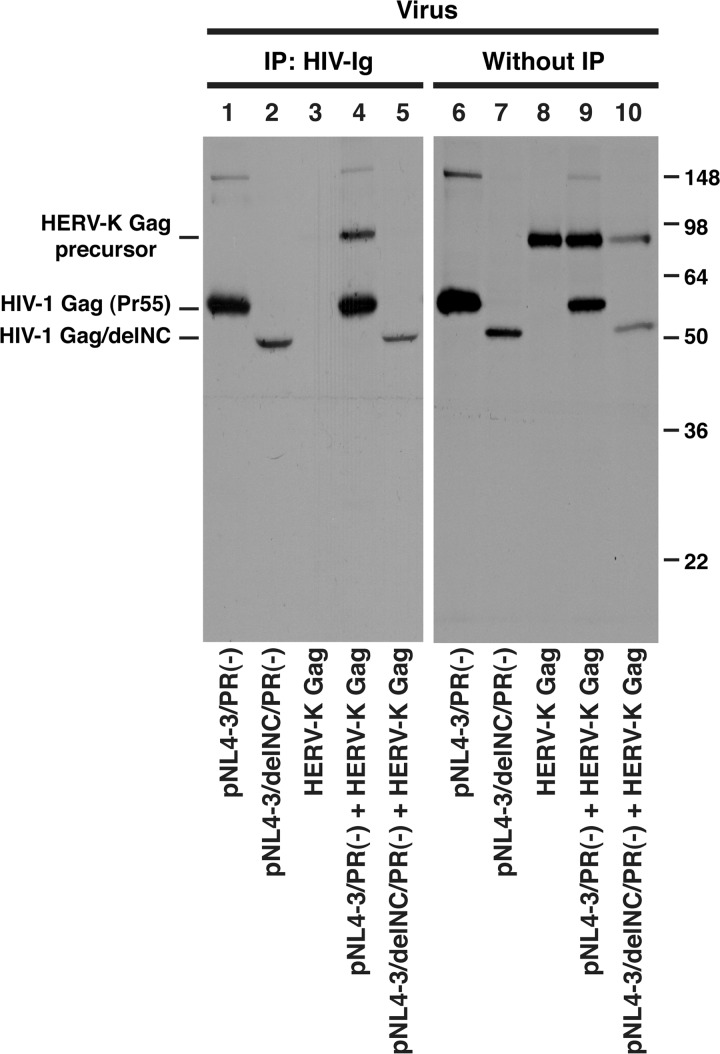
To assess the coassembly of HERV-K Gag and HIV-1 Gag into the same virions unambiguously using a biochemical approach, we examined HERV-K Gag processing by HIV-1 protease in virus particles. A previous report showed that HIV-1 protease is capable of cleaving HERV-K Gag in vitro (33). Therefore, if HERV-K Gag coassembles with HIV-1 virion components, HIV-1 protease is likely to cleave the HERV-K Gag in the same virion. When the supernatants of cells individually transfected with pNL4-3 or a plasmid encoding HERV-K Gag were mixed postharvest, HERV-K Gag in the resulting virus lysate was not cleaved by HIV-1 protease (Fig. 3A, lane 8). This result indicates that HIV-1 protease does not process HERV-K Gag after addition of the lysis buffer. In contrast, when HeLa cells were cotransfected with pNL4-3 and a plasmid encoding HERV-K Gag, HERV-K Gag in VLP pellets was processed by HIV-1 protease (Fig. 3A, lane 7) as indicated by the presence of the p30 band that represents mature HERV-K Gag (Fig. 3A, lanes 2 and 6). These results indicate that HERV-K Gag is copackaged with HIV-1 GagPol into the same virions.
Fig 3.
HIV-1 Gag coassembles with HERV-K Gag into the same virions and rescues the release of HERV-K Gag/PTAP(−). (A) HeLa cells were cotransfected with HIV-1 molecular clones and/or pCRVI plasmids encoding the indicated HERV-K constructs and incubated for 14 h. After 2 h of metabolic labeling with [35S]Met/Cys, the viral lysates were prepared as described in Materials and Methods and subjected to SDS-PAGE. pNL4-3/PR(−) is a protease-deficient HIV-1NL4-3 molecular clone. Note that coexpression of pNL4-3 (lane 7) but not pNL4-3/PR(−) (lane 5) leads to processing of HERV-K Gag. (B and C) For panel B, HeLa cells were cotransfected with pNL4-3/PR(−) and pCRVI (lanes 1 and 4), pUC19 and pCRVI/HERV-K Gag (lanes 2 and 5), or pNL4-3/PR(−) and pCRVI/HERV-K Gag (lanes 3 and 6). For panel C, HeLa cells were cotransfected with pNL4-3/PR(−) and the pCRVI plasmids encoding either MLV Gag (lanes 1 and 3) or HERV-K Gag (lanes 2 and 4). Viral lysates were prepared as for panel A. The viral lysates were subjected to immunoprecipitation (IP) using HIV Ig (left) prior to SDS-PAGE or directly subjected to SDS-PAGE (right). Gels were exposed to X-ray films for 2 days (left) or 1 day (right). (D and E) HeLa cells were cotransfected with HA-tagged WT Gag and Flag-tagged Gag/PTAP(−) lacking the late domain sequence. At 16 h posttransfection, the cell and viral lysates were subjected to SDS-PAGE and analyzed by immunoblotting with anti-HA antibody (D) and anti-Flag antibody (E). The results shown are representative of data from three independent experiments. Note that release of HERV-K PTAP(−) Gag is rescued by coexpression of WT HIV-1 Gag (compare lane 10 with lane 8).
