Abstract
Adaptor protein complex 3 (AP-3) is a heterotetramer that is involved in signal-mediated protein sorting to endosomal-lysosomal organelles. AP-3 deficiency in humans, induced by mutations in the AP3B1 gene, which encodes the β3A subunit of the AP-3 complex, results in Hermansky-Pudlak syndrome 2 (HPS2), which is a rare genetic disorder with defective lysosome-related organelles. In a previous study, we identified the AP-3 complex as an important contributor to HIV-1 assembly and release. We hypothesized that cells from patients affected by HPS2 should demonstrate abnormalities of HIV-1 assembly. Here we report that HIV-1 particle assembly and release are indeed diminished in HPS2 fibroblast cultures. Transient or stable expression of the full-length wild-type β3A subunit in HPS2 fibroblasts restored the impaired virus assembly and release. In contrast, virus-like particle release mediated by MA-deficient Gag mutants lacking the AP-3 binding site was not altered in HPS2 cells, indicating that the MA domain serves as the major viral determinant required for the recruitment of the AP-3 complex. AP-3 deficiency decreased HIV-1 Gag localization at the plasma membrane and late endosomes and increased the accumulation of HIV-1 Gag at an intermediate step between early and late endosomes. Blockage of the clathrin-mediated endocytic pathway in HPS2 cells did not reverse the inhibited virus assembly and release imposed by the AP-3 deficiency. These results demonstrate that the intact and stable AP-3 complex is required for HIV-1 assembly and release, and the involvement of the AP-3 complex in late stages of the HIV-1 replication cycle is independent of clathrin-mediated endocytosis.
INTRODUCTION
The highly orchestrated process of HIV-1 assembly and release is driven by the Gag precursor protein (Pr55-Gag), which is synthesized on free cytosolic ribosomes and modified cotranslationally by the N-terminal attachment of a myristyl group (27, 73). In the absence of other viral proteins, the HIV-1 Gag protein alone forms noninfectious virus-like particles (VLPs), indicating that it plays a central role in particle assembly and release (31). During or shortly after virus release, viral protease (PR)-mediated cleavage of the Gag precursor leads to virus maturation, a morphological transition essential for virus infectivity. The HIV-1 Gag protein contains four structural domains (from the N to C terminus): matrix (MA), capsid (CA), nucleocapsid (NC), and p6, as well as two spacer peptides, SP1 (located between CA and NC) and SP2 (located between NC and p6). The myristylated MA domain is required for proper targeting of Gag to distinct microdomains (such as lipid rafts or tetraspanin-enriched microdomains) of the plasma membrane (PM) through its direct interaction with phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], located at the cytoplasmic leaflet of the PM, resulting in increased exposure of the N-terminal myristate moiety (49, 61). The CA domain mediates Gag multimerization (28). In addition to contributing to Gag-Gag interactions, the NC domain is also responsible for packaging of the viral RNA into budding virions (16). The p6 domain is involved in virus pinching off from the cell surface by recruiting the host ESCRT (endosomal sorting complex required for transport) machinery (8, 19, 47).
In addition to viral determinants required for productive particle assembly and release, some cellular factors have been identified to be involved in these events (11, 14, 30, 37, 38, 71, 75). One example of these host factors is the AP-3 complex (22). The heterotetrameric AP-3 complex is composed of two large subunits (δ and β3), a medium subunit, μ3, and a small subunit, σ3 (58, 59). In mammals, the AP-3 complex is considered to mediate the sorting and transport of membrane proteins from the trans-Golgi network (TGN), early endosomes (EEs), and tubular sorting endosomes to late endosomes (LEs), multivesicular bodies (MVBs), lysosomes, and lysosome-related organelles (43, 53, 59, 60). AP-3 deficiency results in the impaired vacuolar delivery of alkaline phosphatase in yeast (15, 67), the defective pigment granule biogenesis in fruit flies (51, 66), and abnormal melanosomes and platelet-dense granules in mammals (18, 40, 44). Hermansky-Pudlak syndrome (HPS) is a group of autosomal recessive genetic disorders in humans that is characterized by hypopigmentation and platelet dysfunction (21). HPS2, a rare subtype of HPS, is caused by an impaired AP-3 sorting machinery due to mutations in the AP3B1 gene encoding the β3A subunit (12, 13, 18, 25, 26, 32, 35, 39, 72). In HPS2 fibroblasts, missorting of lysosomal membrane proteins to the cell surface results in the increased surface expression of these proteins (18). Similarly, in HPS2 B-lymphoblastoid cells, mistargeting of CD1B to the PM and/or early endosomes induces a defect in antigen presentation (68). In HPS2 cytotoxic T cells (CTL), lytic granules fail to transit along microtubules to the immunologic synapse, resulting in dysfunctional CTL-mediated killing (13).
Experiments of nature can help to validate and extend findings derived in laboratory studies. To further evaluate the role of the AP-3 sorting machinery in late stages of the HIV-1 replication cycle, in the present study we examined HIV-1 particle assembly and release in primary fibroblasts derived from HPS2 patients. We demonstrate that HIV-1 assembly and release are diminished significantly in AP-3-deficient HPS2 fibroblasts and that the impairment in HIV-1 assembly and release could be restored by reconstituting the functional AP-3 complex in HPS2 fibroblasts. AP-3 deficiency in HPS2 fibroblasts reduced HIV-1 Gag localization at the plasma membrane and late endosomes and increased HIV-1 Gag accumulation at transport intermediates between early and late endosomes. Dominant inhibition of early stages of the clathrin-mediated endocytic pathway in HPS2 fibroblasts did not rescue the impairment of virus assembly and release. These data, derived in cells from a naturally occurring genetic deficiency of the AP-3 complex, further support a role for the AP-3 sorting machinery during late stages of the HIV-1 replication cycle.
MATERIALS AND METHODS
Plasmids.
Full-length cDNA of the β3A subunit of the AP-3 complex (AP3B1) was purchased from OriGene Technologies (Rockville, MD) and used as a template in the PCR amplification. PCR products were cloned into pcDNA3.1 (Invitrogen) or pBABE-hygro (Addgene, Cambridge, MA) to generate pcDNA-AP3B1 and pBabe-AP3B1 expression plasmids. pEYFP-Golgi and pECFP-Nuc plasmids were obtained from Clontech Laboratories, Inc. The pEYFP-Golgi plasmid encodes a fusion of enhanced yellow fluorescent protein (EYFP) and the N-terminal 81 amino acids of the human β1,4-galactosyltransferase, targeting the fluorescent protein to the trans-medial region of the Golgi apparatus. The pECFP-Nuc plasmid encodes the cyan fluorescent protein with three tandem repeats of the nuclear localization signal from simian virus large T-antigen. The GFP-Dynamin (K44) plasmid expressing the green fluorescent protein (GFP)-tagged dominant-negative (DN) mutant (K44A) of dynamin was purchased from Addgene (Cambridge, MA) (45). The pNL4-3 expression construct was obtained from Malcolm Martin through the NIH AIDS Reference and Reagent Program. The HIV-1 Gag and protease expression plasmid 3-CCCC was a generous gift from Hans-Georg Kräusslich (University of Heidelberg, Heidelburg, Germany) (74), the HIV-1 Gag-Pol expression plasmid pGPCINS came from Xiao-Fang Yu (Johns Hopkins University, Baltimore, MD) (46), the dynamin (K44A) expression plasmid was from Marc Caron (Duke University, Durham, NC) (2), the FLAG-Eps15 (ΔUIM) expression plasmid encoding the FLAG-tagged dominant-negative mutant of Eps15 (epidermal growth factor receptor substrate 15) with the deletion of the ubiquitin-interacting motif (70) was obtained from Simonia Polo (IFOM-IEO Campus, Milan, Italy) (55), the Rab5-GFP expression plasmid was from Ruth Collins (Cornell University, Ithaca, NY), and the EGFP-AP3B1 plasmid expressing the enhanced GFP (EGFP)-tagged β3A subunit of the AP-3 complex was from Adolfo Saiardi (University College London, London, United Kingdom) (5). Gag, SrcΔMAGag, Gag-CFP, Gag-YFP, and pNL4-3/ΔEnv expression plasmids have been previously described (20, 22, 41).
