Abstract
Stress granules (SGs) are cytoplasmic structures that are induced in response to environmental stress, including viral infections. Here we report that hepatitis C virus (HCV) triggers the appearance of SGs in a PKR- and interferon (IFN)-dependent manner. Moreover, we show an inverse correlation between the presence of stress granules and the induction of IFN-stimulated proteins, i.e., MxA and USP18, in HCV-infected cells despite high-level expression of the corresponding MxA and USP18 mRNAs, suggesting that interferon-stimulated gene translation is inhibited in stress granule-containing HCV-infected cells. Finally, in short hairpin RNA (shRNA) knockdown experiments, we found that the stress granule proteins T-cell-restricted intracellular antigen 1 (TIA-1), TIA1-related protein (TIAR), and RasGAP-SH3 domain binding protein 1 (G3BP1) are required for efficient HCV RNA and protein accumulation at early time points in the infection and that G3BP1 and TIA-1 are required for intracellular and extracellular infectious virus production late in the infection, suggesting that they are required for virus assembly. In contrast, TIAR downregulation decreases extracellular infectious virus titers with little effect on intracellular RNA content or infectivity late in the infection, suggesting that it is required for infectious particle release. Collectively, these results illustrate that HCV exploits the stress granule machinery at least two ways: by inducing the formation of SGs by triggering PKR phosphorylation, thereby downregulating the translation of antiviral interferon-stimulated genes, and by co-opting SG proteins for its replication, assembly, and egress.
INTRODUCTION
Hepatitis C virus (HCV) is a major human pathogen. Over 170 million people are chronically infected, many of whom develop chronic liver disease and hepatocellular carcinoma (1). There is no vaccine against HCV, and the most widely used therapy, pegylated alpha interferon (IFN-α) combined with ribavirin with or without small-molecule protease inhibitors, is not universally curative and has significant side effects (46).
HCV, the sole member of the genus Hepacivirus within the Flaviviridae family (33), is an enveloped, single-stranded, positive-sense RNA virus (10). The HCV genome contains a long open reading frame (ORF) that encodes a single polyprotein of approximately 3,000 amino acids (10). The ORF is flanked by 5′ and 3′ nontranslated regions (NTR) that regulate RNA translation and replication (14, 15, 21). Polyprotein translation is driven by a highly structured internal ribosome entry site (IRES) located in the 5′ NTR (21). The polyprotein is co- and posttranslationally processed by cellular and viral proteases, leading to the expression of the structural (core, E1, and E2) and nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins (37). HCV triggers the autophosphorylation of protein kinase R (PKR), an interferon-induced, double-stranded RNA-activated protein kinase (47) that phosphorylates the α subunit of eukaryotic translation initiation factor (eIF2α), thereby inhibiting initiation of translation of capped cellular mRNA (42) and triggering the formation of stress granules (9).
Stress granules (SGs) are large (50 to 200 nm), dynamic cytoplasmic structures that are induced in response to environmental stress, including viral infections (8). These granules contain stalled translation preinitiation complexes characterized by the presence of cellular mRNAs, translational initiation factors (i.e., eIF4E, eIF4G, eIF4A, eIF4B, eIF3, and the small subunit of the ribosome), and RNA binding proteins, including T-cell-restricted intracellular antigen 1 (TIA-1), the homologous TIA1-related protein TIAR, and RasGAP-SH3 domain binding protein 1 (G3BP1) (2, 3, 9, 28).
Although the physiological function of stress granules is uncertain, it has been suggested that during environmental stresses, they serve as a “way station” through which nontranslated mRNAs pass before being translated or degraded (4). Nonetheless, there is growing evidence that several viruses modulate SG assembly (8, 43). For example, Semliki Forest virus (34), rotavirus (36), West Nile virus (12, 29), and dengue virus (12) inhibit SG formation presumably to blunt their ability to interfere and/or limit viral growth. Poliovirus modulates the composition of SGs by cleaving G3BP1 (49) without interfering with their assembly (38). In contrast, respiratory syncytial virus (20, 31, 32), mouse hepatitis coronavirus (40), and reovirus (39) transiently induce SGs and, in some cases, benefit from them (31). Using an exogenously expressed enhanced green fluorescent protein (EGFP)-G3BP1 construct and live-imaging technology, Jones et al. have recently shown that the assembly and disassembly of G3BP1 containing granules is a dynamic process in HCV-infected cells (23). Ariumi et al. have reported the relocalization of G3BP1 protein as well as P-body components around lipid droplets in HCV-infected cells (6). Finally, Yi et al. have shown that G3BP1 interacts with the HCV NS5B protein and is required for RNA replication in the HCV life cycle (51). Despite these observations, the mechanism by which stress granules are induced during HCV infection and the function of stress granule proteins in the HCV life cycle remain largely unknown.
In this study, we addressed some of those issues using the in vitro HCV cell culture infection model (30, 48, 52). Our results indicate that HCV induces bona fide stress granule formation in infected cells by a PKR kinase-dependent process that is augmented by IFN, since PKR itself is an interferon-stimulated gene product. We also demonstrate that interferon-stimulated proteins like MxA are inversely correlated with the presence of SGs at the single-cell level in IFN-treated HCV-infected cells. Finally, we confirmed that G3BP1 protein is required for efficient HCV infection, extended that requirement to TIA-1 and TIAR proteins, and demonstrated that TIA-1, TIAR, and G3BP1 play a dual role in the HCV life cycle: first at the level of HCV RNA replication and later at the level of HCV particle assembly (TIA-1 and G3BP1) and egress (TIAR).
MATERIALS AND METHODS
Cells, plasmids, antibodies, and reagents.
The origins of Huh-7 (52), Huh-7.5.1 (52), Huh-7.5.1 subclone 2 (Huh-7.5.1c2) (17), and HEK-293T (19) cells have been described previously. All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Cellgro; Mediatech, Herndon, VA) supplemented with 10% fetal calf serum (FCS) (Cellgro), 10 mM HEPES (Invitrogen, Carlsbad, CA), 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM l-glutamine (Invitrogen) in 5% CO2 at 37°C. The subgenomic and full-length JFH-1 stable replicon Huh-7 cell lines have been previously described (17) and were cultured in medium supplemented with G418 (400 and 250 μg/ml, respectively). The plasmids containing the JFH-1 (25) and H77S genomes were kindly provided by T. Wakita and S. Lemon, respectively. Lentiviral vectors encoding short hairpin RNAs (shRNAs) targeting PKR (16), G3BP1, TIA-1, TIAR, and the nontargeting control (shCtrl) were commercially available (Sigma-Aldrich, St. Louis, MO), and their sequences are shown below. Vectors encoding compatible packaging proteins and vesicular stomatitis virus glycoprotein (VSV-G) were kindly provided by Inder Verma (Salk Institute, La Jolla, CA). Rabbit polyclonal antibodies for the detection of cellular MxA and PKR proteins, goat polyclonal antibodies for the detection of cellular TIA-1 and TIAR proteins, and the mouse monoclonal antibody against the HCV core protein were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal antibodies for the detection of USP18, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and phospho-eIF2α (Ser 51) proteins were purchased from Cell Signaling Technology (Danvers, MA). Monoclonal antibodies against G3BP1 and EEA1 were obtained from BD Transduction Laboratories (Franklin Lakes, NJ), and antibody against β-actin protein was purchased from Sigma-Aldrich. Recombinant human IgG anti-E2 and rabbit MS5 anti-NS5A antibodies were kindly provided by D. Burton (The Scripps Research Institute, La Jolla, CA) and M. Houghton (Chiron), respectively. Recombinant human beta interferon 1a (IFN-β) was purchased from PBL Interferon Source (Piscataway, NJ). Sodium arsenite and 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich. Protease and phosphatase inhibitors were purchased from Roche (Indianapolis, IN).
Preparation of viral stocks and infections.
The original JFH-1 virus was generated by transfection of an in vitro-transcribed full-length JFH-1 HCV RNA into Huh-7 cells, and viral stocks were produced by inoculation of Huh-7 cells at a multiplicity of infection (MOI) of 0.01 as described previously (52). High-titer virus stocks of cell culture-adapted JFH-1 day 183 virus (D183) (53) were produced by inoculation of the highly susceptible Huh-7.5.1c2 at a low MOI and used for single-step virus infection experiments (MOI = 0.2 or 5) as described previously (17). Cell extracts containing intracellular infectious HCV particles were prepared by 4 freeze-thaw cycles of infected cells as described previously (18). Infectivity titers of cell culture supernatants and cell extracts were determined by endpoint dilution using Huh-7.5.1 cells as described previously (52).
Lentiviral particle production and Huh-7 cell transduction.