To determine whether HERV-K Gag interacts with HIV-1 Gag in coassembled VLPs, coimmunoprecipitation of HERV-K Gag with HIV-1 Gag in the virus lysates was measured using HIV Ig (Fig. 3B and C). In the absence of HIV-1 Gag, HERV-K Gag was not immunoprecipitated by HIV Ig (Fig. 3B, lane 2). However, in the virus lysates from HeLa cells coexpressing both HIV-1 Gag and HERV-K Gag, HERV-K Gag was coimmunoprecipitated along with HIV-1 Gag by HIV Ig (Fig. 3B, lane 3). Nonspecific precipitation of HIV-1 Gag was observed with a preparation of normal human IgG available to us, which made it difficult to assess the specificity of coimmunoprecipitation between HIV-1 Gag and HERV-K Gag (data not shown). Thus, to assess the specificity, we examined whether MLV Gag is coimmunoprecipitated with HIV-1 Gag. As shown in Fig. 3C, in contrast to HERV-K Gag, MLV Gag was not detected in coimmunoprecipitated materials, suggesting that HERV-K Gag but not MLV Gag coassembles and interacts with HIV-1 Gag in the same virions.
The release of HERV-K Gag/PTAP(−) is rescued by HIV-1 Gag coexpression.
To analyze functionally whether HIV-1 Gag interacts with HERV-K Gag in cells, we examined the impact of HIV-1 Gag coexpression on a HERV-K Gag mutant [HERV-K Gag/PTAP(−)] that is expected to be defective in virus release. The HERV-K Gag p15 domain contains a late domain motif, Pro-Thr-Ala-Pro (PTAP) (21, 32), which, as in HIV-1 Gag (20, 25, 28), is likely to be important for efficient budding. We reasoned that if HIV-1 Gag interacts with HERV-K Gag at the PM, the release of HERV-K Gag/PTAP(−) would be rescued by wild-type (WT) HIV-1 Gag. In the cells, expression and release levels of WT HIV-1 Gag or WT HERV-K Gag were similar regardless of coexpression of HIV-1 Gag/PTAP(−) or HERV-K Gag/PTAP(−) (Fig. 3D). As expected, HERV-K Gag/PTAP(−) was defective in virus release (Fig. 3E, lane 8), and this defect was reversed upon coexpression of WT HERV-K Gag (Fig. 3E, lane 12). Similarly, HIV-1 Gag/PTAP(−) was also rescued by WT HIV-1 Gag (Fig. 3E, lane 9). Notably, we found that HERV-K Gag/PTAP(−) release was also rescued by coexpression of WT HIV-1 Gag (Fig. 3E, lane 10). WT HERV-K Gag, which rescued the release efficiency of the HERV-K mutant, did not rescue that of the HIV-1 mutant (Fig. 3E, lane 11 and 12). These results indicate that WT HIV-1 Gag interacts with, and rescues release of, the HERV-K Gag PTAP mutant at the PM.
Myristoylation and the NC domain are required for colocalization between HIV-1 Gag and HERV-K Gag.