Antibodies.
Rabbit anti-human β3A antibodies were kindly provided by Margaret Robinson (University of Cambridge, Cambridge, United Kingdom). HIV Ig was obtained from Luiz Barbosa through the NIH AIDS Reference and Research Reagent Program. Mouse anti-human δ, μ3A, and σ3A antibodies and mouse anti-EEA1, anti-Vti1b, and anti-syntaxin 8 antibodies were purchased from BD Biosciences, mouse anti-FLAG antibodies were obtained from Sigma-Aldrich (St. Louis, MO), mouse anti-lysosome-associated membrane protein 1 (anti-LAMP-1), anti-LAMP-3, and anti-TGN38 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), and mouse anti-GFP antibodies were from Roche Applied Science. Fluorescently labeled secondary antibodies included goat anti-rabbit or anti-mouse IgG conjugated to allophycocyanin (APC) or fluorescein isothiocyanate (Jackson ImmunoResearch Laboratories, West Grove, PA) or Alexa Fluor 488, 546, and 647 (Invitrogen).
Cell culture and transfections.
Primary cultures of skin fibroblasts from individuals with HPS2 and normal individuals were obtained from the Coriell Cell Repositories (Camden, NJ). HPS2 fibroblasts (GM17890) were maintained in Dulbecco modified Eagle's medium (DMEM, high glucose) with 2 mM l-glutamine, 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 10% CO2. Normal fibroblasts (GM05381) were cultured in DMEM (high glucose) supplemented with 2 mM l-glutamine, 20% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. Normal and HPS2 fibroblasts were transfected using Lipofectamine 2000 (Invitrogen) or X-tremeGENE HP DNA transfection reagent (Roche).
Virus production and infection.
HIV-1 virus stocks pseudotyped with the vesicular stomatitis virus G glycoprotein (VSV-G) were obtained by calcium phosphate or polyethylenimine (PEI) transfection of 293T cells with pNL4-3/ΔEnv and pHCMV-G expression plasmids. At 48 h after transfection, supernatants were collected, filtered through 0.45-μm filters, and centrifuged at 28,000 × g for 2 h at 4°C through a 20% sucrose cushion. Purified VSV-G-pseudotyped NL4-3 viruses were used to infect normal or HPS2 fibroblasts at a higher multiplicity of infection (MOI). At 40 to 48 h after infection, cells were lysed for Western blot analysis. VSV-G-pseudotyped retrovirus stocks encoding the β3A subunit of the AP-3 complex were prepared from 293T cells cotransfected with pBabe-AP3B1, pCL-Ampho, and pHCMV-G expression plasmids. Supernatants were harvested at 40 to 48 h after transfection, filtered through 0.45-μm filters, aliquoted into cryovials, and stored.
Confocal microscopy.
Normal and HPS2 fibroblasts were grown overnight on glass coverslips in 6-well plates and then fixed with 3.8% formaldehyde in a sodium phosphate buffer at room temperature for 10 to 15 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with 5% bovine serum albumin in phosphate-buffered saline (PBS) for 1 h. Fibroblasts were then immunostained with the indicated primary antibodies and fluorescently labeled secondary antibodies. CD63 or LAMP-1 was revealed by mouse anti-LAMP-3 or mouse anti-LAMP-1 antibodies, respectively, followed by goat anti-mouse IgG conjugated to Alexa Fluor 546. To examine the distribution pattern of the endogenous β3A subunit, normal and HPS2 fibroblasts were transfected with a pECFP-Nuc plasmid. At 24 h after transfection, cells were fixed, permeabilized, and immunostained with rabbit anti-β3A antibodies, followed by goat anti-rabbit IgG conjugated to Alex Fluor 546. To observe the subcellular localization of the cyan fluorescent protein (CFP)-tagged Gag, normal and HPS2 fibroblasts were transfected with a Gag-CFP expression plasmid. At 20 to 24 h or 30 to 36 h after transfection, fibroblasts were fixed for examination. To observe the subcellular localization of the Gag protein, normal and HPS2 fibroblasts were transfected with the Gag expression plasmid. At 20 to 24 h or 30 to 36 h posttransfection, fixed fibroblasts were subjected to immunostaining with rabbit anti-p17 antisera and goat anti-rabbit IgG conjugated to Alexa Fluor 546. In some colocalization experiments, normal and HPS2 fibroblasts were cotransfected with Gag-GFP and HcRed-CD63, Gag-CFP and YFP-Golgi, or Gag-HcRed and Rab5-GFP expression plasmids. At 30 to 36 h posttransfection, fibroblasts were fixed for examining the colocalization of two fluorescent labels. In other colocalization experiments, HPS2 fibroblasts were transfected with Gag expression plasmids. At 30 to 36 h after transfection, fibroblasts were immunostained with rabbit anti-p17 antisera and either mouse anti-TGN38, anit-EEA1, anti-Vti1b, or anti-syntaxin 8 antibodies, followed by goat anti-rabbit IgG conjugated to Alex Fluor 546 and goat anti-mouse IgG conjugated to Alex Fluor 488. Confocal images were acquired using a Nikon TE2000-U laser-scanning confocal microscope or a Nikon A1R confocal microscope. Data analysis was performed with NIS-Elements AR 3.0 software. The Pearson's correlation coefficient (R) was used to quantify the levels of colocalization between two fluorescent probes.
Flow cytometry analysis.
Cell surface staining of CD63 and LAMP-1 was performed in normal fibroblasts, HPS2 fibroblasts, and HPS2 (β3A+) populations. Samples were immunostained with mouse anti-LAMP-3 or mouse anti-LAMP-1 antibodies, followed by APC-conjugated goat anti-mouse IgG secondary antibodies. Samples were then examined on a BD FACSCalibur flow cytometer, and the data were analyzed with FlowJo software.
Transferrin uptake assay.
HPS2 fibroblasts were transfected with EGFP, GFP-dynamin (K44A), or FLAG-Eps15 (ΔUIM) expression plasmids. At 24 h after transfection, cells were incubated in serum-free medium at 37°C for 1 h. Transferrin (Tf) conjugated to Alexa Fluor 546 (Invitrogen) was then added to medium at 20 μg/ml. After 1 h of uptake, HPS2 fibroblasts were washed extensively in PBS, fixed, permeabilized, and immunostained with mouse anti-GFP or mouse anti-FLAG antibodies and goat anti-mouse IgG conjugated to Alex Fluor 488.
RESULTS
The AP-3 complex is impaired in HPS2 fibroblast cultures.
Our previous studies indicated that the AP-3 sorting machinery is recruited by HIV-1 Gag for particle assembly and release (22). To better understand the role for the AP-3 complex in late stages of the HIV-1 replication cycle, we examined primary cultures of skin fibroblasts derived from individuals with HPS2. To confirm that AP-3 deficiency appeared in HPS2 fibroblasts, we compared AP-3 expression in normal and HPS2 fibroblast cultures (Fig. 1A). Immunoblot analysis was performed with whole-cell extracts from each of the two primary fibroblast cultures. AP-3 expression in HPS2 fibroblasts was much lower than in normal fibroblasts: the δ-subunit was lower by approximately 3-fold, the σ3A subunit by almost 4-fold, and μ3A and β3A subunits were lower by more than approximately 10-fold. We also performed immunofluorescence microscopy to examine the distribution pattern of the AP-3 complex in the two primary fibroblast cultures by immunostaining with anti-β3A antibodies. In normal fibroblast cultures, the AP-3 complex localized at peripheral and perinuclear regions and displayed a punctate distribution pattern (Fig. 1B, left). In contrast, in HPS2 fibroblast cultures the AP-3 complex was less stained, consistent with Western blot assay results showing reduced expression of this complex (Fig. 1B, right).
Fig 1.