Lentiviral particles were produced in HEK-293T cells by cotransfection of plasmids encoding short hairpin RNAs (shRNAs) targeting the indicated gene products (see below) and the plasmids necessary for vesicular stomatitis virus glycoprotein-pseudotyped lentivirus production as described previously (16). Cell supernatants were collected at 40 h posttransfection, filtered through 0.45-μm filters, aliquoted, and kept at −80°C till needed. Twofold serial dilutions of lentiviral particles were used to inoculate Huh-7 cells, and the downregulation of target proteins was determined 7 days posttransduction by Western blotting. Proliferation of the cells (measured by cell counting) was monitored, and cell viability was confirmed by MTT cytotoxicity assays. Lentiviruses displaying no cytotoxic effect were selected for further experiments, and their sequences are as follows: for PKR, TCCTGGCTCATCTCTTTATTC; for shCtrl (nontargeting control shRNA), sequence not available; for TIA-1, no. 1, GCCGTTGTTTACTTAAAGATT, no. 3, GCCAGTATATGCCTAATGGTT, and no. 4, CGGAAGATAATGGGTAAGGAA; for TIAR, no. 1, CCCATATTGCTTTGTGGAATT, no. 2, CCACAACAGTATGGACAGTAT, and no. 6, ATTCATGATAGGCTTCGATTT; and for G3BP1, no. 1, GCCTGTAAGAAATACAGGATT, and no. 3, CCACCTCATGTTGTTAAAGTA.
Protein analyses.
For confocal immunofluorescence experiments, Huh-7 cells were grown in glass bottom 96-well plates (Nunc; Thermo Scientific, Rochester, NY) and treated or infected as indicated in the figure legends. At various time points, cells were washed with phosphate-buffered saline (PBS) and fixed for 20 min with 4% paraformaldehyde (PFA) at room temperature. After extensive washing with PBS, blocking buffer (1× PBS, 3% bovine serum albumin [BSA], 10% fetal bovine serum [FBS], 0.3% Triton X-100) was added for 1 h at room temperature, after which the cells were incubated for 1 h at room temperature with a mix of primary antibodies at the appropriate dilutions in binding buffer (1× PBS, 3% BSA, 0.3% Triton X-100). After the cells were washed with PBS, binding buffer containing a mixture of fluorophore-conjugated secondary antibodies (1:1,000 dilution) (Invitrogen) and Hoechst dye (0.5 mg/ml) (Invitrogen), for nucleus staining, was added for 1 h at room temperature. For lipid droplet staining, LipidTOX reagent (Invitrogen) was added at a 1:1,000 dilution in PBS and images were obtained using a Zeiss LSM 710 laser scanning confocal microscope. For the combined in situ hybridization and conventional immunofluorescence experiments, Huh-7 cells were infected with HCV JFH-1 D183 virus at a high multiplicity of infection (MOI = 5). Forty-eight hours later, the infected and uninfected cells were seeded in glass bottom 96-well plates at a density of 15,000 cells in each well. The next day, the cells were treated with 1,000 U/ml of IFN-β for 7 h, after which they were fixed in 4% paraformaldehyde and processed for the detection of MxA, USP18, or GAPDH mRNA using sequence-specific probes provided by Affymetrix (Santa Clara, CA) that were used according to the manufacturer's instructions. MxA, USP18, or GAPDH and G3BP1 proteins were detected by conventional immunofluorescence as described above. The Image-Pro Plus package software was used to perform single-cell quantitation analysis of confocal images followed by statistical analysis (Pearson correlation coefficients and P values) using GraphPad Prism software.
For Western blot analysis, cell extracts were prepared in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease and phosphatase inhibitors, and after protein quantitation by bicinchoninic acid (BCA) assay, equivalent amounts of total protein for each sample (typically 30 μg) were separated by polyacrylamide-SDS gel electrophoresis and transferred to Immobilon (Millipore; Billerica, MA) membranes. The membranes were blocked for 1 h at room temperature with PBS-5% milk and incubated with the primary antibodies diluted in 1% milk-0.1% Tween 20 in PBS at room temperature for various periods of time, depending on each antibody. After being washed at least 4 times for 15 min each with 0.1% Tween 20 in PBS, the filters were incubated with a dilution of goat anti-rabbit, goat anti-mouse, or mouse anti-goat IgG conjugated to horseradish peroxidase in 1% milk-0.1% Tween 20 in PBS. After being washed at least 4 more times with 0.1% Tween 20 in PBS, the filters were developed using the SuperSignal-West-Pico or -Femto substrate purchased from Thermo Scientific (Rockford, IL). Images were obtained in a Molecular Imager ChemiDocXRS+ and processed with ImageLab software (Bio-Rad, Hercules, CA). Densitometry of nonsaturated Western blots was performed using Image J analysis software (NIH), and the results are expressed relative to those for control cells using EEA1 or β-actin protein expression for normalization and as loading controls.
For immunoprecipitation experiments, Huh-7 cells were grown in 10-cm dishes and were infected with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5). Seventy-two hours after infection, when the cultures reached 80% confluence, cells were washed twice with 10 ml of ice-cold PBS and were lysed in 1 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.5% NP-40, supplemented with a cocktail of serine and cysteine protease inhibitors [catalog number 04 693 132 001; Roche] and a cocktail of phosphatase inhibitors [catalog number 04 906 845 001; Roche]). After cell debris was removed by centrifugation, the cell lysates were incubated with antibodies to G3BP1, TIA-1, and TIAR or with the corresponding isotype control antibodies at 4°C. After an overnight incubation, equal amounts of protein G-agarose beads (Roche) were added to each tube and were incubated for another 4 h at 4°C, after which the tubes were centrifuged and the supernatants containing the unbound material were collected. The beads were washed 3 times with lysis buffer, boiled at 100°C for 15 min in 2× loading buffer, and finally analyzed by SDS-PAGE and Western blotting as indicated above.
RNA analyses.
Total RNA was extracted from the cells using the guanidinium isothiocyanate extraction method (52) after 20 μg of glycogen (Roche) per sample was added as a carrier. HCV, MxA, USP18, and GAPDH (for normalization) RNA levels were measured by reverse transcription–real-time quantitative PCR (RT-qPCR) as described previously (52) using the following primers: for HCV, 5′-TCTGCGGAACCGGTGAGTA-3′ and 5′-TCAGGCAGTACCACAAGG-3′; for GAPDH, 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′; for MxA, (5′-AGAGGACCATCGGAATCTTG-3′ and 5′-CCCTTCTTCAGGTGGAACAC-3′); and for USP18, 5′-CTCAGTCCCGACGTGGAACT-3′ and 5′-ATCTCTCAAGCGCCATGCA-3′.
Downregulation experiments in persistently infected cells.
Huh-7 cells were infected with JFH-1 virus at a low multiplicity of infection (MOI = 0.1) as previously described (52). After 3 weeks of infection, the cells were inoculated with the corresponding lentiviruses. Cells were split on days 2 and 4 posttransduction. Downregulation of the corresponding target proteins was confirmed by Western blotting. On day 5, the cells were washed extensively with PBS and the medium was replaced. Forty-eight hours later, the supernatants were collected and extracellular infectivity was determined by endpoint dilution using Huh-7.5.1 cells as previously described (52). Cell extracts were harvested and the intracellular infectivity and HCV RNA were analyzed by titration and RT-qPCR, respectively, as described previously (18).
Downregulation experiments in acutely infected cells.
Huh-7 cells were inoculated with the corresponding lentiviruses. On day 7 the downregulation of each target protein was determined by Western blotting. Downregulated cells were infected with JFH-1 D183 virus at a multiplicity of infection of 0.2 (low MOI) or 5 (high MOI) as described before (52). Five hours after inoculation of the virus, the cells were washed twice with PBS and the medium was replaced. At the indicated times after infection, the supernatants were collected and extracellular infectivity was determined by endpoint dilution using Huh-7.5.1 cells as previously described (52). Cell extracts were harvested and the intracellular infectivity, HCV RNA, and viral proteins were analyzed by titration, RT-qPCR, and Western blotting, respectively, as described previously (18).
Electroporation experiments with in vitro-transcribed RNAs.
The H77S, Jc1, and Jc1 GND mutant RNAs were produced using the T7 MEGAscript (Ambion) kit by following the manufacturer's instructions. In vitro-transcribed RNAs were introduced into Huh-7.5.1c2 cells by electroporation as previously reported (52). At various time points, the cells were fixed and processed for immunofluorescence analysis or harvested for HCV RNA quantitation by RT-qPCR as described above.
RESULTS
HCV induces stress granule (SG) formation in HCV-infected cells.