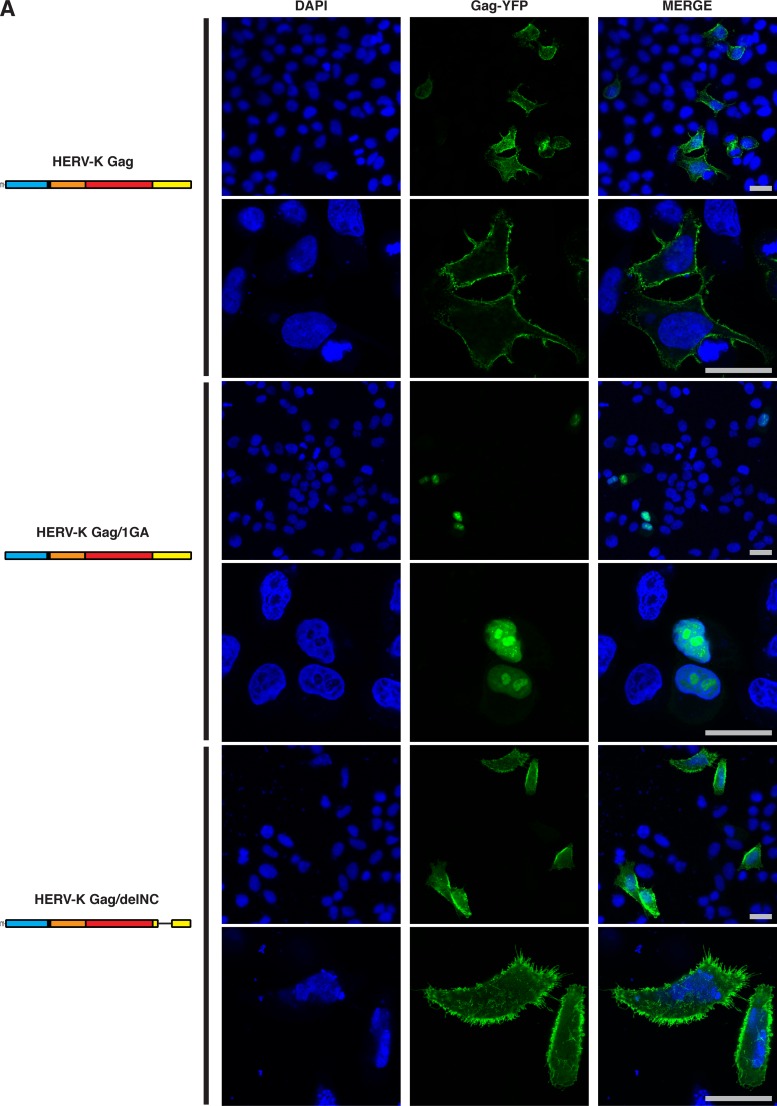
Coassembly between phylogenetically distant retroviruses may be mediated by a general mechanism such as scaffolding by membrane or RNA. To determine the mechanism by which HERV-K Gag colocalizes with HIV-1 Gag at the PM, we examined two HERV-K Gag mutants, Gag/1GA-YFP and Gag/delNC-YFP. HERV-K Gag/1GA-YFP lacks the N-terminal myristoylation site and is likely to be defective in membrane binding. HERV-K Gag/delNC-YFP lacks two zinc finger domains and basic amino acids in the NC domain and, therefore, is expected to be defective in Gag multimerization promoted by NC-RNA binding. Interestingly, we found that HERV-K Gag/1GA-YFP not only was not membrane bound but also was localized to the nucleus (Fig. 4A). This observation suggests that HERV-K Gag may have a cryptic nuclear localization phase, as observed for Rous sarcoma virus Gag (48). When WT HERV-K Gag was cotransfected with HERV-K Gag/1GA-YFP, a population of HERV-K Gag/1GA-YFP was recruited to the PM and colocalized with WT HERV-K Gag (Fig. 4B [middle] and D). In contrast, WT HIV-1 Gag did not rescue the localization of HERV-K Gag/1GA-YFP to the PM (Fig. 4C [middle] and D). These results suggest that membrane binding of HERV-K Gag is essential for colocalization of HIV-1 Gag and HERV-K Gag.
Fig 4.
Myristoylation- or NC-deficient HERV-K Gag mutants do not colocalize with HIV-1 Gag efficiently. (A) HeLa cells singly expressing HERV-K WT Gag-YFP, HERV-K Gag/1GA-YFP, or HERV-K Gag/delNC-YFP were examined using fluorescence microscopy at 16 h after cotransfection as for Fig. 2. Low (top; original magnification, ×20) and high (bottom; original magnification, ×63) magnifications of the same field are shown for each condition. (B) HeLa cells coexpressing HERV-K WT Gag-mRFP with HERV-K WT Gag-YFP (top), Gag/1GA-YFP (middle), or Gag/delNC-YFP (bottom) were examined as described for panel A. (C) HeLa cells coexpressing HIV-1 WT Gag-mRFP with HERV-K WT Gag-YFP (top), Gag/1GA-YFP (middle), or Gag/delNC-YFP (bottom) were examined as described for panel A. Bars, 10 μm. (D) The R strength of correlation between fluorescence intensities of the indicated Gag-fluorescent protein chimeras was calculated for cells coexpressing these Gag derivatives. Data from 14 to 50 cells are shown as means ± SEM. P values were determined using Student's t test. *, P < 0.01; **, P < 0.001; ***, P < 0.0001.
Unlike the 1GA mutant, HERV-K Gag/delNC-YFP localized to the entire cell surface (Fig. 4A). WT HERV-K Gag-mRFP colocalized with HERV-K Gag/delNC-YFP (Fig. 4B, bottom) nearly as efficiently as with WT HERV-K Gag-YFP (Fig. 4D). In contrast, colocalization between WT HIV-1 Gag-mRFP and YFP-tagged HERV-K Gag was substantially reduced by the deletion in the HERV-K Gag NC domain (Fig. 4C and D). These results indicate that not only myristoylation of HERV-K Gag but also the NC domain of HERV-K Gag is important for colocalization between HIV-1 Gag and HERV-K Gag.
The NC domain is required for the interaction between HIV-1 Gag and HERV-K Gag.
