Abstract
Simian immunodeficiency virus (SIV) infection of natural hosts is characterized by nonpathogenic chronic viremia, maintenance of gastrointestinal epithelial barrier integrity, and low numbers of target cells. Assessment of cell-associated virus load in T cell subsets in multiple anatomic compartments of chronically SIV-infected sabeus African green monkeys (AGMs) revealed that gastrointestinal memory CD4+ T lymphocytes are a major source of cell-associated virus and a significant contributor to SIV viremia in AGMs.
TEXT
Natural host primate species of simian immunodeficiency virus (SIV) have evolved to escape the immunopathogenic consequences of SIV infection, despite sustaining chronic SIV viremia. Moreover, they appear to maintain normal gastrointestinal (GI) epithelial barrier integrity, avoiding the consequences of chronic microbial translocation that is associated with the chronic immune activation of pathogenic SIV/human immunodeficiency virus (HIV) infection (3–5, 7, 22). It is well described that the effector memory CD4+ T lymphocyte population in the GI tract is a major source of cell-associated virus in HIV infection (2, 5). In natural SIV hosts, the main source of cell-associated virus in the periphery appears to be the CD4+ T lymphocytes (1, 9, 11, 18, 20); however, the anatomic distribution of cell-associated virus in nonpathogenic SIV infections is not well defined. Both SIV-infected and uninfected natural SIV hosts characteristically have low numbers of CD4+ CCR5+ T lymphocytes, target cells of SIV, in peripheral blood and mucosal compartments (12, 15, 17, 21), yet maintain substantial plasma viral RNA levels. While the systemic CD4+ T lymphocyte population is typically maintained in the periphery of natural hosts, there is an acute depletion of CD4+ T cells in the GI tract during acute infection (8, 21), analogous to that of pathogenic hosts. Additionally, relatively large proportions of CD4− CD8− (double-negative [DN]) T lymphocytes have been described in the natural SIV hosts (1, 10, 12, 16, 17): these DN cells are a T cell population which may maintain some functions typical of CD4+ T lymphocytes (10, 13, 25). Furthermore, downregulation of the CD4 molecule, the primary HIV receptor, may be a mechanism that allows natural SIV hosts to evade the pathogenicity of SIV infection (1). However, only a very small proportion of the DN T lymphocytes in the peripheral blood of natural SIV hosts appear to harbor the virus (1). Additionally, the central memory CD4+ T lymphocyte population in the peripheral blood of the natural host species sooty mangabeys appears to be resistant to SIV infection (15) and therefore is also unlikely to be a major contributor to viral replication in this species. Definition of the anatomic distribution of cell-associated virus of nonpathogenic SIV infection may be important to understanding the mechanisms that evolved to protect these natural SIV host species from immune consequences of infection and informing the quest to extinguish active viral reservoirs in HIV-infected humans.
To better define the cell-associated virus pools in natural SIV hosts, we investigated the T-cell-associated virus population in multiple tissue compartments of sabeus African green monkeys (AGMs) who were intravenously inoculated with the SIVsab9351BR virus stock 3 years and 11 months (27, 28) prior to terminal necropsy. All animals had an unremarkable clinical course following infection, despite being part of a previously described cohort who underwent temporary immune depletion of T and/or B cell subsets within the first year of infection (28). The animals had no significant changes in their CD4+ T lymphocyte counts throughout infection and maintained set point viremia ranging from 5,610 to 22,062 viral RNA copies/ml of plasma (Table 1). Moreover, the SIV-infected AGMs all had normal levels of lipopolysaccharide (LPS) in peripheral plasma (range, 2.87 to 3.5 pg/ml), measured as previously described (4), indicating the expected lack of microbial translocation in this natural host species (3, 4). All animals used in this study were maintained according to the Guide for the Care and Use of Laboratory Animals (14).
Table 1.