It was previously reported that cells expressing an ectopic EGFP-G3BP construct would relocalize the EGFP-G3BP protein into discrete cytoplasmic aggregates upon HCV infection (23), suggesting that HCV induces the formation of SGs. In order to determine the nature of these cytoplasmic foci, we first examined the localization of endogenously expressed SG markers (G3BP1, TIA-1, and TIAR) in persistently HCV-infected Huh-7 cells and uninfected cells. As expected, endogenous G3BP1 displayed diffuse cytoplasmic distribution in uninfected Huh-7 cells, while TIA-1 and TIAR were located both in the nucleus and cytoplasm (Fig. 1A, Mock). Notably, all three markers relocalized in cytoplasmic granules in a subset of HCV-positive cells (Fig. 1A, HCV-infected), suggesting that HCV infection triggers the formation of SGs in ∼14% of the infected cells. Further confocal analysis revealed that G3BP1 colocalized with TIA-1 and TIAR in some of the infected cells, confirming the stress granule nature of those foci (Fig. 1B). Next, Huh-7 cells were infected with the cell culture-adapted JFH-1 day 183 virus (D183) at a high multiplicity of infection (MOI = 5), and the number of total infected cells and SG-positive infected cells was quantitated at various time points after infection. As shown in Fig. 1C, the fraction of HCV E2-positive cells increased over the time course of the experiment, from ∼18% at 24 h to ∼85% at the end of the experiment. Interestingly, the fraction of HCV E2-positive cells that contain SGs increased as the infection proceeded and peaked at 72 h postinfection, when ∼15% of the HCV E2-positive cells contained SGs in their cytoplasm. Importantly, SG formation was never observed in HCV-negative cells. To rule out the possibility that the SG-negative cells might be constitutively unable to respond to any stress stimulus, HCV-infected and uninfected Huh-7 cells were treated with sodium arsenite, a widely used and well-known SG inducer (5, 22, 50), at different times after infection, and the formation of SGs was monitored and quantitated. As shown in Fig. 1D, sodium arsenite induced SGs in most of the infected and uninfected cells, suggesting that the lack of response in the majority (85%) of HCV-infected cells (Fig. 1C) is not due to an intrinsic defect on the stress response pathway. These results are compatible with recent results suggesting that SG formation is an oscillatory phenomenon during HCV infection (23). To address whether SG formation is virus genotype specific, we electroporated the highly permissive Huh-7.5.1c2 cells with in vitro-transcribed H77S (genotype 1a), Jc1 (intragenotypic chimeric genotype 2a), and Jc1-GND mutant (nonreplicative) RNAs and monitored the formation of stress granules and the intracellular HCV RNA content at several times thereafter. As expected, both H77S and Jc1, but not Jc1-GND, RNA-electroporated cells were able to sustain HCV RNA replication (Fig. 1E, lines), and SGs were induced in a fraction of the H77S and Jc1 (Fig. 1E, bars). Note that electroporation of Jc1-GND RNA did not trigger any SG formation, suggesting that active HCV RNA replication is required for SG formation.
Fig 1.
HCV triggers stress granule (SG) formation in infected cells. (A) Persistently HCV-infected (at an MOI of 0.1 for 3 weeks) and uninfected control (Mock) Huh-7 cells were seeded in glass bottom 96-well plates, fixed with 4% PFA, and processed for immunofluorescence analysis. In green are stress granule markers (G3BP1, TIA-1, and TIAR); in red, viral E2 or NS5A proteins; and in blue, nuclei stained with Hoechst. Images displayed are examples of 2-μm sections of confocal microscope 40× field snapshots. These results were confirmed in 3 independent experiments. (B) Colocalization of G3BP1 with TIA-1 and TIAR in persistently HCV-infected (at an MOI of 0.1 for 3 weeks) cells. Left panels show staining of TIA-1 (upper panel) and TIAR (lower panel) in green, middle panels show staining of G3BP1 in red, and right panels show the merge of the corresponding images. Yellow dots represent colocalization of G3BP1 with TIA-1 or TIAR protein in SG structures. Images displayed are projections of 5 consecutive 0.4-μm z-series images taken with a confocal microscope. These results were confirmed in 3 independent experiments. (C) Quantitation of SG induction kinetics in HCV-infected Huh-7 cells. Huh-7 cells seeded in a glass bottom 96-well plate format were infected at an MOI of 5 with JFH-1 D183 virus and fixed at the indicated times after infection. After the last time point, all the wells were processed for the detection of cellular G3BP1 and viral E2 proteins by immunofluorescence and pictures were taken in a confocal microscope. Three to five 40× fields were randomly selected, and the presence or absence of SGs was determined in E2-positive cells. The results display the percentage of total cells that are E2 positive (blue bars) and the percentage of E2-positive cells that are also SG positive (red bars). Data are presented as averages and standard deviations (n = 3 to 5). Similar results were obtained in another two independent experiments. (D) Quantitation of sodium arsenite-induced SG formation at different times after HCV infection. Huh-7 cells were infected as described for panel C or left uninfected as a control. At the indicated times after infection, the cells were treated with 0.5 mM sodium arsenite for 45 min and the induction of SGs was quantitated by immunofluorescence as described above. The same treatment of persistently infected cells was done in parallel for comparison. The results are displayed as percentages of cells containing SGs (mean and SD; n = 5). These results are representative of two independent experiments, each one performed in duplicate. (E) Quantitation of SG induction and the intracellular HCV RNA levels in H77S, Jc1, or Jc1 GND RNA-containing cells at the indicated times after electroporation. Huh-7.5.1c2 cells were electroporated as described in Materials and Methods, and the presence of HCV-replicating cells and SGs was determined by staining with antibodies against E2 and G3BP1, respectively. Results are displayed as SG-containing E2-positive cells (left y axis; bars). Asterisks indicate absence of detection. The HCV RNA content at each time point was determined by RT-qPCR and normalized by the GAPDH mRNA levels (right y axis; lines). RNA results are displayed as averages and standard deviations of HCV RNA copy numbers per microgram of total RNA (means and SDs; n = 3). These results are representative of two independent electroporation experiments, each one performed in duplicate.
HCV-induced stress granule formation is dependent on PKR and is greatly enhanced by interferon.
We have previously reported that HCV infection triggers PKR and eIF2α phosphorylation, leading to a decrease in the cellular protein synthesis without inhibiting HCV translation or replication (16). Since SG induction is triggered by translational inhibition (9), HCV-induced PKR activation could be responsible for the SG formation observed in HCV-infected cells. To test this hypothesis, cells in which PKR had been downregulated and control cells were infected with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5) and the presence of SGs was monitored and quantitated at different times after infection. As expected (16), HCV infection triggered the phosphorylation of eIF2α in a PKR-dependent manner (Fig. 2A). Importantly, PKR downregulation almost completely abolished the induction of SG in HCV-infected cells (Fig. 2B), with no effect on the stress granule protein (G3BP1, TIA-1, and TIAR) content (Fig. 2A) or HCV RNA accumulation (data not shown) over the time course of the experiment. Together, these results indicated that PKR is the key kinase involved in HCV-induced SG formation.
Fig 2.
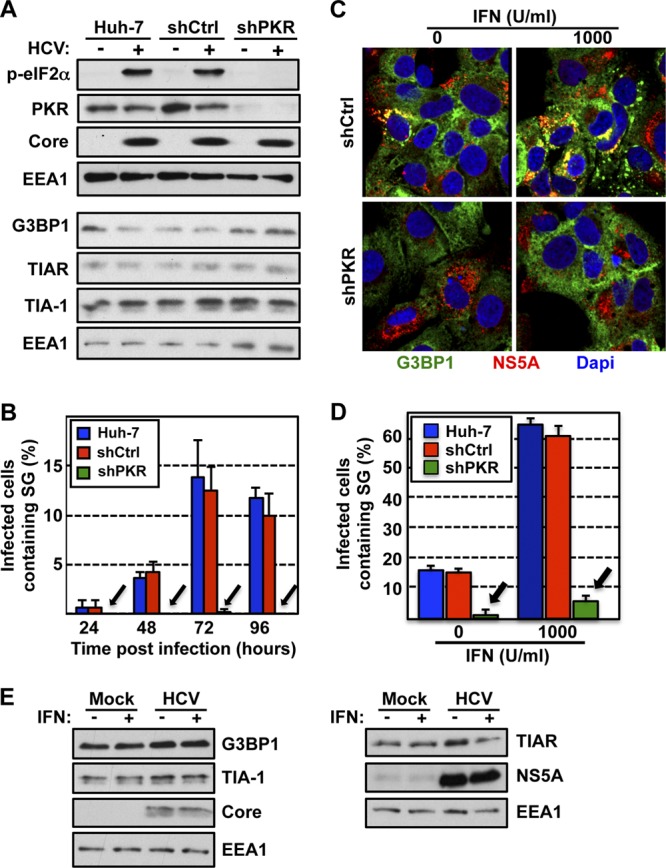
Stress granule induction in HCV-infected cells is dependent on PKR and is increased by IFN treatment. (A) Huh-7 cells were transduced with lentiviruses expressing shRNAs against PKR (shPKR) or a nontargeting shRNA control (shCtrl). Seven days after transduction, the cells were infected (+) or not (−) with JFH-1 D183 virus at an MOI of 5 and PKR, phospho-eIF2α, HCV core, G3BP1, TIAR, and TIA-1 protein expression was determined 72 h later. EEA1 expression is shown as a loading control. (B) Huh-7 cells that were transduced and infected as described for panel A were fixed with 4% PFA for 30 min at the indicated times after infection. After the last time point, all the wells were processed for the detection of cellular G3BP1 and viral E2 proteins by immunofluorescence, and pictures were taken in a confocal microscope. Three to five 40× fields were randomly selected, and the cells present in those fields (n > 200) were scored for the presence or absence of stress granules. Results are displayed as the percentage of infected cells containing SG. Data are displayed as averages and standard deviations (mean and SD; n = 3 to 5). Arrows point to the reduced SG formation in cells in which PKR had been downregulated. The results displayed are representative of three independent experiments. (C) Interferon enhances SG induction in HCV-infected cells. Huh-7 cells transduced as for panel A were infected with JFH-1 D183 virus at an MOI of 5 and 72 h later were treated with 1,000 U/ml of interferon β (IFN). Seven hours after IFN treatment, the cells were fixed and processed for immunofluorescence for the detection of G3BP1 and viral NS5A proteins. In green is G3BP1, in red NS5A, and in blue the nuclei stained with Hoechst. Images displayed are representative examples of 2-μm-thick sections of confocal microscope 40× field snapshots. (D) Three to five fields were randomly selected, and the E2-positive cells present in those fields (n > 200) were scored for the presence or absence of stress granules (based on G3BP1 staining). Results shown in the graph are displayed as percentages of infected cells containing SGs. Data are displayed as averages and standard deviations (mean and SD; n = 3 to 5). Arrows indicate the reduced SG formation in cells in which PKR had been downregulated. These results were confirmed in three independent experiments. (E) Western blotting of G3BP1, TIA-1, and TIAR expression in Huh-7 cells that were infected or not and treated or not with IFN as for panel C. HCV core and NS5A and cellular EEA1 protein expression is shown as an infection marker and a loading control, respectively.