To analyze whether the NC domain is required for coassembly between HIV-1 Gag and HERV-K Gag, we examined the release of the HERV-K Gag mutant (HERV-K Gag/delNC) upon WT HIV-1 Gag coexpression. HERV-K Gag/delNC was not capable of virus release, and consistent with the role for NC in coassembly, the release of HERV-K Gag/delNC was not rescued by WT HIV-1 Gag (data not shown). For further analysis of the role of NC in coassembly between HIV-1 Gag and HERV-K Gag, we next examined the impact of HIV-1 NC deletion on coassembly. Since HIV-1 Gag/delNC alone can be released extracellularly in the absence of active PR (Fig. 5, lane 7) (45), rescue of this HIV-1 Gag derivative by WT HERV-K Gag cannot be readily assessed. Therefore, we examined coimmunoprecipitation of HERV-K Gag with HIV-1 Gag/delNC using HIV Ig. As observed in Fig. 3B, HERV-K Gag was coimmunoprecipitated with WT HIV-1 Gag (Fig. 5, lane 4). However, when coexpressed with HIV-1 Gag/delNC instead of WT HIV-1 Gag, HERV-K Gag was not coimmunoprecipitated by HIV Ig (Fig. 5, lane 5). These results suggest that the NC domain is required for the interaction between HIV-1 Gag and HERV-K Gag.
Fig 5.
HIV-1 Gag/delNC does not heteromultimerize with HERV-K Gag in virions. HeLa cells were cotransfected with the indicated plasmids and incubated for 14 h. pNL4-3 Gag/delNC lacks a majority of the NC sequence. After 2 h of metabolic labeling with [35S]Met/Cys, the viral lysates were analyzed as for Fig. 3B. The results shown are representative of data from three independent experiments. Note that the delNC change in HIV-1 Gag abolishes coimmunoprecipitation of HERV-K Gag with HIV-1 Gag.
Myristoylation and the NC domain of HERV-K are important for reduction of HIV-1 release efficiency and infectivity.
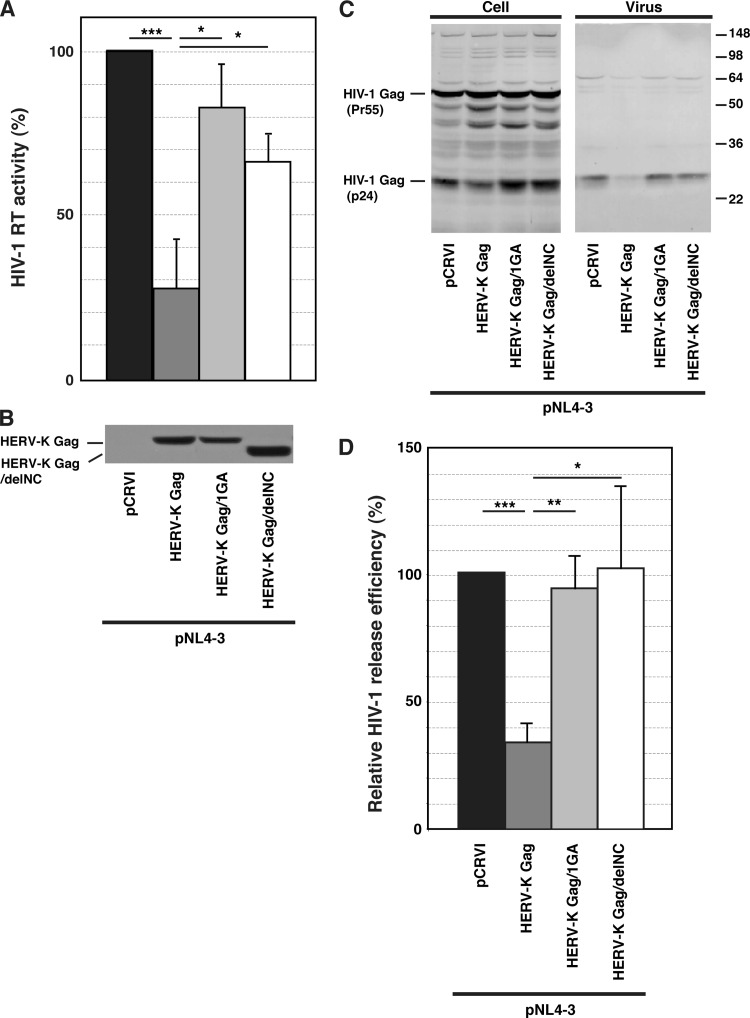
To address whether coassembly of HERV-K Gag with HIV-1 Gag is required for the negative impact on the HIV-1 release efficiency (Fig. 1), we measured the effect of HERV-K Gag mutants defective in coassembly on release of HIV-1 (Fig. 6). Expression levels of HERV-K Gag mutants were similar to that of WT HERV-K Gag in transfected HeLa cells (Fig. 6B). As observed in Fig. 1, both the amount of released HIV-1 measured by supernatant RT activity (Fig. 6A) and the HIV-1 release efficiency (Fig. 6C and D) were significantly reduced by WT HERV-K Gag coexpression. In contrast, both 1GA and delNC mutations in HERV-K Gag attenuated or reversed inhibition of HIV-1 release (Fig. 6A, C, and D). These results strongly suggest that coassembly of HERV-K Gag is required for reduction of HIV-1 release.