AP-3 sorting machinery is impaired in HPS2 fibroblast cultures. (A) Western blot analysis of AP-3 expression in primary fibroblast cultures. The same number of normal and HPS2 fibroblast cultures was lysed. Four subunits of the AP-3 complex and actin were detected by immunoblotting using anti-δ, -β3A, -μ3A, -σ3A, and -actin antibodies. (B) Immunofluorescence microscopy analysis of the AP-3 complex in primary fibroblast cultures. Normal and HPS2 fibroblast cultures were transfected with a pECFP-Nuc plasmid. At 24 h after transfection, cells were fixed, permeabilized, and immunostained for the β3A subunit. pECFP-Nuc is shown in cyan, and the β3A subunit is in red. Bars, 10 μm. (C) Immunofluorescence microscopy analysis of subcellular localization of CD63 and LAMP-1. Normal (left) and HPS2 (right) fibroblast cultures were immunostained using anti-CD63 (top) and anti-LAMP-1 (bottom) antibodies. Bars, 10 μm. (D) Flow cytometric analysis of cell surface CD63 (top) and LAMP-1 (bottom). Unpermeabilized normal and HPS2 fibroblasts were immunostained with mouse anti-LAMP-3 or anti-LAMP-1 antibodies, followed by goat anti-mouse IgG conjugated to APC. The outlined plot represents the isotype control, the filled plot represents HPS2 fibroblast cultures, and the dash-lined plot represents normal fibroblast cultures.
The AP-3 deficiency results in increased surface expression of lysosomal membrane proteins (18). To examine whether defects in the AP-3 complex alter trafficking pathways of lysosomal membrane proteins, we observed by confocal microscopy the distribution patterns of CD63 and LAMP-1 in normal and HPS2 fibroblast cultures. Immunofluorescence staining of permeabilized fibroblast cultures revealed that CD63 and/or LAMP-1 in normal (Fig. 1C, left) and HPS2 fibroblast (Fig. 1C, right) cultures exhibited the perinuclear and punctate distribution pattern. Next, we performed flow cytometry assays to compare cell surface expression of CD63 and LAMP-1 in the two fibroblast cultures. The levels of CD63 and LAMP-1 on the surfaces of HPS2 fibroblast cells were increased by 2- to 3-fold, compared to normal fibroblast cultures (Fig. 1D). As expected, these results confirm that AP-3-dependent trafficking is impaired in HPS2 fibroblast cultures resulting in missorting of CD63 and LAMP-1 to the cell surface.
HIV-1 assembly is inhibited in HPS2 fibroblast cultures.
We reasoned that HPS2 fibroblast cultures may demonstrate a deficiency in assembly of HIV-1 particles. To test this hypothesis, normal and HPS2 fibroblast cultures were transfected with pNL4-3 proviral DNA. At 40 to 48 h after transfection, cell-associated and released p24 antigens were measured in enzyme-linked immunosorbent assays (ELISAs). The percentage of p24 antigen in supernatants as a fraction of p24 antigen in supernatant plus cell lysate was then determined and used as a standard to compare particle release efficiency from HPS2 fibroblasts versus normal fibroblasts. As expected, virus release efficiency from HPS2 fibroblast cultures was significantly lower than that from normal fibroblast cultures (Fig. 2A). To further examine HIV-1 assembly in HPS2 fibroblasts, we used VSV-G-pseudotyped NL4-3 viruses to infect the two fibroblast cultures at a higher MOI. Virus release from HPS2 fibroblast cultures was markedly inhibited, while the levels of the cell-associated Gag proteins in the two fibroblast cultures were comparable, confirming that HPS2 fibroblasts do not support efficient particle assembly and release (Fig. 2B). Similarly, VLP release efficiency in HPS2 fibroblasts expressing Gag alone was lower than that in normal fibroblasts expressing Gag alone (Fig. 2C).
Fig 2.
HIV-1 particle release is inhibited in HPS2 fibroblast cultures. (A) Normal and HPS2 fibroblast cultures were transfected with pNL4-3 proviral DNA. At 40 to 48 h after transfection, p24 levels in cell lysates and supernatants were measured in ELISAs. Particle release efficiency was calculated by determining the percentage of p24 antigen in supernatant as a fraction of p24 antigen in supernatant plus cell lysate. Data from three independent experiments are shown (means ± SD). P values were determined by using a paired Student's t test. **, P < 0.01. (B) Normal and HPS2 fibroblast cultures were infected with VSV-G-pseudotyped NL4-3 viruses at a higher MOI. At 40 to 48 h after infection, Western blot analysis of cell lysates and pelleted virion lysates using anti-HIV Ig and anti-actin antibodies was performed. (C and D) Normal and HPS2 fibroblast cultures were transfected with Gag (C) or SrcΔMAGag (D) expression plasmids. At 40 to 48 h posttransfection, a p24 antigen ELISA was performed with cell lysates and supernatants. Particle release efficiency was calculated by determining the percentage of p24 antigen in supernatant as a fraction of p24 antigen in supernatant plus cell lysate. Data represent means ± SD from three independent experiments. ns, not significant; *, P < 0.05.
It is well known that the treatment of the fungal metabolite brefeldin A (BFA) disassociates the AP-3 complex from membranes and impairs AP-3 function in protein sorting (50). We examined the effect of BFA on HIV particle release from normal fibroblasts. We treated normal fibroblasts with BFA for 1 h and observed a redistribution of endogenous AP-3 from punctate intracellular membrane-bound structures to a somewhat more diffuse appearance (see Fig. S1A in the supplemental material). As expected, 1-h BFA treatment increased cell surface CD63 levels (see Fig. S1B). Similar to previous observations (38, 52, 63), the results of our Western blot assay of HIV-1 Gag-Pol VLP did not show an inhibitory effect on particle release induced by 1-h BFA treatment (Fig. S1C). We interpreted this to indicate that either the incomplete dissociation of AP-3 from membranes by BFA (50) was not sufficient to disrupt AP-3-mediated transport of Gag, or that the brief disruption of AP-3 function by BFA was dissociated from the relevant time of Gag transport in the treated cells.
A previously described MA-deleted Gag mutant (SrcΔMAGag), which bears the v-src signaling sequence at its N terminus and supports efficient VLP assembly and release, was next examined in our studies. In contrast to the reduced Gag VLP release efficiency in HPS2 fibroblasts, SrcΔMAGag VLP release efficiency in HPS2 fibroblasts was comparable to that in normal fibroblasts (Fig. 2D), consistent with previous studies that used a small interfering RNA (siRNA) approach to show that the MA domain is the viral determinant required for the Gag-AP3 interaction engaged in the productive HIV-1 assembly and release (22). Taken together, our results indicate that AP-3-deficient fibroblasts derived from HPS2 patients do not support efficient assembly and release of HIV-1.
Expression of the full-length wild-type β3A subunit in HPS2 fibroblast cultures reverses the inhibited assembly and release of HIV-1.