We have also previously reported that interferon (IFN) treatment of HCV-infected cells strongly enhances HCV-induced phosphorylation of PKR and eIF2α, thereby inhibiting de novo cellular protein synthesis, including translation of antiviral interferon-stimulated genes, without inhibiting the translation of HCV proteins in the cell (16, 41, 44, 45). Since PKR and eIF2α phosphorylation appear to be required for SG induction in HCV-infected cells, we predicted that IFN would increase SG formation in HCV-infected cells. To test this hypothesis, cells in which PKR had been downregulated and control cells were infected with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5), and 72 h later, the cells were treated with 1,000 U/ml of IFN-β for 7 h, after which SG formation was detected by immunofluorescence. As shown in Fig. 2C and D, the number of HCV-infected SG-containing cells was greatly enhanced by IFN treatment (∼65% in IFN-treated versus 15% in untreated cells) in a PKR-dependent manner (∼6% in shPKR-treated cells versus 65% in shCtrl-treated cells). This effect was not due to an induction in the expression of any of the stress granule proteins G3BP1, TIA-1, and TIAR by IFN (Fig. 2E), suggesting that IFN induced the formation of SGs in the infected cells without increasing the intracellular content of these SG proteins. This increase was specific for HCV-infected cells, since uninfected cells did not contain any SGs even after IFN treatment, while IFN treatment enhanced SG formation in subgenomic replicon-containing Huh-7 cells (∼25% in IFN-treated versus 5% in untreated cells) and full-length replicon-containing Huh-7 cells (∼40% in IFN-treated versus 7% in untreated cells), although to a lower extent (data not shown). Collectively, these results demonstrate that IFN greatly enhances HCV-induced stress granule formation in a PKR-dependent manner.
Interferon-stimulated protein expression is suppressed in stress granule-positive HCV-infected Huh-7 cells.
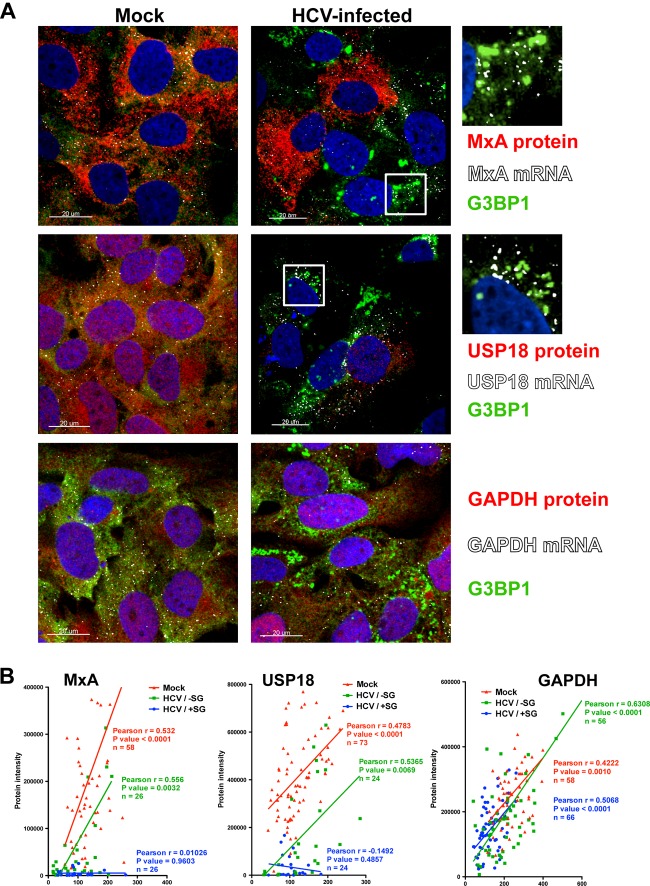
We have previously shown that interferon-stimulated protein expression is suppressed in IFN-treated HCV-infected cultures relative to that in IFN-treated uninfected cells (16). Since stress granules are known to contain stalled translation preinitiation complexes (26, 27) and are strongly induced by IFN in HCV-infected cells (see Fig. 2C and D), we examined the induction of interferon-stimulated mRNA and protein expression in HCV-infected cells at the population (Fig. 3) and single-cell (Fig. 4) levels. JFH-1 D183-infected (MOI = 5) and uninfected (mock) control Huh-7 cells were treated with IFN for 7 h, after which MxA and USP18 mRNA and protein content were determined by RT-qPCR and Western blotting, respectively. As we have previously reported (16), IFN treatment induced the expression of MxA and USP18 mRNAs to similar degrees at the population level in uninfected and HCV-infected cells (Fig. 3A). In contrast, MxA and USP18 proteins were highly induced in uninfected cells but poorly or not induced in HCV-infected cells (Fig. 3B). Confocal microscopic analysis (Fig. 4A) of parallel wells was used to study the expression of interferon-stimulated gene (MxA [upper panels] and USP18 [middle panels]) and GAPDH (lower panels) mRNAs (in white), their corresponding proteins (in red), and G3BP1 (in green) protein, as a stress granule marker, by fluorescence in situ hybridization (FISH) and conventional immunofluorescence, respectively. As shown in Fig. 4A, MxA and USP18 proteins (red stain) were highly induced by IFN in uninfected cells (Mock) and SG-negative HCV-infected cells. However, SG-positive HCV-infected cells showed reduced expression of MxA and USP18 proteins despite high-level expression of the corresponding mRNA (white dots in Fig. 4A). As shown in Fig. 4B, single-cell quantitative analysis of MxA, USP18, and GAPDH mRNA dots and protein staining revealed a highly significant positive correlation between the amount of mRNA and corresponding protein in uninfected (Mock [in red]) and SG-negative HCV-infected (HCV/−SG [in green]) cells. The same analysis performed in G3BP1-positive SG-containing HCV-infected (HCV/+SG [in blue]) cells showed a significant positive correlation for GAPDH, while there was no correlation at all for MxA or USP18. These results suggest that de novo translation of short-lived mRNAs, such as the potentially antiviral interferon-stimulated gene mRNAs, is impaired in stress granule-containing HCV-infected cells.
Fig 3.

ISG mRNA and protein induction by IFN in HCV-infected cells. Huh-7 cells were infected (HCV-infected) or not (Mock) with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5). Forty-eight hours later, cells were treated or not with 1,000 U/ml of IFN-β for 7 h. Shown are results of MxA and USP18 mRNA (A) and protein (B) analysis by RT-qPCR and Western blotting, respectively. RNA results were normalized to GAPDH mRNA and are displayed as copy numbers per microgram of total RNA. Data are displayed as the average and standard deviation (mean and SD; n = 3). Core and EEA1 protein expression is shown as an infection marker and a loading control, respectively. These results are representative of 3 independent experiments.
Fig 4.