Fig 6.
Coexpression of myristoylation- or NC-deficient HERV-K Gag mutants does not reduce HIV-1 release efficiency. (A) HeLa cells were cotransfected with pNL4-3 and the indicated pCRVI plasmids. HIV-1 RT activity of virus pellets from transfected cells was measured at 16 h posttransfection. (B) Lysates of cotransfected cells were subjected to SDS-PAGE and analyzed by immunoblotting with anti-Flag antibody. (C) Cell and viral lysates from cotransfected cells were subjected to SDS-PAGE and analyzed by immunoblotting with HIV Ig. (D) Virus release efficiency was calculated by dividing the amount of p24CA in the viral lysates by the total amount of Gag in the cell and viral lysates. Data from four independent experiments are shown as means ± standard deviations. P values were determined using Student's t test. *, P < 0.01; **, P < 0.001; ***, P < 0.0001.
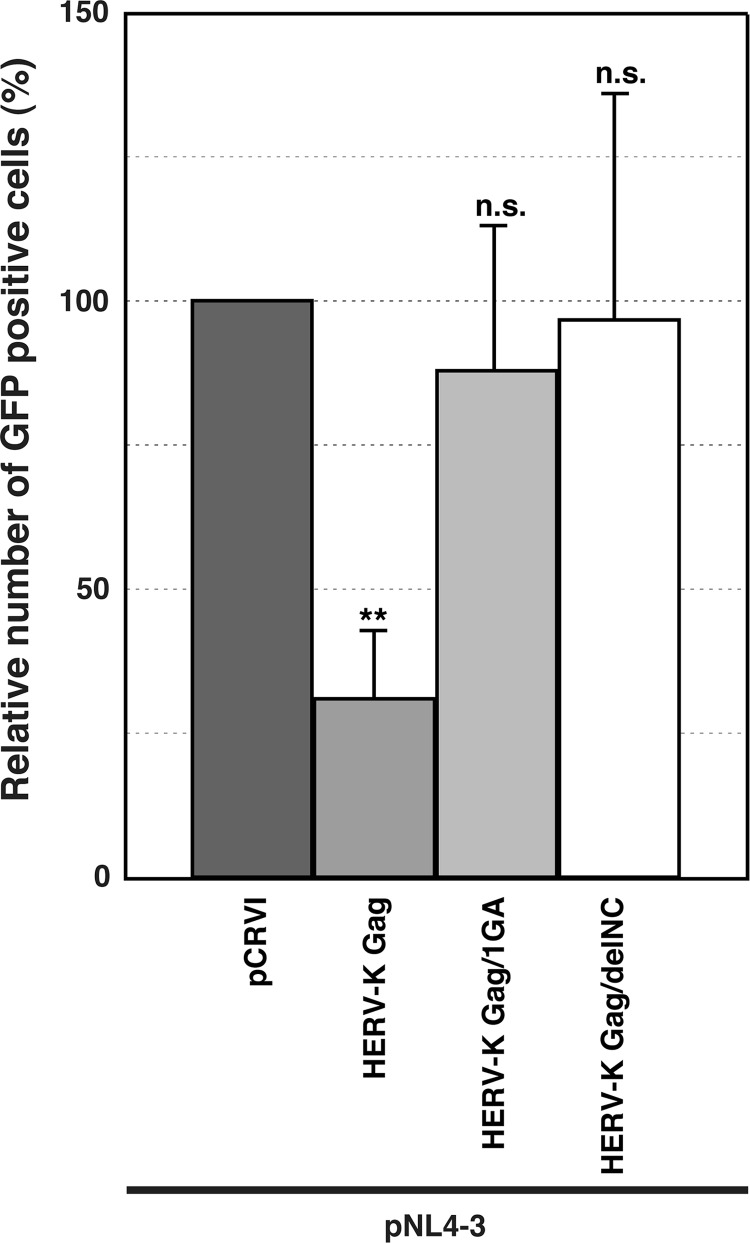
We further examined the infectivity of HIV-1 produced from HeLa cells cotransfected with both pNL4-3 and HERV-K-Gag-encoding plasmids (Fig. 7). CEM-GFP cells were infected with preparations of each virus that had been normalized by RT activity. The relative infectivity of HIV-1 was reduced when viruses were prepared from supernatants of cells that coexpress WT HERV-K Gag but not 1GA or delNC mutant HERV-K Gag (Fig. 7). These results suggest that coassembly of HERV-K Gag also interferes with the early phase in the replication of progeny HIV-1.
Fig 7.