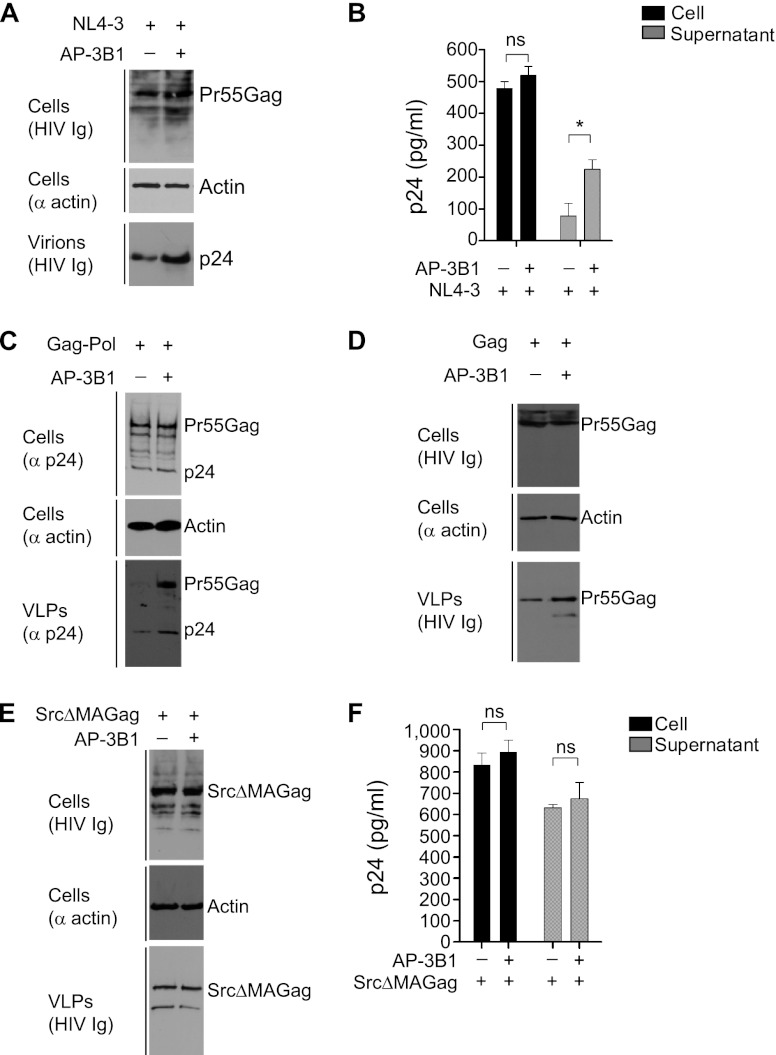
To validate the specific role for the AP-3 complex in HIV-1 particle assembly and release, we next performed experiments to restore AP-3 function in HPS2 fibroblasts. pNL4-3 proviral DNA, along with either control empty pcDNA vectors or AP-3B1 expression plasmids, was transfected into HPS2 fibroblast cultures at a ratio of 1:1. Western blot assays and p24 ELISAs indicated that transient expression of β3A in HPS2 cells increased virus release (Fig. 3A and B). An HIV-1 Gag-Pol expression plasmid was also used in our studies to evaluate the impact of β3A on Gag-Pol VLP release from HPS2 fibroblasts. As shown in Fig. 3C, β3A expression in HPS2 cells did not alter the levels of Gag expression and processing (top). However, β3A expression in HPS2 cells increased the yield of extracellular Gag-Pol VLP by about 4-fold (bottom). Similarly, the increased yield of extracellular VLPs mediated by Gag alone was also detected in HPS2 cells with β3A expression (Fig. 3D). In contrast, SrcΔMAGag-mediated VLP release from β3A-expressed HPS2 fibroblasts was not altered (Fig. 3E and F). These data, together with the results shown in Fig. 2, indicated that no virion components other than HIV-1 Gag proteins are involved in AP-3-dependent particle assembly and release and that the MA domain plays an important role in the recruitment of the AP-3 sorting machinery.
Fig 3.
Transient expression of the full-length wild-type β3A subunit in HPS2 fibroblast cultures rescues particle release. (A) HPS2 fibroblast cultures were cotransfected with pNL4-3 proviral DNA and empty pcDNA3.1 vectors (-) or AP-3B1 (+) expression plasmids at a ratio of 1:1. At 40 to 48 h after transfection, cell lysates and pelleted virion lysates were subjected to immunoblot analysis with the indicated antibodies (anti-actin and anti-HIV Ig). (B) Similar experiments to those described for panel A were performed. At 40 to 48 h after transfection, p24 levels in cell lysates and supernatants were determined by ELISAs. ns, not significant; *, P < 0.05. Data from three independent experiments are shown as means ± SD. (C to E) HIV-1 Gag-Pol (C), Gag (D), or SrcΔMAGag expression plasmids (E) were transfected into HPS2 fibroblast cultures, as well as either empty pcDNA3.1 vectors (-) or AP-3B1 expression plasmids (+). Western blot analysis of cell lysates and pelleted VLP lysates using the indicated antibodies (anti-p24, anti-actin, and anti-HIV Ig) was performed 40 to 48 h after transfection. (F) Similar experiments as those described for panel E were performed. At 40 to 48 h after transfection, p24 levels in cell lysates and supernatants were determined by ELISAs. ns, not significant. Data from three transfections are shown as means ± SD.
Complementary to transient reconstitution of the AP-3 complex, we used an alternative approach to generate HPS2 fibroblast populations stably expressing full-length wild-type β3A. In our experiments, HPS2 fibroblast cultures were transduced with a pBABE retroviral vector encoding the β3A subunit. Pooled populations, referred to as HPS2 (β3A+) populations, were selected for hygromycin resistance. To test whether expression levels of the AP-3 complex are restored in HPS2 (β3A+) populations, we performed Western blot assays on samples from HPS2 fibroblast cultures and HPS2 (β3A+) populations. HPS2 (β3A+) populations exhibited increased levels of the two large subunits of the AP-3 complex, δ and β3A, compared to HPS2 fibroblasts (Fig. 4A). We subsequently examined whether the impaired function of AP-3 in the sorting of lysosomal membrane proteins was restored in HPS2 (β3A+) populations by quantifying cell surface levels of CD63 and LAMP-1. Our flow cytometry analysis indicated that the surface expression levels of CD63 and LAMP-1 were decreased in HPS2 (β3A+) populations compared to those in HPS2 fibroblasts (Fig. 4B). The levels of CD63 and LAMP-1 surface expression in HPS2 (β3A+) populations were similar to those in normal fibroblasts, demonstrating that the defect in the AP3-dependent trafficking pathway seen in HPS2 fibroblasts is restored in HPS2 (β3A+) populations. We then evaluated virus release from HPS2 (β3A+) populations infected with VSV-G-pseudotyped NL4-3 viruses. Western blot analysis indicated that virus release from HPS2 (β3A+) populations was increased compared to HPS2 fibroblast cultures (Fig. 4C). To gain quantitative data, similar experiments were repeatedly performed, and p24 levels in cell lysates and supernatants were determined in ELISAs. Consistent with Western blotting results, virus release from HPS2 (β3A+) populations was increased by about 3-fold compared to HPS2 fibroblasts (Fig. 4D). Collectively, these gain-of-function studies demonstrated a specific role for the AP-3 complex in HIV-1 assembly and release.
Fig 4.
HPS2 (β3A+) fibroblast populations support efficient particle assembly and release. (A) Western blot analysis of AP-3 expression. HPS2 fibroblast cultures and HPS2 (β3A+) populations were lysed and subjected to immunostaining for δ and β3A subunits and actin. (B) Flow cytometry analysis of cell surface expression of CD63 (left) and LAMP-1 (right). Unpermeabilized normal fibroblasts, HPS2 fibroblasts, and HPS2 (β3A+) fibroblast populations were immunostained to detect surface CD63 and LAMP-1. (C) pNL4-3 proviral DNA was transfected into HPS2 fibroblast cultures and HPS2 (β3A+) populations. After 2 days, cell and virion lysates were subjected to Western blotting using anti-HIV Ig. (D) Similar experiments as those described for panel C were performed. At 40 to 48 h after transfection, cell lysates and supernatants were quantified in p24 ELISAs. ns, not significant; **, P < 0.01. Data from three independent experiments are shown as means ± SD.
AP-3 deficiency alters HIV-1 Gag subcellular localization.