Viral load of chronically SIV-infected AGMs
| Animal no. | SIV RNA load in plasma (copies/ml) |
|---|---|
| 242-04 | 22,062 |
| 361-06 | 9,416 |
| 362-06 | 6,455 |
| 367-06 | 5,610 |
| 369-06 | 13,631 |
We utilized four-way simultaneous cell sorting to obtain purified T cell subsets from multiple lymphoid organs and the GI tract following necropsy of these five chronically SIV-infected AGMs and then determined the level of viral DNA in each T cell subset. Animals were sacrificed, and blood, ileum, mesenteric and axillary lymph nodes, spleen, and bone marrow were harvested. Tissues were digested with collagenase, and mononuclear cells were isolated on a Percoll gradient, as previously described (26). Isolated cells were stained with anti-CD3 Pacific Blue (SP34.2), anti-CD4 peridinin chlorophyll protein (PerCP) Cy5.5 (L200), anti-CD8 allophycocyanin (APC) (SK1), anti-CD95 phycoerythrin (PE) (DX2) (all BD Biosciences), and amine live-dead stain (yellow, Invitrogen). Memory CD4+ T lymphocytes (CD4+ CD95+), naive CD4+ T lymphocytes (CD4+ CD95−), DN T lymphocytes, and memory CD8+ T lymphocytes (CD8+ CD95+) were isolated according to the gating strategy shown in Fig. 1A. We aimed to obtain >2.0 × 105 cells from each sorted population and achieved that goal in each T cell subset isolated, except for the CD4+ CD95+ T cells in the ileum (median, 1.6 × 105 cells; range, 3.3 × 103 to 2.0 × 105 cells) and all T cell populations isolated from the bone marrow (BM) (median, 1.6 × 105 cells; range, 3 × 103 to 2.7 × 105 cells). Sorted cells were pelleted and stored at −80°C until analysis. Naive CD4+ CD95− T lymphocytes were not isolated from the ileum due to the limited number of naive cells in that compartment. All comparisons of cell-associated viral load and CD4 mRNA in T cell subpopulations were performed using the Wilcoxon rank sum test (GraphPad Prism Software, version 5), and the lowest obtainable P value with our sample size is 0.06.
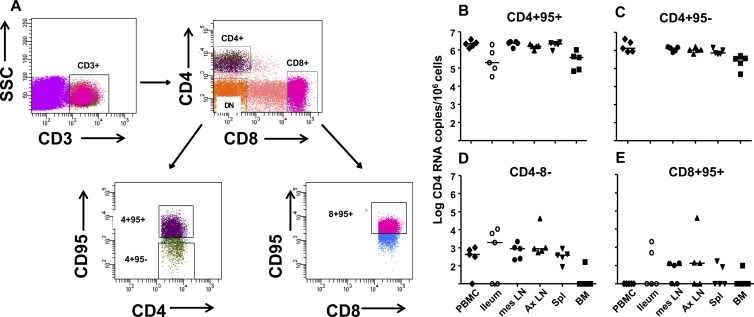
Fig 1.
Gating strategy for and CD4 mRNA expression in T cell populations sorted from multiple anatomic compartments in chronically SIV-infected AGMs. (A) After gating on singlets and the lymphocyte populations on forward and side scatter plots, CD3+ T lymphocytes were first separated into CD4+, CD8+, and CD4− CD8− (DN) populations. The DN T lymphocytes were collected without further subdivision, whereas the CD4+ lymphocyte population was subdivided into CD95+ (memory) and CD95− (naïve) populations prior to collection. CD95+ (memory) cells of the CD8+ lymphocyte population were also collected. Postsort purities determined on the PBMC fractions were routinely >98%. CD4 RNA copy numbers are shown per 106 cells in panel B for CD4+ CD95+, panel C for CD4+ CD95−, panel D for DN, and panel E for CD8+ CD95+ T lymphocyte populations isolated from multiple anatomic compartments of SIV-infected AGMs. mes LN, mesenteric lymph nodes; Ax LN, axillary LNs; Spl, spleen. Horizontal lines represent medians.