Interferon-stimulated protein expression is suppressed in stress granule-positive HCV-infected cells. Huh-7 cells were infected (HCV-infected) or not (Mock) with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5). Forty-eight hours later, the cells were treated or not with 1,000 U/ml of IFN-β for 7 h. (A) Visualization of MxA (upper panels), USP18 (middle panels), and GAPDH (lower panels) mRNAs (white dots) and the corresponding proteins (in red) as well as G3BP1 protein (in green) by fluorescence in situ hybridization (FISH) and conventional immunofluorescence after IFN treatment of HCV-infected and uninfected control (Mock) cells. Images displayed are of single 0.3-μm z-sections taken in a confocal microscope at a magnification of ×40. Nuclei are displayed in blue. Pictures on the very right are enlargements of the boxed areas showing a partial localization of MxA or USP18 mRNAs in stress granules. (B) Single-cell quantitation analysis of the number of MxA (left graph), USP18 (middle graph), and GAPDH (right graph) mRNA dots and the corresponding protein intensity per cell in samples that were treated with IFN as described above. Three data sets are displayed in each graph: in red, uninfected cells; in green, stress granule-negative HCV-infected cells; and in blue, stress granule-positive HCV-infected cells. Each data point in the graphs represents a single cell. Pearson correlation coefficients (r), their P values, and the number of cells analyzed (n) are displayed for each data set. The results displayed are representative of 2 (USP18 and GAPDH) or 3 (MxA) independent experiments.
Stress granule proteins TIA-1, TIAR, and G3BP1 are required for efficient HCV infection.
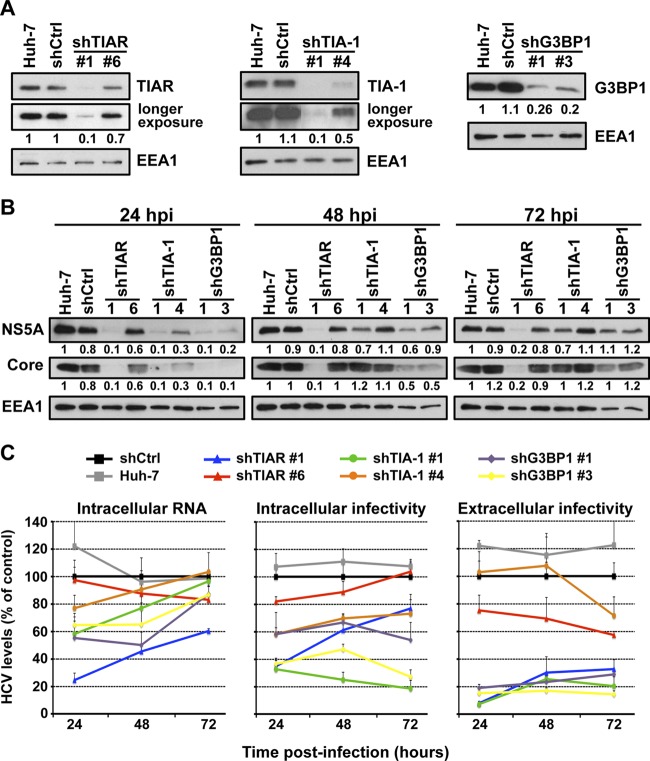
It was recently shown that the stress granule protein G3BP1 is required for efficient HCV infection (51). To extend those findings, we wished to examine if other stress granule proteins, e.g., TIA-1 and TIAR, are also required for HCV infection. We transduced Huh-7 cells with lentiviruses expressing shRNAs targeting TIA-1, TIAR, and G3BP1 and a nontargeting shRNA control (shCtrl). One week after transduction, the downregulation of the target proteins was confirmed by Western blotting (Fig. 5A), and the absence of cytotoxic effects was demonstrated by MTT assay (Fig. 5B). In order to study the SG protein requirement on HCV infection in a more sensitive system (multiple rounds of infection), downregulated and control cells were inoculated with JFH-1 D183 virus at a low multiplicity of infection (MOI = 0.2). Six days after infection, the intracellular HCV RNA content and extracellular infectivity were determined by RT-qPCR and titration, respectively. Downregulation of each of the three proteins had a negative impact on HCV infection, reducing both intracellular HCV RNA and the extracellular infectivity (Fig. 5C). These results confirmed the requirement of G3BP1 and demonstrated that TIA-1 and TIAR are also required for efficient HCV infection.
Fig 5.

Stress granule proteins TIA-1, TIAR, and G3BP1 are required for efficient HCV infection. Huh-7 cells were transduced with lentiviruses that express shRNAs targeting TIA-1, TIAR, or G3BP1 or a nontargeting shRNA control (shCtrl). (A) After 7 days, the downregulation of each protein was confirmed by Western blotting and the relative quantitation was determined by densitometry (shown below each gel). The intensity of TIA-1, TIAR, and G3BP1 proteins relative to EEA1 protein (loading control) in Huh-7 cells was set as 1 and used to calculate the relative amount of each protein. (B) Downregulated and control cells were seeded in 96-well plates at 5,000 cells per well. Four days later, cytotoxicity assays (MTT) were performed by following the manufacturer's instructions. Results are displayed as percentages of the control (shCtrl). Assays were run with 6 replicate wells per cell type in two independent experiments. (C) Downregulated cells were infected with JFH-1 D183 virus at a low multiplicity of infection (MOI = 0.2), and the intracellular HCV RNA levels and extracellular infectivity were determined on day 6 postinfection by RT-qPCR and titration assays, respectively. GAPDH mRNA quantitation of the same cellular extracts was used for normalization of the HCV RNA levels. Results are displayed as percentages of the control (Huh-7 cells). Data are displayed as averages and standard deviations (mean and SD; n = 3). These results were confirmed in 2 independent experiments performed in triplicate.
Viral proteins colocalize and/or directly interact with TIA-1, TIAR, or G3BP1 in HCV-infected cells.
To gain insight into the mechanism by which stress granule proteins TIA-1, TIAR, and G3BP1 can regulate HCV infection, colocalization and coimmunoprecipitation studies were performed. Huh-7 cells were infected with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5), and 72 h later, the cells were processed for immunofluorescence and confocal analysis. As shown in Fig. 6A, all three SG proteins (in orange) colocalized with the HCV NS5A (in red) protein to some extent (see colocalization mask at the extreme right side of Fig. 6A). Interestingly, NS5A–TIA-1, NS5A-TIAR, and NS5A-G3BP1 colocalization takes place predominantly in association with lipid droplets (in green), a very important organelle involved in viral assembly (35). These observations confirm and extend a prior report demonstrating the colocalization of the HCV core protein and G3BP1 in lipid droplets (6). These results suggest that the stress granule proteins might play a direct role in HCV infection by interacting with NS5A, core, and possibly other HCV proteins. To address this hypothesis, G3BP1, TIA-1, and TIAR proteins were immunoprecipitated from HCV-infected cells using specific and isotype control antibodies followed by Western blot analysis for the presence of HCV proteins (core, NS4A, NS4B, NS5A, and NS5B) in the immunoprecipitate and supernatant. As shown in Fig. 6B, each of the specific antibodies immunoprecipitated the corresponding target protein very efficiently, while the isotype controls did not. Importantly, a small but readily detectable fraction of the HCV NS5A and NS5B proteins was specifically immunoprecipitated by antibodies to TIA-1 and TIAR, but not G3BP1 or the corresponding isotype control antibodies.
Fig 6.
Colocalization and coimmunoprecipitation of stress granule proteins with HCV viral proteins during infection. (A) Huh-7 cells were infected with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5). Seventy-two hours later, the cells were fixed and processed by immunofluorescence for the detection of G3BP1, TIA-1, or TIAR (in orange) and viral NS5A (in red) proteins. Lipid droplets (in green) were stained using LipidTOX from Invitrogen. Images displayed are representative pictures of single 0.35-μm z-sections taken in a confocal microscope at a magnification of ×63. Nuclei are displayed in blue. The staining of each channel and the merge and the colocalization mask channel between stress granule and NS5A proteins are shown. These results were confirmed in at least 2 independent experiments. (B) Huh-7 cells were infected as described for panel A and were subjected to immunoprecipitations using G3BP1-, TIA-1-, and TIAR-specific antibodies and isotype controls 72 h later, as indicated in Materials and Methods. Detection of target proteins as well as viral core, NS4A, NS4B, NS5A, and NS5B proteins was carried out in the immunoprecipitated material (IP) and the unbound material in the supernatant (S). Gels were loaded using 5% of the starting material (Input), 20% of the immunoprecipitated material (IP), and 5% of the unbound supernatant (S). These results were confirmed in 2 independent experiments.
TIA-1, TIAR, and G3BP1 are not required to maintain HCV RNA replication once it has been established.