Expression of WT HERV-K Gag but not myristoylation- or NC-deficient mutants reduces HIV-1 infectivity. (A) Virus stocks were prepared from HeLa cells cotransfected with pNL4-3 and the indicated pCRVI plasmids. The viruses were normalized by RT activity. CEM-GFP cells, which harbor an HIV-1 LTR-driven GFP reporter gene, were infected with these virus stocks. At 2 days postinfection, GFP-positive cells were counted by flow cytometry. Data from four independent experiments are shown as means ± standard deviations. P values were determined using Student's t test. **, P < 0.001; ns, not significant.
DISCUSSION
In this study, we found that HERV-K Gag coassembles with HIV-1 Gag into the same virions at the PM. This is supported by the following observations: (i) HERV-K Gag was processed by HIV-1 protease in virions, (ii) virion-associated HERV-K Gag was coimmunoprecipitated with HIV-1 Gag, and (iii) a HERV-K Gag mutant defective in virus release was rescued by HIV-1 Gag. To our knowledge, this is the first example of coassembly between lenti- and betaretroviruses. Importantly, coassembly of HERV-K Gag correlates with reduction in the release efficiency and infectivity of HIV-1. Therefore, if HIV-1 infection induces HERV-K coexpression in the infected cell, as observed for primary T cells infected in vitro (17), HERV-K Gag potentially impairs the HIV-1 replication via coassembly.
The release efficiency of the HERV-K late domain mutant was rescued by WT HIV-1 Gag coexpression (Fig. 3E, lane 10). However, WT HERV-K Gag did not rescue the release efficiency of the HIV-1 Gag/PTAP(−) mutant (Fig. 3E, lane 11). There are at least two possible reasons for low or no functional complementation by WT HERV-K Gag. First, even though WT HERV-K Gag might recruit the ESCRT machinery via its PTAP motif to assembly sites of HIV-1 Gag/PTAP(−) as it does to HERV-K Gag/PTAP(−) assembly sites, the restored release efficiency of HIV-1 Gag/PTAP(−) might be offset by the suppressive effect of WT HERV-K Gag on HIV-1 release. Second, the ESCRT machinery recruited by the HERV-K Gag PTAP motif might not be optimally positioned or stably retained in assembling HIV-1 particles, since the HERV-K Gag PTAP motif is located between the MA and CA domains, unlike the HIV-1 PTAP motif. While late domain motifs can be interchanged regardless of the position in the context of some retroviral Gag proteins (46), the context/position dependence of these motifs has also been documented (see, for example, references 39 and 42). In the context of mixed virions, ESCRT-bound HERV-K Gag may not partition into HIV-1 Gag multimers due to steric hindrance. Alternatively, the HERV-K Gag PTAP motif may be not exposed in mixed multimers.
Coassembly of HERV-K Gag with HIV-1 Gag required both myristoylation and the NC domain (Fig. 4 and 5). Both myristate-dependent Gag membrane binding and the NC-RNA interaction are thought to function as scaffoldings, thereby promoting retrovirus assembly. Previous cell-based studies showed that while both play overlapping roles in promoting tight interactions between HIV-1 Gag molecules (27), both are necessary for higher-order Gag multimerization (34, 44). However, while necessary, interactions through membrane and RNA scaffolding are unlikely to be sufficient for heteromultimerization of native HIV-1 and HERV-K Gag proteins. Indeed, MLV Gag, which has NC and can bind to RNA, did not coassemble with HIV-1 Gag at the PM (Fig. 2C) (1). If MA-membrane binding and the NC-RNA interaction are sufficient for heteromultimerization, MLV Gag should have coassembled with HIV-1 Gag at the PM like HERV-K Gag. Previous studies established that the presence of homologous CA is necessary for coassembly of Gag proteins, not only between retroviruses of different genera (e.g., MLV [a gammaretrovirus] and HIV-1 [a lentivirus]) but also between those within the same genera (e.g., MLV and spleen necrosis virus [SNV]) (1, 36). Within primate lentiviruses, which share substantial homology in their CA sequences, coassembly was observed for HIV-1 and HIV-2 and for HIV-1 and SIVmac (10, 11). In this regard, it is intriguing that even though HIV-1 and HERV-K belong to different genera, the CA C-terminal domain (CTD) of HERV-K is more similar to the CA CTD of HIV-1 Gag than to the CA CTDs of other betaretroviruses such as MMTV (26). More specifically, both the major homology region (MHR) (V161RQGSKEPYPDFV173) and a downstream region (E200NANPEC206) in the HERV-K CA CTD have substantial similarities to their counterparts in the HIV-1 CA CTD (MHR [I153RQGPKEPFRDYV165], identity of 62% and similarity of 85%; downstream region [Q192NANPDC198], identity of 71% and similarity of 100%) (26). Such high sequence similarity is not present between the HIV-1 CA CTD and the MLV CA CTD. Therefore, it is possible that heterodimerization between CA CTDs may occur and promote coassembly between HIV-1 Gag and HERV-K Gag, together with scaffolding functions provided by myristoylation and the NC domain. Further investigation is necessary to determine whether the HERV-K Gag CA CTD is required for HIV-1 Gag and HERV-K Gag heterodimerization.