To determine the effect of AP-3 deficiency on HIV-1 Gag subcellular localization, we observed the distribution pattern of fluorescently tagged Gag in normal and HPS2 fibroblasts. As shown in Fig. 5A and B, Gag-CFP in the two fibroblast cultures exhibited three main patterns: a diffuse pattern (left), an intracellular punctate pattern (middle), and a dual intracellular punctate and PM pattern (right). To provide a meaningful comparison of Gag localization in normal and HPS2 fibroblast cultures, about 50 fibroblasts transfected with Gag-CFP were included in our experiments. The number of fibroblasts displaying the individual distribution patterns of Gag-CFP was recorded, and percentages of fibroblasts exhibiting the individual distributions were determined. At 20 h after transfection, more than 80% of normal and HPS2 fibroblasts displayed a weak and diffuse pattern of fluorescence (data not shown). At 30 to 36 h after transfection, 46% of normal fibroblasts showed the intracellular punctate and PM pattern, and 32% of normal fibroblasts exhibited an intracellular punctate pattern. In contrast to normal fibroblasts, only 16% of HPS2 fibroblasts showed the intracellular punctate and PM pattern, and 72% of HPS2 fibroblasts displayed an intracellular punctate pattern (Fig. 5C). We also observed the subcellular localization of the Gag protein without a fluorescent tag in the two fibroblast cultures. Gag protein was detected by immunostaining with rabbit anti-p17 antisera. Similarly, the reduced PM localization of Gag was also detected in HPS2 fibroblasts compared to normal fibroblasts (data not shown). These data suggest that AP-3 deficiency decreases the PM localization of the HIV-1 Gag.
Fig 5.
AP3 deficiency alters HIV-1 Gag subcellular localization. (A and B) Normal (A) or HPS2 (B) fibroblast cultures were transfected with Gag-CFP expression plasmids. At 30 to 36 h posttransfection, cells were fixed for confocal microscopy. The diffuse pattern of Gag-CFP is shown on the left, the intracellular punctate pattern is in the middle, and the intracellular punctate and plasma membrane patterns are on the right. Bars, 10 μm. (C) The percentage of cells displaying the individual distribution patterns of Gag-CFP as a fraction of 50 cells expressing Gag-CFP was calculated. Representative data from three independent experiments are shown. (D and E) Normal (D) or HPS2 (E) fibroblast cultures were cotransfected with Gag-GFP and HcRed-CD63 expression plasmids. At 30 to 36 h after transfection, cells were fixed for confocal microscopy. Gag-GFP is shown in green (far left), HcRed-CD63 is in red (left), and the colocalized pixels are shown in yellow (right). Enlarged views of the boxed regions in the merged images are shown on the far right (D′ and E′). Bars, 10 μm. (F) The degree of colocalization between Gag-GFP and HcRed-CD63 in normal and HPS2 cells was quantified using the Pearson correlation coefficient. Data are as means ± SD for 15 to 20 cells. Three independent experiments were performed.
Our previous colocalization studies reveal that, in HeLa cells, dominant inhibition of the Gag-AP-3 interaction eliminated the colocalization of HIV-1 Gag with the LE/MVB marker CD63 (22). To examine whether AP-3 deficiency also affects Gag-CD63 colocalization, we performed quantitative fluorescence colocalization studies. In our experiments, Gag-GFP and HcRed-CD63 were cotransfected into normal and HPS2 fibroblasts. At 30 to 36 h after transfection, cells were fixed for confocal microscopy. In normal fibroblasts, a higher level of the colocalization of Gag-GFP with HcRed-CD63 was observed (R, 0.88 ± 0.02; mean ± standard deviation [SD]) (Fig. 5D). In contrast, a lower level of Gag-GFP/HcRed-CD63 colocalization was detected in HPS2 fibroblasts (R, 0.38 ± 0.03) (Fig. 5E). The difference in levels of Gag-GFP/HcRed-CD63 colocalization was found between normal and HPS2 fibroblasts, suggesting that AP-3 deficiency reduces HIV-1 Gag localization at the LE/MVB compartments (Fig. 5F).
The reduced Gag localization at the LE/MVB seen in HPS2 fibroblasts implied that Gag is entrapped in other subcellular compartments. To determine the identity of subcellular compartments where HIV-1 Gag is enriched in HPS2 fibroblasts, additional markers for the different subcellular compartments were employed in our experiments. Recent studies suggest that the AP-3 complex mediates membrane protein trafficking from the TGN or early endosomes to the LE/MVB compartments. As an AP-3 cargo protein, HIV-1 Gag may accumulate within the TGN or early endosomes in AP-3-deficient HPS2 fibroblasts. To test this hypothesis, Gag-CFP and YFP-Golgi, or Gag-HcRed and Rab5-GFP, were cotransfected into HPS2 fibroblasts (Fig. 6A and B). However, either Gag-CFP did not colocalize with the marker for the trans-medial region of the Golgi apparatus (Fig. 6A, right and far right), or Gag-HcRed did not colocalize with the Rab5-positive early endosomes (Fig. 6B, right and far right). We next examined whether HIV-1 Gag without a fluorescent tag colocalizes with endogenous TGN38 (a marker for the TGN) or EEA1 (early endosome antigen 1, a marker for early endosomes). Similarly, Gag proteins did not accumulate at either TGN38-associated trans-Golgi network (Fig. 6C, right and far right) or EEA1-positive early endosomal compartments (Fig. 6D, right and far right). We also assessed the colocalization of Gag with the endosomal SNARE (soluble N-ethylmaleimide-sensitive factor-activating protein receptor) proteins Vti1b (vesicle transport through interaction with t-SNAREs homolog 1B) and syntaxin 8, two potential markers for the transport intermediates between early and late endosomes (1, 4, 7, 57). In normal fibroblast cultures, Gag exhibited lower levels of colocalization with Vti1b (R, 0.13 ± 0.03) (Fig. 6E) or syntaxin 8 (R, 0.18 ± 0.02) (Fig. 6G). However, in HPS2 fibroblasts, the degree of colocalization between Gag and Vti1b (R, 0.55 ± 0.05) (Fig. 6F) or syntaxin 8 (R, 0.43 ± 0.01) (Fig. 6H) was increased by 2- to 4-fold. The increased accumulations of HIV-1 Gag at the Vti1b- and syntaxin 8-positive endosomes in HPS2 cells suggest that AP-3 deficiency impairs the endosomal trafficking of the HIV-1 Gag protein.
Fig 6.
Defective AP-3 increases the accumulation of HIV-1 Gag at the transport intermediates between early and late endosomes. (A and B) HPS2 fibroblast cultures were cotransfected with Gag-CFP and YFP-Golgi (A) or Gag-HcRed and Rab5-GFP (B) expression plasmids. Gag-CFP or Rab5-GFP is shown in green (far left). YFP-Golgi or Gag-HcRed is shown in red (left). Merged images are shown on the right. Enlarged views of the boxed regions in merged images are shown in far right panels (A′ and B′). Bars, 10 μm. (C and D) HPS2 fibroblast cultures were transfected with Gag expression plasmids. Transfected cells were fixed, permeabilized, and immunostained with poly-clonal anti-p17 antisera and either anti-TGN38 antibodies (C) or anti-EEA1 antibodies (D). Gag is shown in green (far left), and TGN38 or EEA1 is shown in red (left). Merged images are shown on the right. Enlarged views of the boxed regions in the merged images are shown in the far right panels (C′ and D′). Bars, 10 μm. (E to H) Normal (E and G) and HPS2 (F and H) fibroblast cultures were transfected with Gag expression plasmids. Transfected cells were immunostained with polyclonal anti-p17 antisera and either anti-Vti1b antibodies (E and F) or anti-syntaxin 8 antibodies (G and H). Gag is shown in green (far left), and Vti1b or syntaxin 8 is shown in red (left). Merged images are shown on the right. Enlarged views of the boxed regions in the merged images are shown in the far right panels (E′, F′, G′, and H′). Bars, 10 μm. The degrees of colocalization between Gag and Vti1b or syntaxin 8 in normal and HPS2 cells were quantified using the Pearson correlation coefficient. Data are shown as means ± SD from 15 to 20 cells. Three independent experiments were performed.
Blockage of clathrin-mediated endocytosis in HPS2 fibroblasts does not restore the impaired virus release.