The multiple-cycle experiments (i.e., low-MOI infections) whose results are shown in Fig. 5 revealed that TIA-1, TIAR, and G3BP1 are required for efficient HCV infection, and the colocalization and coimmunoprecipitation studies suggested a direct interaction between SG and viral proteins, but they did not identify the step in the HCV life cycle at which they are required. Since TIA-1, TIAR, and G3BP1 are RNA binding proteins, we hypothesized that they could be required for HCV RNA replication. To test this hypothesis, we took advantage of an Huh-7 cell line that stably replicates an HCV JFH-1 subgenomic RNA that encodes all of the nonstructural HCV proteins and contains all of the cis elements required for HCV RNA replication and translation. JFH-1 subgenomic replicon cells were transduced with lentiviruses expressing shRNAs targeting TIA-1, TIAR, and G3BP1 proteins as described above. Eight days after transduction, cells were harvested, and the intracellular content of the target proteins and the HCV RNA and NS5A protein content were determined by RT-qPCR and Western blotting. As shown in Fig. 7A and B, downregulation of TIA-1, TIAR, and G3BP1 had no impact on the steady-state levels of HCV RNA or NS5A protein, suggesting that these SG proteins are not required to maintain HCV RNA replication once it has been established.
Fig 7.
TIA-1, TIAR, and G3BP1 proteins are not required for maintenance of HCV RNA replication, but they are required at a postreplication step in persistently infected cells. (A and B) Huh-7 cells bearing a subgenomic JFH-1 replicon were transduced with lentiviruses expressing shRNAs targeting TIA-1, TIAR, or G3BP1 or a nontargeting shRNA control (shCtrl). Eight days after transduction, the cells were harvested and the expression and relative quantitation of target proteins and HCV NS5A protein were determined by Western blotting (A). Densitometry results are shown under each gel image. β-Actin was used for normalization. (B) The intracellular HCV RNA content of the same samples was determined by RT-qPCR. GAPDH mRNA quantitation was used for normalization. Results are shown as percentages of the control (shCtrl). Data are displayed as averages and standard deviations (mean and SD; n = 3). These results were confirmed in 3 independent experiments performed in triplicate. (C and D) Persistently infected cells were transduced with lentiviruses expressing shRNA that target TIA-1, TIAR, or G3BP1 or an irrelevant shRNA control (shCtrl). Seven days after transduction, the supernatants were collected and the cells were harvested for further analysis. (C) The expression and relative quantitation of target proteins and HCV NS5A protein were determined by Western blotting. Densitometry results are shown under each gel image. EEA1 protein was used as a loading control. (D) The intracellular HCV RNA and infectivity titer as well as the extracellular infectivity titer were determined by RT-qPCR and titration assays, respectively. GAPDH mRNA quantitation of the same samples was used for normalization of the HCV RNA levels. Black bars, intracellular HCV RNA; gray bars, intracellular infectivity; light gray bars with black dots, extracellular infectivity. Results are shown as percentages of the control (shCtrl). Data are displayed as averages and standard deviations (n = 3). These results are representative of 3 independent experiments, each one performed in triplicate.
TIA-1, TIAR, and G3BP1 are required at early and late steps in the HCV life cycle.
Next, we analyzed the impact of TIA-1, TIAR, and G3BP1 protein downregulation on HCV RNA replication and infectious virus production and secretion in two different HCV infection models: persistently (ongoing infection) and acutely (de novo infection) infected cells. First, we used the persistently HCV JFH-1-infected Huh-7 cell model, which allows us to analyze not only HCV RNA replication but also intracellular and extracellular infectivity in an ongoing infection. Downregulation of TIA-1, TIAR, and G3BP1 proteins in persistently infected cells had little or no impact on the steady-state levels of intracellular HCV RNA (Fig. 7D) or NS5A protein (Fig. 7C) compared to the levels in nontargeting shRNA control cells. These results confirm the subgenomic replicon results shown in Fig. 7B, and they suggest that TIA-1, TIAR, and G3BP1 are not required to maintain HCV RNA replication once it has been established. Interestingly, downregulation of the three SG proteins reduced the production of both intracellular and extracellular infectious virus (Fig. 7D) suggesting that TIA-1, TIAR, and G3BP1 might regulate late steps in the HCV life cycle.
Next, we determined the impact of TIA-1, TIAR, and G3BP1 protein downregulation in acutely infected cells. This system allows us to manipulate the expression level of those proteins prior to infection and analyze its impact on HCV infection at different times postinfection. Huh-7 cells were transduced as described above, and downregulation of the target proteins was examined by Western blotting 5 days later (Fig. 8A). Cells in which the proteins had been downregulated and control cells were infected at a high multiplicity of infection (MOI = 5) with JFH-1 D183 virus, and the accumulation of intracellular HCV RNA, the accumulation of HCV NS5A and core proteins, and intracellular and extracellular viral infectivity were determined by RT-qPCR, Western blotting, and virus titration, respectively, at various times after infection. As shown in Fig. 8B, the accumulation of HCV NS5A and core proteins was reduced in cells in which TIAR, TIA-1, and G3BP1 had been downregulated compared to control cells early after infection (24 hours postinfection [hpi]) and was restored to nearly normal levels in TIA-1 and G3BP1 downregulated cells late in the infection (72 hpi). Similarly, as shown in Fig. 8C, the intracellular HCV RNA content was also reduced in TIAR, TIA-1, and G3BP1 downregulated cells early in the infection (24 hpi) and recovered to normal levels in almost all cells in which the proteins had been downregulated over the time course of the experiment (72 hpi). The reduction in HCV NS5A and core protein expression and HCV RNA content correlated with the downregulation efficiency achieved by each shRNA. These results are consistent with previous reports describing a functional requirement of G3BP1 protein in HCV RNA replication (6, 51) and suggested that TIAR and TIA-1 proteins are also required at early steps in the HCV life cycle. Moreover, our results demonstrate that the negative effect on the HCV RNA and protein content observed early after infection is overcome at later time points, suggesting that there is a delay in viral replication.
Fig 8.
TIA-1, TIAR, and G3BP1 proteins are required in early and late steps in the HCV life cycle. Huh-7 cells were transduced with lentiviruses that express shRNAs targeting TIA-1, TIAR, or G3BP1 or a nontargeting shRNA control (shCtrl). (A) After 5 days, the downregulation of the corresponding proteins was confirmed by Western blotting. EEA1 protein expression is shown as a loading control and was used to perform relative quantitation of each band by densitometry analysis. Longer-exposure images are shown for TIAR and TIA-1 gels. (B and C) Downregulated cells were infected with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5), and the accumulation of NS5A and core protein (B) and the accumulation of the intracellular HCV RNA and intracellular and extracellular infectivity (C) were determined at the indicated times postinfection by Western blotting, RT-qPCR, and titration assays, respectively. Relative quantitation of NS5A and core proteins is shown below each gel. EEA1 protein expression was used as a loading control. GAPDH mRNA quantitation of the same cellular extracts was used for normalization of the HCV RNA levels. The RNA and infectivity results are displayed as percentages of the control (shCtrl) at each time point. Data are displayed as averages and standard deviations (mean and SD; n = 3). These results are representative of 3 independent experiments performed in triplicate.
Next we examined the impact of TIAR, TIA-1, and G3BP1 downregulation on late steps of infection such as assembly and release of HCV infectious virions. Both intracellular and extracellular infectivity titers were reduced at 24 hpi in cells in which TIAR, TIA-1, and G3BP1 had been downregulated (Fig. 8C), most probably reflecting the reduced intracellular HCV RNA and viral protein content achieved in the early stage of infection. Interestingly, the extracellular infectivity titers did not increase proportionally to the expansion of HCV RNA in the downregulated cells at the 48- and 72-h time points. These results suggested that TIAR, TIA-1, and G3BP1 proteins are also required at late steps in the HCV life cycle, most likely in assembly or release. To distinguish between assembly and release, we determined the effect of TIAR, TIA-1, and G3BP1 downregulation on the accumulation of intracellular infectious virus. The level of intracellular infectious virus increased proportionally to the expansion of HCV RNA in cells in which TIAR (blue and red lines) had been downregulated, suggesting that TIAR protein is required for the release of infectious virus but not its assembly. However, the level of intracellular infectious virus did not increase proportionally to the expansion of HCV RNA in cells in which TIA-1 (green and orange lines) and G3BP1 (purple and yellow lines) had been downregulated over time. These results suggest that TIA-1 and G3BP1 are required for the assembly of infectious virus. Although different pairs of shRNAs had different degrees of effect on the HCV infection, they correlate with the degree of downregulation of the specific target protein achieved by each shRNA.
Finally, to test whether TIA-1, TIAR, and G3BP1 must be contained in SGs in order to promote efficient HCV infection, their expression was downregulated in PKR-downregulated cells in which SGs are not induced by HCV infection (see Fig. 2). After confirming the degree of downregulation of each target protein by Western blotting (Fig. 9A), the cells were infected with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5) and the intracellular HCV RNA and infectivity and the extracellular infectivity were analyzed 24 h after infection. As shown in Fig. 9B, TIA-1, TIAR, and G3BP1 downregulation had a negative effect on HCV RNA and infectivity irrespective of the presence or absence of PKR. Quantification of SG formation in all those cells demonstrated a profound reduction of SGs in all PKR downregulated cell types (Fig. 9C). These results demonstrate that TIA-1, TIAR, and G3BP1 proteins are required for efficient HCV infection in the absence of SG formation.