Interestingly, coexpression of HERV-K Gag inhibited HIV-1 release (Fig. 1 and 6). Based on the following results, we surmise that HERV-K Gag interferes with the HIV-1 release after both HIV-1 Gag and HERV-K Gag bind to, and heteromultimerize at, the PM. First, HIV-1 Gag bound to and colocalized with HERV-K Gag at the PM (Fig. 2A and B). Second, HERV-K Gag/1GA, which does not bind membrane, failed to inhibit HIV-1 release (Fig. 6). Third, HERV-K Gag/delNC, which is capable of membrane binding but not efficient colocalization or coassembly with HIV-1 Gag, did not inhibit the HIV-1 release (Fig. 6). Of note, the role for the HERV-K Gag NC domain in inhibition of HIV-1 release is not specific to HERV-KCON, since HERV-K GagPro containing an NC sequence derived from Jurkat (GagPro#09) was also capable of suppressing HIV-1 release (Fig. 1A and B). The mechanisms for reduction of HIV-1 release are still unclear, but HERV-K Gag could interfere with HIV-1 assembly steps following Gag multimerization, such as the membrane curvature and/or pinching off, through the heteromultimerization between HIV-1 Gag and HERV-K Gag. Furthermore, the infectivity of HIV-1 was inhibited by HERV-K Gag coexpression (Fig. 7). It is possible that interaction between HIV-1 Gag and HERV-K Gag interferes with maturation of HIV-1 particles and viral core formation. Alternatively, coassembled particles may be deficient in packaging HIV-1 genomic RNA. Further studies are necessary to identify the stages of HIV-1 budding and acquisition of infectivity that are inhibited by heteromultimerization between HIV-1 Gag and HERV-K Gag.
Previous studies observed that HERV-K expression is increased upon HIV-1 infection in T cells. Our study demonstrates that HERV-K Gag interferes with the release efficiency and infectivity of HIV-1 through coassembly with HIV-1 Gag, at least in tissue culture models. In future studies, it will be important to confirm these observations using virions and T cells derived from HIV-1-infected patients. Meanwhile, it is tempting to speculate that endogenous retroviruses such as HERV-K, which have existed for a long time in the human genome, might protect the host cells from the threat of exogenous retroviruses, as is the case with Fv1, a remnant of mouse endogenous retrovirus Gag that inhibits the postentry process of MLV infection (29, 30, 50). In this regard, it will be interesting to determine the mechanism of enhancement of HERV-K expression upon HIV-1 infection and whether HERV-K expression regulates HIV-1 replication in vivo.
ACKNOWLEDGMENTS
We thank the members of our laboratories for helpful discussions and critical reviews of the manuscript. We also thank Paul D. Bieniasz and David Ott for providing plasmids. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV Ig from NABI and NHLBI and CEM-GFP from Jacques Corbeil.
This work was supported by grants from the National Institutes of Health, primarily by R01 CA144043 to D.M.M. and partly by R56 AI089282 to A.O.
Footnotes
Published ahead of print 1 August 2012
REFERENCES
- 1. Ako-Adjei D, Johnson MC, Vogt VM. 2005. The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J. Virol. 79:13463–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arien KK, et al. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailes E, et al. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 4. Balasubramaniam M, Freed EO. 2011. New insights into HIV assembly and trafficking. Physiology (Bethesda) 26:236–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 101(Suppl. 2):14572–14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beimforde N, Hanke K, Ammar I, Kurth R, Bannert N. 2008. Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology 371:216–225 [DOI] [PubMed] [Google Scholar]
- 7. Bennett RP, Wills JW. 1999. Conditions for copackaging Rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding. J. Virol. 73:2045–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boller K, Frank H, Lower J, Lower R, Kurth R. 1983. Structural organization of unique retrovirus-like particles budding from human teratocarcinoma cell lines. J. Gen. Virol. 64:2549–2559 [DOI] [PubMed] [Google Scholar]
- 9. Boller K, et al. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89:567–572 [DOI] [PubMed] [Google Scholar]
- 10. Boyko V, et al. 2006. Coassembly and complementation of Gag proteins from HIV-1 and HIV-2, two distinct human pathogens. Mol. Cell 23:281–287 [DOI] [PubMed] [Google Scholar]
- 11. Chen J, Pathak VK, Peng W, Hu WS. 2008. Capsid proteins from human immunodeficiency virus type 1 and simian immunodeficiency virus SIVmac can coassemble into mature cores of infectious viruses. J. Virol. 82:8253–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colicelli J, Goff SP. 1988. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 199:47–59 [DOI] [PubMed] [Google Scholar]
- 13. Contreras-Galindo R, Almodovar-Camacho S, Gonzalez-Ramirez S, Lorenzo E, Yamamura Y. 2007. Comparative longitudinal studies of HERV-K and HIV-1 RNA titers in HIV-1-infected patients receiving successful versus unsuccessful highly active antiretroviral therapy. AIDS Res. Hum. Retroviruses 23:1083–1086 [DOI] [PubMed] [Google Scholar]
- 14. Contreras-Galindo R, et al. 2006. A new real-time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J. Virol. Methods 136:51–57 [DOI] [PubMed] [Google Scholar]
- 15. Contreras-Galindo R, et al. 2012. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J. Virol. 86:262–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y. 2006. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retroviruses 22:979–984 [DOI] [PubMed] [Google Scholar]
- 17. Contreras-Galindo R, Lopez P, Velez R, Yamamura Y. 2007. HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res. Hum. Retroviruses 23:116–122 [DOI] [PubMed] [Google Scholar]