Previous studies proposed that AP-3 deficiency results in increased lysosomal membrane protein trafficking to late endocytic compartments through the indirect pathway via clathrin-mediated endocytosis from the PM (18, 36). Thus, it is possible that HIV-1 Gag follows a similar indirect pathway in AP-3-deficient cells. If so, the decreased particle release from HPS2 fibroblasts might result from the enhanced endocytic events occurring at the PM. To test this hypothesis, we used a DN approach to specifically interfere with early stages of the clathrin-dependent endocytic pathway in HPS2 fibroblasts and examined whether the inhibited particle assembly and release were reversed. In our experiments, dominant-negative mutants of dynamin (K44A) or Eps15 (ΔUIM) were employed to disrupt clathrin-coated vesicle budding from the PM or clathrin-coated vesicle formation at the PM, respectively (56, 62). Our transferrin uptake experiments indicated that HPS2 fibroblasts in the absence of DN dynamin or DN Eps15 exhibited the highly efficient cellular uptake of Alexa Fluor 546-conjugated transferrin from the surface (Fig. 7A). In contrast, expression of DN dynamin or DN Eps15 in HPS2 fibroblasts eliminated or greatly reduced the uptake of fluorescently labeled transferrin from the surface (Fig. 7B and C). These studies confirmed that either dynamin (K44A) or Eps15 (ΔUIM) could function as the endocytosis inhibitor.
Fig 7.
Blockage of clathrin-mediated endocytosis does not restore the inhibited virus release from HPS2 fibroblast cultures. (A to C) HPS2 fibroblast cultures were transfected with pEGFP (A), GFP-dynamin (K44A) (B), or FLAG-Eps15 (ΔUIM) (C) expression plasmids. At 24 h after transfection, HPS2 fibroblast cultures were subjected to a transferrin uptake assay, followed by fixation, permeabilization, and immunostaining with anti-GFP antibodies (A and B) or anti-FLAG antibodies (C). EGFP, GFP-dynamin (K44A), or FLAG-Eps15 (ΔUIM) is shown in green. Transferrin (Tf) is shown in red. Bars, 10 μm. (D) Normal (left) and HPS2 (right) fibroblast cultures were cotransfected with pNL4-3 proviral DNA and empty pcDNA3.1 vectors (Ctrl), dynamin (K44A), or Eps15 (ΔUIM) expression plasmids at a ratio of 1:1. At 48 h after transfection, cell and pelleted virion lysates were subjected to Western blot analysis using anti-HIV Ig and anti-actin antibodies. (E) HPS2 fibroblast cultures were cotransfected with Gag-protease expression plasmids (Gag-Pr) and empty pcDNA3.1 vectors (Ctrl), dynamin (K44A), or Eps15 (ΔUIM) expression plasmids at a ratio of 1:1. At 48 h after transfection, Western blot analysis of cell and pelleted virion lysates was performed using anti-HIV Ig and anti-actin antibodies.
We then examined the effects of the indicated endocytosis inhibitor on virus or VLP release from HPS2 fibroblasts. In our experiments, normal or HPS2 fibroblasts were transfected with NL4-3 as well as either dynamin (K44A) or Eps15 (ΔUIM). Immunoblot analysis of cell and virion lysates using anti-HIV Ig indicated that expression of either endocytosis inhibitor in HPS2 fibroblasts did not alter virus assembly and release (Fig. 7D). To confirm the role of the clathrin-mediated endocytic pathway in AP-3-dependent particle assembly and release, Gag-protease expression plasmids, along with dynamin (K44A) or Eps15 (ΔUIM) expression plasmids, were transfected into HPS2 cells. Similarly, expression of either endocytosis inhibitor in HPS2 cells did not increase or decrease Gag VLP release (Fig. 7E). Together, these results indicated that impaired particle assembly and release imposed by AP-3 deficiency were not restored by blocking the clathrin-mediated endocytic pathway.
DISCUSSION
In this study, we characterized HIV-1 particle assembly and release in AP-3-deficient cells derived from a naturally occurring deficiency in this pathway, HPS2. We described previously that siRNA-mediated knockdown of the AP-3 complex impaired HIV-1 particle assembly and release (22). Here, we showed that HPS2 cells were inefficient in particle assembly and release and that the impaired particle assembly and release were rescued by reconstituting the functional AP-3 complex. Confocal microscopy revealed that AP-3 deficiency increased the accumulation of HIV-1 Gag at the Vti1b- and syntaxin 8-positive endosomes, resulting in a decrease in Gag localization to the PM. These results confirm an active role for the AP-3 complex in late stages of the HIV-1 replication cycle. Furthermore, AP-3 deficiency-mediated impairment of virus assembly and release was not restored by inhibition of the clathrin-mediated endocytic pathway.
To date, only 12 HPS2 cases were reported. Genetic studies have revealed that HPS2 patients carry mutations in the AP3B1 gene, which encodes the β3A subunit of the AP-3 complex (18, 26, 35, 64, 72). In cells derived from HPS2 patients, all four subunits of the AP-3 complex become highly unstable and more sensitive to proteasome-mediated degradation (18, 26, 35). As a consequence, the AP-3-mediated transport pathway is impaired, as characterized by increased trafficking of lysosomal membrane proteins to the PM (13, 18, 26). Indeed, our flow cytometric assays indicated that, in the absence of AP-3, the cell surface expression levels of lysosomal membrane proteins CD63 and LAMP-1 were enhanced by 2- to 3-fold. However, our immunofluorescence microscopy studies showed that AP-3 deficiency did not alter the steady-state distribution pattern of CD63 and LAMP-1, confirming that the two missorted proteins follow an AP-3-independent pathway for transport to late endocytic compartments by the recruitment of the AP-2 sorting machinery at the PM (9, 18).
Our p24 antigen ELISA and Western blotting results, showing reduced particle release from HPS2 cells relative to normal cells, are consistent with results reported for studies in transformed cells when using an siRNA approach to deplete endogenous AP-3 (11, 22). Similar observations were also documented in HIV-1-infected dendritic cells (cis-infection pathway) (29). Reconstitution of the AP-3 complex in HPS2 cells by transiently or stably expressing the full-length wild-type β3A subunit not only restored the impaired AP-3-dependent trafficking of CD63 and LAMP-1 but also rescued the inhibited HIV-1 particle assembly and release, emphasizing a specific role for the AP-3 sorting machinery in late stages of the HIV-1 replication cycle (5, 11, 22, 29).
Expressing wild-type β3A in HPS2 cells improved the yields of extracellular Gag-Pol VLPs or Gag VLPs other than SrcΔMAGag VLPs, demonstrating that the MA domain is the key viral determinant required for the recruitment of the AP-3 sorting machinery. Indeed, our yeast two-hybrid studies further identified that a sequence of Lys-Trp-Glu-Lys-Ile-Arg at amino acids 15 to 20 within a predicted helical structure at the N terminus of the MA domain is required for binding to the AP-3δ subunit (data not shown). Double alanine substitutions within this sequence failed to bind to the AP-3δ subunit and impaired particle assembly and release (data not shown), suggesting that the KWEKIR motif of the MA protein may be involved in the recruitment of AP-3 to facilitate HIV-1 assembly and release. We should note, however, that evidence for direct binding between the δ-subunit and MA was not reproducible by nuclear magnetic resonance methods that employed recombinant proteins in solution (42). This may indicate that the interaction requires cellular membranes or other factors present when examined in yeast or mammalian cells, or potentially that a direct interaction between MA and AP-3δ does not adequately explain the role of AP-3 on HIV-1 Gag trafficking and assembly.
In addition to the Gag–AP-3δ interaction required for HIV-1 particle assembly and release, the interaction of β3A with the microtubule-based anterograde motor Kif3A (kinesin family member 3A) has been reported to be involved in HIV-1 Gag release (5). In this study, Azevedo et al. proposed that IP7 (diphosphoinositol pentakisphosphate)-mediated pyrophosphorylation of β3A is involved in regulating the β3A-Kif3A interaction (5). The mechanisms of Kif3A-mediated linkage of the microtubule cytoskeleton to the AP3-dependent trafficking of HIV-1 Gag warrant further investigation.