Fig 9.
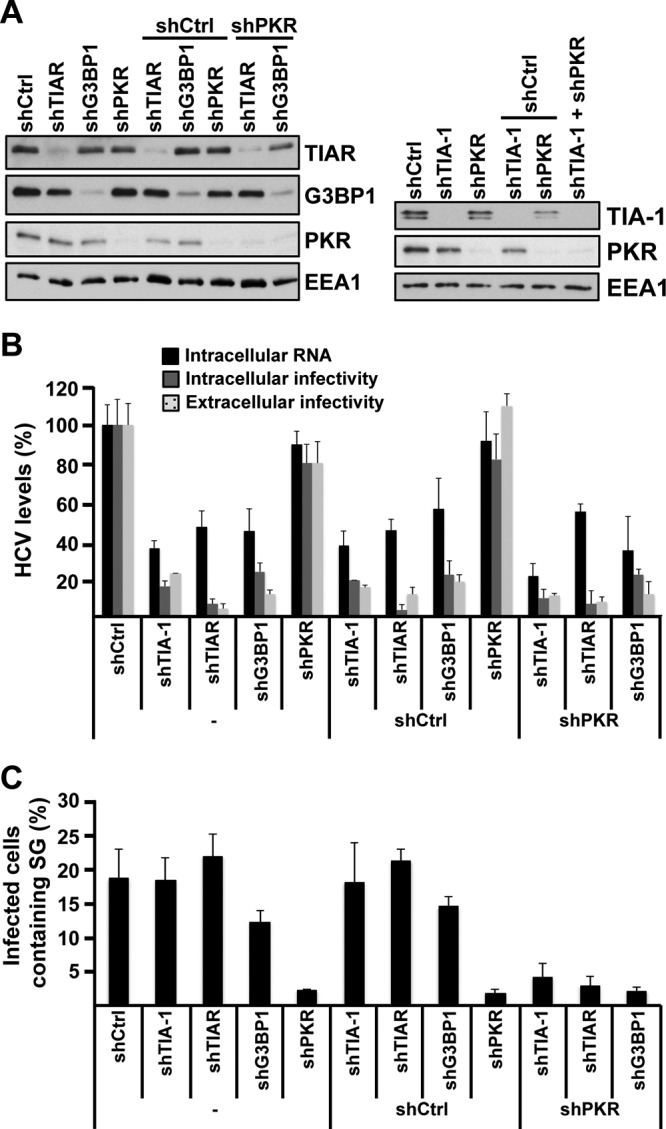
The requirement of TIA-1, TIAR, and G3BP1 proteins in HCV infection is independent of their presence in stress granules. (A) Huh-7 cells were transduced with one or two lentiviruses as indicated, and 5 days later, the expression of target proteins was determined by Western blotting. (B) After downregulation was confirmed, the cells were infected with JFH-1 D183 virus at a high multiplicity of infection (MOI = 5), and the intracellular HCV RNA and infectivity titers and extracellular infectivity titers were determined 24 h later as indicated in Materials and Methods. The results shown are averages and standard deviations (mean and SD; n = 3) (C) Cells infected in parallel were fixed 72 h after inoculation, and the number of HCV-infected cells containing stress granules was determined by immunofluorescence and counting as described for Fig. 2B. The results displayed were confirmed in 2 independent experiments, each one performed in triplicate. The shRNA sequences used in these experiments were shTIA-1 no. 1, shTIAR no. 1, shG3BP1 no. 1, shPKR, and shCtrl.
Collectively, the experiments performed in acutely infected cells suggest that TIAR, TIA-1, and G3BP1 proteins regulate early steps that affect HCV RNA and protein accumulation and that TIA-1 and G3BP1 are also required for efficient infectious virus particle assembly, while TIAR is required for infectious virus particle release.
DISCUSSION
The recent development of an in vitro HCV infection system that recapitulates all the steps of the HCV life cycle permits identification and characterization of cellular factors and pathways that regulate the infection. Using this infection model, we have previously identified cellular factors required for HCV assembly and secretion (17), and we have shown that autophagy proteins are required for the initiation of HCV replication by controlling the initiation of translation of incoming genomic RNA (11). In this study, we showed that stress granules (SGs) are induced in HCV-infected cells, we characterized their mechanism of induction, and we showed that SG proteins G3BP1, TIA-1, and TIAR are required at both early and late steps during HCV infection.
Confocal microscopic analysis demonstrated the formation of cytoplasmic structures containing SG proteins (G3BP1, TIA-1, and TIAR) in a fraction of HCV-infected cells (Fig. 1). These results are consistent with previous observations by Jones et al. in which dynamic assembly and disassembly of ectopically expressed EGFP-G3BP-containing foci were observed by live-cell imaging (23). In this study, we confirmed the formation of SGs by looking at endogenously expressed G3BP1 protein and extended the observation to two other SG proteins, i.e., TIA-1 and TIAR. Bona fide SGs are characterized by the presence of cellular mRNAs, RNA binding proteins (e.g., G3BP1, TIA-1, and TIAR), and translation initiation factors. Using fluorescence in situ hybridization and conventional immunofluorescence analysis, we showed that cellular MxA and USP18 mRNAs partially colocalized with G3BP1-containing granules in IFN-treated HCV-infected cells, suggesting that HCV triggers the formation of bona fide SGs (Fig. 4A). A recent report by Ariumi et al. described the transient induction of G3BP1 containing SGs early after HCV infection (36 to 48 hpi) and G3BP1 relocalization around lipid droplets at later time points (6). Although we did observe relocalization of G3BP1, TIA-1 or TIAR protein around structures resembling lipid droplets in some of the infected cells late in the infection (Fig. 6A), in other infected cells G3BP1 was still localized in SG (Fig. 1), suggesting a dynamic and/or variable localization of these proteins during HCV infection.
It has been previously shown that SG formation is triggered by events that inhibit cellular mRNA translation (9). We (16) and others (7, 24) have previously reported that HCV infection triggers PKR activation and subsequent eIF2α phosphorylation, leading to a small decrease in global protein synthesis that is further suppressed by treatment with type I interferon (IFN). Therefore, we asked if PKR was required for the induction of SGs in HCV infected cells. PKR downregulation almost completely abolished the formation of SGs, indicating that PKR plays a critical role in HCV-induced SG formation (Fig. 2). Consistent with the higher PKR expression level and the increased PKR and eIF2α phosphorylation levels in HCV-infected IFN-treated cells (16), IFN greatly enhanced the number of SG containing HCV-infected cells. However, IFN did not trigger SG formation in uninfected cells despite higher levels of PKR expression (data not shown), suggesting that the PKR activation state rather than the total PKR protein level itself is important for SG induction. Quantitation of the immunofluorescence and fluorescence in situ hybridization images suggests that HCV induces translational inhibition in SG-containing cells, thereby reducing interferon-stimulated protein expression without decreasing the corresponding mRNA content (Fig. 4). Analysis of a non-interferon-stimulated gene, such as that for GAPDH, did not show any reduction of GAPDH protein accumulation in SG-containing HCV-infected cells as was observed for MxA and USP18 proteins. This difference can be explained by the relatively long half-life (∼72 h) and abundance of preexisting GAPDH protein (13) compared to the typical absent or low interferon-stimulated protein content under basal conditions in Huh-7 cells (Fig. 3). Collectively, these results suggest that SG formation in HCV-infected cells reflects the ability of HCV to induce PKR and eIF2α phosphorylation and translational inhibition at the single-cell level. The dynamic oscillatory process of SG assembly and disassembly described by others (23) and the function of PKR in the formation of SG described herein might reflect a balance between the ability of HCV to trigger PKR phosphorylation and the capacity of the cell to dephosphorylate PKR in order to maintain the cellular translation. Alternatively, transient SG induction might allow the survival of the cells that otherwise could die as a consequence of prolonged inhibition of translation. This could indirectly promote viral persistence, since it has been previously demonstrated that SG induction prevents apoptosis (5).
In this study, we also obtained evidence that SG proteins G3BP1, TIA-1, and TIAR are required for efficient HCV infection affecting both early (initiation of HCV RNA replication) and late (virus particle assembly and release) events in the HCV life cycle (Fig. 5 to 9), confirming and extending results of a recent study in which G3BP1 was shown to be required for HCV infection at the level of HCV RNA amplification (51). Our results show that G3BP1 is required to initiate viral RNA replication (Fig. 8) in the early stages of infection but not to maintain it (Fig. 7), and they suggest that G3BP1 also regulates the assembly of HCV infectious virions (Fig. 7 and 8) later in the infection. Interestingly, the partial relocalization of G3BP1 around lipid droplets observed by others (6) and us (Fig. 6A) is compatible with a late function during infection, since lipid droplets have been shown to be an important platform during HCV assembly (35).