- 18. Dewannieux M, et al. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garrison KE, et al. 2007. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 3:e165 doi:10.1371/journal.ppat.0030165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garrus JE, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 21. George M, et al. 2011. Identification of the protease cleavage sites in a reconstituted Gag polyprotein of an HERV-K(HML-2) element. Retrovirology 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gervaix A, et al. 1997. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. U. S. A. 94:4653–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gonzalez-Hernandez MJ, et al. 2012. Expression of human endogenous retrovirus type-K (HML-2) is activated by the Tat protein of HIV-1. J. Virol. 86:7790–7805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gottlieb GS, et al. 2003. Molecular epidemiology of dual HIV-1/HIV-2 seropositive adults from Senegal, West Africa. AIDS Res. Hum. Retroviruses 19:575–584 [DOI] [PubMed] [Google Scholar]
- 25. Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U. S. A. 88:3195–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heslin DJ, et al. 2009. A single amino acid substitution in a segment of the CA protein within Gag that has similarity to human immunodeficiency virus type 1 blocks infectivity of a human endogenous retrovirus K provirus in the human genome. J. Virol. 83:1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hogue IB, Hoppe A, Ono A. 2009. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding. J. Virol. 83:7322–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang M, Orenstein JM, Martin MA, Freed EO. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42:709–732 [DOI] [PubMed] [Google Scholar]
- 30. Jolicoeur P, Baltimore D. 1976. Effect of Fv-1 gene product on proviral DNA formation and integration in cells infected with murine leukemia viruses. Proc. Natl. Acad. Sci. U. S. A. 73:2236–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kannangai R, et al. 2003. HIV-2 subtype circulating in India (south). J. Acquir. Immune Defic. Syndr. 33:219–222 [DOI] [PubMed] [Google Scholar]
- 32. Kraus B, Boller K, Reuter A, Schnierle BS. 2011. Characterization of the human endogenous retrovirus K Gag protein: identification of protease cleavage sites. Retrovirology 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuhelj R, et al. 2001. Inhibition of human endogenous retrovirus-K10 protease in cell-free and cell-based assays. J. Biol. Chem. 276:16674–16682 [DOI] [PubMed] [Google Scholar]
- 34. Kutluay SB, Bieniasz PD. 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 6:e1001200 doi:10.1371/journal.ppat.1001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- 36. Lee SK, Boyko V, Hu WS. 2007. Capsid is an important determinant for functional complementation of murine leukemia virus and spleen necrosis virus Gag proteins. Virology 360:388–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3:e10 doi:10.1371/journal.ppat.0030010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lefebvre G, et al. 2011. Analysis of HIV-1 expression level and sense of transcription by high-throughput sequencing of the infected cell. J. Virol. 85:6205–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li F, Chen C, Puffer BA, Montelaro RC. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li SK, et al. Detection and identification of plasma bacterial and viral elements in HIV/AIDS patients in comparison to healthy adults. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2011.03690.x [DOI] [PubMed] [Google Scholar]
- 41. Lower R, Lower J, Kurth R. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 93:5177–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin-Serrano J, Perez-Caballero D, Bieniasz PD. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ono A, Freed EO. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ono A, Waheed AA, Joshi A, Freed EO. 2005. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 79:14131–14140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ott DE, et al. 2003. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 77:5547–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parent LJ, et al. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salemi M, et al. 2003. Mosaic genomes of the six major primate lentivirus lineages revealed by phylogenetic analyses. J. Virol. 77:7202–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. U. S. A. 99:3944–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. SenGupta D, et al. 2011. Strong human endogenous retrovirus-specific T cell responses are associated with control of HIV-1 in chronic infection. J. Virol. 85:6977–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sveda MM, Soeiro R. 1976. Host restriction of Friend leukemia virus: synthesis and integration of the provirus. Proc. Natl. Acad. Sci. U. S. A. 73:2356–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tandon R, et al. 2011. Identification of human endogenous retrovirus-specific T cell responses in vertically HIV-1-infected subjects. J. Virol. 85:11526–11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Turner G, et al. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531–1535 [DOI] [PubMed] [Google Scholar]
- 53. Venter JC, et al. 2001. The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- 54. Vogetseder W, Dumfahrt A, Mayersbach P, Schonitzer D, Dierich MP. 1993. Antibodies in human sera recognizing a recombinant outer membrane protein encoded by the envelope gene of the human endogenous retrovirus K. AIDS Res. Hum. Retroviruses 9:687–694 [DOI] [PubMed] [Google Scholar]
- 55. Willey RL, et al. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]