Consistent with the impaired virus or VLP release from HPS2 cells, our confocal microscopy studies of subcellular localization of Gag-CFP or Gag in normal and HPS2 cells revealed that AP-3 deficiency prevented HIV-1 Gag localization to the PM. Nevertheless, decreased instead of increased Gag localization to the PM in HPS2 cells is in contrast to lysosomal membrane proteins, whose trafficking is affected by AP-3 deficiency through indirect mechanisms. As mentioned above, in AP-3-deficient cells, lysosomal membrane proteins are mistargeted to the PM, followed by the recruitment of the AP-2 sorting machinery at the PM to transport them to late endosomal compartments. As a consequence of altered trafficking of these proteins in AP-3-deficient cells, cell surface expression of lysosomal membrane proteins is increased (18). We consider the possibility that AP-3 deficiency may promote clathrin-mediated endocytic events, resulting in reduced PM localization of HIV-1 Gag. Indeed, a previous study from the Thali laboratory reported a negative role for clathrin-associated AP-2 complex in HIV-1 assembly and release through the reciprocal interaction of Gag with AP-2 (6). However, dominant inhibition of early stages of the clathrin-mediated endocytic pathway in HPS2 cells did not significantly increase NL4-3 or Gag VLP release, excluding the PM origin for intracellular compartment-enriched Gag populations in HPS2 cells.
In mammalian cells, the AP-3 sorting machinery mediates membrane protein trafficking to late endocytic compartments (17). AP-3 deficiency, therefore, may prevent Gag from reaching LE/MVB compartments. Our Gag-GFP/HcRed-CD63 colocalization studies in normal and HPS2 cells support that AP-3 deficiency decreases HIV-1 Gag trafficking to LE/MVB compartments. However, HPS2 cells also exhibited low to moderate levels of Gag-GFP/HcRed-CD63 colocalization (R, 0.38 ± 0.03), raising the possibility that other host factors might complement AP-3 function in Gag trafficking to LE/MVB compartments. Indeed, AP-1 has been identified to mediate retroviral Gag trafficking to LE/MVB compartments and promote retroviral Gag release (11). AP-1 silencing eliminated the colocalization of retroviral Gag with LE/MVB compartments and inhibited virus budding and release, suggesting that AP-1 may cooperate with AP-3 on mediating retroviral Gag trafficking to LE/MVB compartments. This hypothesis was further supported by their studies showing that depletion of AP-1 in combination with AP-3 induced an increased inhibition of HIV-1 release compared to depletion of AP-1 alone or depletion of AP-3 alone (11).
Previous immunofluorescence and immuno-electron microscopy studies of AP-3 localization in mammalian cells indicated that AP-3 recruits cargo proteins on the TGN or early endosomes (53, 65). As the AP-3 cargo protein, HIV-1 Gag in AP-3-deficient HPS2 cells was supposed to be accumulated within the TGN or early endosomes. Surprisingly, our fluorescence microscopy studies of HPS2 cells did not reveal a significant colocalization of Gag-CFP, Gag-HcRed, or Gag with either the trans-medial region of the Golgi apparatus, TGN38-positive trans-Golgi network, Rab5-positive early endosomes, or EEA1-positive early endosomes. In contrast, the levels of colocalization of Gag with the endosomal SNARE proteins Vti1b and syntaxin 8 were increased compared to normal cells, suggesting that AP-3 may be involved in the trafficking of HIV-1 Gag between early and late endosomes. A previous study with AP-3 δ−/− mocha mouse fibroblasts also reported that AP-3 deficiency increased the accumulation of the AP-3 cargo protein NPC1 (Niemann-Pick type C protein 1) at Vti1b- and syntaxin 8-positive endosomes (7). Another important consideration for the present study is that the colocalization of Gag and the LE/MVB compartment observed by immunofluorescence microscopy in a number of cell lines is at least partly due to retention of particles by tetherin, followed by endocytosis to this compartment (48, 69). Therefore, the static examination of Gag within intracellular locations may not represent outward or productive movement of Gag. Instead, the association of Gag and MVB markers at intracellular sites may represent a surrogate for particle formation at the PM when cells express tetherin and Gag is expressed in the absence of Vpu. This does not negate the fact that productive particle release was diminished upon AP-3 depletion by siRNA or in HPS2 cells and restored when AP-3 was restored.
Of note, in addition to its sorting role within the endocytic pathway, AP-3 has been determined to play a role in sorting protein to the regulated secretory pathway (3, 10). Several previous studies indicated that intracellular Ca2+ levels modulate HIV-1 Gag trafficking and assembly, presumably by regulating exocytic events (23, 24, 33, 54). It is possible that AP-3 mediates HIV-1 Gag trafficking to the PM along the secretory pathway, which is regulated by intracellular Ca2+ signaling. The linkage between Ca2+ signaling and the AP-3-dependent secretory pathway in HIV-1 assembly and release needs further investigation.
In summary, our studies in AP-3-deficient cells derived from HPS2 patients provide support for a productive role of the AP-3 complex in HIV-1 particle assembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marc Caron, Hans-Georg Kräusslich, Simona Polo, Margaret Robinson, Adolfo Saiardi, Xiao-Fang Yu, and Bindong Liu for reagents. We thank the Meharry Morphology Core for assistance with confocal microscopy. We thank Qiujia Shao for assistance with flow cytometry.
This work was supported primarily by NIH grants R21AI089330, G12MD007586, U54MD007593, and P30AI054999 (to X.D.). Additional support was provided by R01AI040338 (to P.S.), SC1AI080580 (to F.V.), and T32HL007737 (to J.S.).
Footnotes
Published ahead of print 8 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Antonin W, et al. 2000. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19:6453–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aramori I, et al. 1997. Molecular mechanism of desensitization of the chemokine receptor CCR-5: receptor signaling and internalization are dissociable from its role as an HIV-1receptor. EMBO J. 16:4606–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asensio CS, Sirkis DW, Edwards RH. 2010. RNAi screen identifies a role for adaptor protein AP-3 in sorting to the regulated secretory pathway. J. Cell Biol. 191:1173–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atlashkin V, et al. 2003. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol. Cell. Biol. 23:5198–5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. 2009. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. U. S. A. 106:21161–21166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Batonick M, et al. 2005. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology 342:190–200 [DOI] [PubMed] [Google Scholar]
- 7. Berger AC, et al. 2007. The subcellular localization of the Niemann-Pick type C proteins depends on the adaptor complex. J. Cell Sci. 120:3640–3652 [DOI] [PubMed] [Google Scholar]
- 8. Bieniasz PD. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55–63 [DOI] [PubMed] [Google Scholar]
- 9. Bonifacino JS. 2004. Insights into the biogenesis of lysosome-related organelles from the study of the Hermansky-Pudlak syndrome. Ann. N. Y. Acad. Sci. 1038:103–114 [DOI] [PubMed] [Google Scholar]
- 10. Bossi G, Griffiths GM. 2005. CTL secretory lysosomes: biogenesis and secretion of a harmful organelle. Semin. Immunol. 17:87–94 [DOI] [PubMed] [Google Scholar]
- 11. Camus G, et al. 2007. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol. Biol. Cell 18:3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiang P-W, Spector E, Thomas M, Frei-Jones M. 2010. Novel mutation causing Hermansky-Pudlak syndrome type 2. Pediatr. Blood Cancer 55:1438. [DOI] [PubMed] [Google Scholar]
- 13. Clark RH, et al. 2003. Adaptor protein 3-dependent microtubule-mediated movement of lytic granules to the immunological synapse. Nat. Immunol. 4:1111–1120 [DOI] [PubMed] [Google Scholar]
- 14. Cooper J, et al. 2011. Filamin a protein interacts with human immunodeficiency virus type 1 Gag protein and contributes to productive particle assembly. J. Biol. Chem. 286:28498–28510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cowles CR, Odorizzi G, Payne GS, Emr SD. 1997. The adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell 91:109–118 [DOI] [PubMed] [Google Scholar]
- 16. De Guzman RN, et al. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384–388 [DOI] [PubMed] [Google Scholar]
- 17. Dell'Angelica EC. 2009. AP-3-dependent trafficking and disease: the first decade. Curr. Opin. Cell Biol. 21:552–559 [DOI] [PubMed] [Google Scholar]
- 18. Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. 1999. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the adaptor. Mol. Cell 3:11–21 [DOI] [PubMed] [Google Scholar]
- 19. Demirov DG, Freed EO. 2004. Retrovirus budding. Virus Res. 106:87–102 [DOI] [PubMed] [Google Scholar]
- 20. Derdowski A, Ding L, Spearman P. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Pietro SM, Dell'Angelica EC. 2005. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic 6:525–533 [DOI] [PubMed] [Google Scholar]
- 22. Dong X, et al. 2005. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120:663–674 [DOI] [PubMed] [Google Scholar]
- 23. Ehrlich LS, Medina GN, Carter CA. 2011. ESCRT machinery potentiates HIV-1 utilization of the PI(4,5)P2-PLC-IP3R-Ca2+ signaling cascade. J. Mol. Biol. 413:347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ehrlich LS, et al. 2010. Activation of the inositol (1,4,5)-triphosphate calcium gate receptor is required for HIV-1 Gag release. J. Virol. 84:6438–6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Enders A, et al. 2006. Lethal hemophagocytic lymphohistiocytosis in Hermansky-Pudlak syndrome type II. Blood 108:81–87 [DOI] [PubMed] [Google Scholar]
- 26. Fontana S, et al. 2006. Innate immunity defects in Hermansky-Pudlak type 2 syndrome. Blood 107:4857–4864 [DOI] [PubMed] [Google Scholar]
- 27. Freed EO. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1–15 [DOI] [PubMed] [Google Scholar]
- 28. Ganser-Pornillos BK, Yeager M, Sundquist WI. 2008. The structural biology of. Curr. Opin. Struct. Biol. 18:203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia E, Nikolic DS, Piguet V. 2008. HIV-1 replication in dendritic cells occurs through a tetraspanin-containing compartment enriched in AP-3. Traffic 9:200–214 [DOI] [PubMed] [Google Scholar]
- 30. Garrus JE, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 31. Gheysen D, et al. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103–112 [DOI] [PubMed] [Google Scholar]
- 32. Gochuico BR, et al. 2012. Interstitial lung disease and pulmonary fibrosis in Hermansky-Pudlak syndrome type 2, an AP-3 complex disease. Mol. Med. 18:56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grigorov B, Arcanger F, Roingeard P, Darlix JL, Muriaux D. 2006. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 359:848–862 [DOI] [PubMed] [Google Scholar]
- 34. Reference deleted.