Like that of G3BP1, the downregulation of TIA-1 and TIAR displayed differential effects in acutely and persistently infected cells. Both proteins were required for efficient HCV RNA and protein accumulation early after de novo infection, but they were dispensable in ongoing infections, suggesting that initiation and maintenance of HCV RNA replication are differentially regulated. Consistent with this idea, we have previously reported the requirement of autophagy proteins for the establishment of HCV replication but not for maintenance of the infection (11). Alternatively, differences in the requirement for G3BP1, TIA-1, and TIAR in persistently infected and acutely infected cells and stable replicon-containing cells could also reflect selection or adaptation of the cells or the virus under these different conditions. In agreement with this idea, we have previously reported the coevolution of cells and virus during persistent infection in vitro (53), Nonetheless, irrespective of their ability to regulate the initiation of the infection, our results demonstrate that TIA-1 and G3BP1 are also required for virus particle assembly, while TIAR is required for release of infectious particles, suggesting that these SG proteins have dual functions during the HCV life cycle. Consistent with this dual function, we found that small fraction (around 1%) of the NS5A and NS5B proteins coimmunoprecipitate with TIA-1 and TIAR during the infection. Since these viral proteins are involved in HCV RNA replication and NS5A also plays a role in viral particle assembly, it is conceivable that TIA-1 and TIAR might regulate those steps by directly binding to NS5A and NS5B. A previous report by Yi et al. showed that NS3, NS5A, and NS5B coimmunoprecipitated with an overexpressed myc-G3BP1 construct in HCV-infected cells, and they suggested that G3BP1 is present in replication complexes (51). In contrast to those results, we were not able to detect any specific coimmunoprecipitation of core, NS4A, NS4B, NS5A, or NS5B protein with the endogenous G3BP1 protein, suggesting either that G3BP1 does not bind to any of those viral proteins or that the binding was too transient or weak to be detected under our experimental conditions. Consistent with the latter hypothesis, we showed that G3BP1 as well as TIA-1 and TIAR proteins colocalized with NS5A protein in some of the infected cells.
Since SGs are induced upon HCV infection and since G3BP1, TIA-1, and TIAR proteins are required for efficient HCV infection, it would be interesting to know if the effects of these proteins are related to their presence in stress granules. Our experiments in cells with double downregulation of PKR and stress granule proteins that do not produce SGs upon HCV infection suggest that the effects of G3BP1, TIA-1, and TIAR may be independent of their aggregation within SGs.
In conclusion, we have shown that HCV triggers the formation of stress granules in a PKR-dependent manner and that IFN greatly increases the number of SG-containing cells. We have confirmed that G3BP1 is required for HCV infection, and we identified two new host factors, TIA-1 and TIAR, that regulate HCV infection by affecting both early (i.e., initiation of HCV RNA replication) and late (i.e., virus particle assembly and release) steps in the HCV life cycle.
ACKNOWLEDGMENTS
We are grateful to Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for providing the infectious JFH-1 molecular clone and replicon constructs, Stanley Lemon (University of North Carolina, Chapel Hill, NC) for the H77S molecular clone, Charles Rice (Rockefeller University, New York, NY) for the Huh-7.5 cells from which Huh-7.5.1 cells and Huh-7.5.1c2 cells were derived, Dennis Burton (The Scripps Research Institute, La Jolla, CA) for recombinant human IgG anti-E2, Michael Houghton (Chiron) for MS5 anti-NS5A antibody, and Inder Verma (Salk Institute, La Jolla, CA) for lentiviral plasmids. We are grateful to Josan Chung and Christina Whitten for excellent technical assistance and to Marlene Dreux, Ariel Rodriguez, Lorena Ver, and Cristina Godio for helpful discussions, advice, and critical reading of the manuscript.
Urtzi Garaigorta was supported by The Irvington Institute Fellowship Program of the Cancer Research Institute. This work was supported by grant AI-079043 from the NIH.
Footnotes
Published ahead of print 1 August 2012
This is paper number 21557 from the Scripps Research Institute.
REFERENCES
- 1. Alter HJ, Seeff LB. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17–35 [DOI] [PubMed] [Google Scholar]
- 2. Anderson P, Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson P, Kedersha N. 2009. Stress granules. Curr. Biol. 19:R397–R398 [DOI] [PubMed] [Google Scholar]
- 4. Anderson P, Kedersha N. 2002. Stressful initiations. J. Cell Sci. 115:3227–3234 [DOI] [PubMed] [Google Scholar]
- 5. Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10:1324–1332 [DOI] [PubMed] [Google Scholar]
- 6. Ariumi Y, et al. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 85:6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arnaud N, et al. 2010. Hepatitis C virus controls interferon production through PKR activation. PLoS One 5:e10575 doi:10.1371/journal.pone.0010575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choo QL, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 88:2451–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dreux M, Gastaminza P, Wieland SF, Chisari FV. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franch HA, Sooparb S, Du J, Brown NS. 2001. A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J. Biol. Chem. 276:19126–19131 [DOI] [PubMed] [Google Scholar]
- 14. Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friebe P, Lohmann V, Krieger N, Bartenschlager R. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047–12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garaigorta U, Chisari FV. 2009. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gastaminza P, et al. 2008. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82:2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gastaminza P, Kapadia SB, Chisari FV. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 20. Hanley LL, et al. 2010. Roles of the respiratory syncytial virus trailer region: effects of mutations on genome production and stress granule formation. Virology 406:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Honda M, Beard MR, Ping LH, Lemon SM. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ivanov PA, Chudinova EM, Nadezhdina ES. 2003. RNP stress-granule formation is inhibited by microtubule disruption. Cell Biol. Int. 27:207–208 [DOI] [PubMed] [Google Scholar]
- 23. Jones CT, et al. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 28:167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang JI, et al. 2009. PKR protein kinase is activated by hepatitis C virus and inhibits viral replication through translational control. Virus Res. 142:51–56 [DOI] [PubMed] [Google Scholar]
- 25. Kato T, et al. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817 [DOI] [PubMed] [Google Scholar]
- 26. Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81 [DOI] [PubMed] [Google Scholar]
- 27. Kedersha N, Anderson P. 2009. Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90:155–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kedersha N, Anderson P. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969 [DOI] [PubMed] [Google Scholar]
- 29. Li W, et al. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989–12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 31. Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE., Jr 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 84:12274–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindquist ME, Mainou BA, Dermody TS, Crowe JE., Jr 2011. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology 413:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maniloff J. 1995. Identification and classification of viruses that have not been propagated. Arch. Virol. 140:1515–1520 [DOI] [PubMed] [Google Scholar]
- 34. McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miyanari Y, et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 36. Montero H, Rojas M, Arias CF, Lopez S. 2008. Rotavirus infection induces the phosphorylation of eIF2alpha but prevents the formation of stress granules. J. Virol. 82:1496–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. 2004. Structural biology of hepatitis C virus. Hepatology 39:5–19 [DOI] [PubMed] [Google Scholar]
- 38. Piotrowska J, et al. 2010. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 84:3654–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qin Q, Carroll K, Hastings C, Miller CL. 2011. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2alpha phosphorylation and PKR. J. Virol. 85:8798–8810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 9:2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robert F, et al. 2006. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2 · GTP · Met-tRNAiMet ternary complex availability. Mol. Biol. Cell 17:4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Samuel CE. 1993. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268:7603–7606 [PubMed] [Google Scholar]
- 43. Schutz S, Sarnow P. 2007. How viruses avoid stress. Cell Host Microbe 2:284–285 [DOI] [PubMed] [Google Scholar]
- 44. Shimoike T, McKenna SA, Lindhout DA, Puglisi JD. 2009. Translational insensitivity to potent activation of PKR by HCV IRES RNA. Antiviral Res. 83:228–237 [DOI] [PubMed] [Google Scholar]
- 45. Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. 2008. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 15:836–841 [DOI] [PubMed] [Google Scholar]
- 46. Thomas DL. 2012. Advances in the treatment of hepatitis C virus infection. Top. Antivir. Med. 20:5–10 [PMC free article] [PubMed] [Google Scholar]
- 47. Thomis DC, Samuel CE. 1993. Mechanism of interferon action: evidence for intermolecular autophosphorylation and autoactivation of the interferon-induced, RNA-dependent protein kinase PKR. J. Virol. 67:7695–7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305 [DOI] [PubMed] [Google Scholar]
- 50. Yang F, et al. 2006. Polysome-bound endonuclease PMR1 is targeted to stress granules via stress-specific binding to TIA-1. Mol. Cell. Biol. 26:8803–8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yi Z, et al. 2011. Hepatitis C virus co-opts Ras-GTPase-activating protein-binding protein 1 for its genome replication. J. Virol. 85:6996–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong J, et al. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhong J, et al. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]