- 35. Huizing M, et al. 2002. Nonsense mutations in ADTB3A cause complete deficiency of the β3A subunit of adaptor complex-3 and severe Hermansky-Pudlak syndrome type 2. Pediatr. Res. 51:150–158 [DOI] [PubMed] [Google Scholar]
- 36. Janvier K, Bonifacino JS. 2005. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell 16:4231–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joshi A, Garg H, Ablan SD, Freed EO. 2011. Evidence of a role for soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) machinery in HIV-1 assembly and release. J. Biol. Chem. 286:29861–29871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joshi A, Garg H, Nagashima K, Bonifacino JS, Freed EO. 2008. GGA and Arf proteins modulate retrovirus assembly and release. Mol. Cell 30:227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jung J, et al. 2006. Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood 108:362–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kantheti P, et al. 1998. Mutation in delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron 21:111–122 [DOI] [PubMed] [Google Scholar]
- 41. Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. 1990. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J. Virol. 64:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kyere SK, Mercredi PY, Dong X, Spearman P, Summers MF. 15 June 2012, posting date The HIV-1 matrix protein does not interact directly with the protein interactive domain of AP-3δ. Virus Res. [Epub ahead of print.] doi:10.1016/j.viruses.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Le Borgne R, Alconada A, Bauer U, Hoflack B. 1998. The mammalian adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J. Biol. Chem. 273:29451–29461 [DOI] [PubMed] [Google Scholar]
- 44. Le Borgne R, Hoflack B. 1998. Mechanisms of protein sorting and coat assembly: insights from the clathrin-coated vesicle pathway. Curr. Opin. Cell Biol. 10:499–503 [DOI] [PubMed] [Google Scholar]
- 45. Lee E, De Camilli P. 2002. Dynamin at actin tails. Proc. Natl. Acad. Sci. U. S. A. 99:161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luo K, et al. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morita E, Sundquist WI. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425 [DOI] [PubMed] [Google Scholar]
- 48. Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 49. Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 101:14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ooi CE, Dell'Angelica EC, Bonifacino JS. 1998. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the adaptor complex to membranes. J. Cell Biol. 142:391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ooi CE, et al. 1997. Altered expression of a novel adaptin leads to defective pigment granule biogenesis in the Drosophila eye color mutant garnet. EMBO J. 16:4508–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pal R, Mumbauer S, Hoke GM, Takatsuki A, Sarngadharan MG. 1991. Brefeldin A inhibits the processing and secretion of envelope glycoproteins of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 7:707–712 [DOI] [PubMed] [Google Scholar]
- 53. Peden AA, et al. 2004. Localization of the adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 164:1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Perlman M, Resh MD. 2006. Identification of an intracellular trafficking and assembly pathway for HIV-1 Gag. Traffic 7:731–745 [DOI] [PubMed] [Google Scholar]
- 55. Polo S, et al. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416:451–455 [DOI] [PubMed] [Google Scholar]
- 56. Praefcke GJ, McMahon HT. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147 [DOI] [PubMed] [Google Scholar]
- 57. Pryor PR, et al. 2004. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 5:590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Robinson MS. 2004. Adaptable adaptors for coated vesicles. Trends Cell Biol. 14:167–174 [DOI] [PubMed] [Google Scholar]
- 59. Robinson MS, Bonifacino JS. 2001. Adaptor-related proteins. Curr. Opin. Cell Biol. 13:444–453 [DOI] [PubMed] [Google Scholar]
- 60. Rous BA, et al. 2002. Role of adaptor complex in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell 13:1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saad JS, et al. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. U. S. A. 103:11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmid EM, McMahon HT. 2007. Integrating molecular and network biology to decode endocytosis. Nature 448:883–888 [DOI] [PubMed] [Google Scholar]
- 63. Schubert U, Strebel K. 1994. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J. Virol. 68:2260–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shotelersuk V, Dell'Angelica EC, Hartnell L, Bonifacino JS, Gahl WA. 2000. A new variant of Hermansky-Pudlak syndrome due to mutations in a gene responsible for vesicle formation. Am. J. Med. 108:423–427 [DOI] [PubMed] [Google Scholar]
- 65. Simpson F, et al. 1996. A novel adaptor-related protein complex. J. Cell Biol. 133:749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Simpson F, Peden AA, Christopoulou L, Robinson MS. 1997. Characterization of the adaptor-related protein complex. J. Cell Biol. 137:835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stepp JD, Huang K, Lemmon SK. 1997. The yeast adaptor protein complex, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol. 139:1761–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sugita M, et al. 2002. Failure of trafficking and antigen presentation by CD1 in Ap-3-deficient cells. Immunity 16:697–706 [DOI] [PubMed] [Google Scholar]
- 69. Van Damme N, et al. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Verli H, Calazans A, Brindeiro R, Tanuri A, Guimaraes JA. 2007. Molecular dynamics analysis of HIV-1 matrix protein: clarifying differences between crystallographic and solution structures. J. Mol. Graph. Model. 26:62–68 [DOI] [PubMed] [Google Scholar]
- 71. VerPlank L, et al. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U. S. A. 98:7724–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wenham M, et al. 2010. Two patients with Hermansky Pudlak syndrome type 2 and novel mutations in AP3B1. Haematologicaae 95:333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wills JW, Craven RC. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639–654 [DOI] [PubMed] [Google Scholar]
- 74. Wodrich H, Schambach A, Kräusslich H-G. 2000. Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 28:901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang F, Zang T, Wilson SJ, Johnson MC, Bieniasz PD. 2011. Clathrin facilitates the morphogenesis of retrovirus particles. PLoS Pathog. 7:e1002119 doi:10.1371/journal.ppat.1002119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.