CD4 mRNA expression in CD4− T lymphocyte populations of AGMs.
To assess the purity of our sorted cell populations, we first determined the level of expression of CD4 in the T lymphocyte subsets by quantitating the amount of CD4 mRNA present. Total DNA or RNA was isolated from thawed cells using the AllPrep DNA/RNA microkit (Qiagen, Inc.) according to the manufacturer's protocol. DNase treatment was used to reduce DNA contamination of isolated RNA prior to cDNA synthesis. Quantitative reverse transcriptase PCR (qRT-PCR) was performed in duplicate to quantify the CD4 mRNA copies in the sorted cells. All reactions were carried out in 96-well optical plates (Applied Biosystems) in a 25-μl reaction volume containing 5 μl cDNA and 20 μl QuantiTect probe PCR master mix (Qiagen) using the ABI 7900 robotic thermal cycler set for 15 min at 95°C, with 45 cycles of 15 s at 95°C and 1 min at 60°C. The following primer pairs and probe were used: CD4 forward primer, 5′-CAA GGA TGC TTT TCC ATG ATC A-3′; CD4 reverse primer, 5′-AGC AGG TGG GTG TCA GAG TTG-3′; and CD4 probe, 5′-56-carboxyfluorescein (56-FAM)–CAG TCA ATC–ZEN–CGA ACA CCA GCA ATT CCA–3′ Iowa Black FQ (3IABkFQ)-3′. The levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA were also determined as a control for RNA integrity and assay conditions. The GAPDH primers and probe were as follows: forward primer, 5′-GCA CCA CCA ACT GCT TAG CAC-3′; reverse primer, 5′-TCT TCT GGG TGG CAG TGA TG-3′; and probe, 5′-VIC–TCG TGG AAG GAC TCA TGA CCA CAG TCC–6-carboxytetramethylrhodamine (TAMRA)-3′ (Integrated DNA Technologies).
The CD4 mRNA copy number was normalized to the number of cells in the same sample, which was determined by CCR5 DNA copy number quantitated by quantitative PCR (qPCR), and divided by 2 to account for two chromosomes per cell. The CCR5 qPCR primers and probe used were as follows: AGMCCR5 forward, 5′-GGA ATC CTG AAA ACT CTG CTT CG-3′; AGMCCR5 reverse, 5′-GAG AAG GAC AAT GTT GTA GGG AG-3′; and AGMCCR5 probe, 5′-56-FAM–AGA AGAGGC–ZEN–ACA GGG CTG TGA GGC T–3IABkFQ-3′. The numbers of both CCR5 DNA and CD4 mRNA copies in a reaction were calculated based on standard curves with plasmids containing the target gene, with the lower detection limit of the all assays being <10 target gene copies/well. Results were analyzed with SDS 7900 system software version 2.3 (Applied Biosystems). Copy numbers of sabeus SIV (SIVsab) gag DNA and CD4 mRNA are reported as the copy number per million cells (Fig. 1B to E).
As expected, the amount of CD4 mRNA present in the CD4-expressing cells (CD4+ CD95+ and CD4+ CD95− T cells) (Fig. 1B and C) from each anatomic compartment was at least 3 logs higher than that in the DN T lymphocytes or CD8+ CD95+ T lymphocytes (P = 0.06 for all). However, it is notable that CD4 mRNA was detected at a low level (median, 874 copies; range, 161 to 4.1 × 104 copies per 106 cells) in the DN T lymphocytes isolated from most tissues of the monkeys, with the exception of bone marrow, where CD4 mRNA was only detected in sorted DN T cells from only one SIV-infected AGM (Fig. 1D). This finding supports the conclusion that a portion of the DN T cell population in SIV-infected AGMs arises from downregulation of the CD4 molecule by SIV-infected CD4+ T cells, as previously reported (1). CD4 mRNA was also detected at a low level (median, 134.2 copies; range, 99 to 4.1 × 104 copies/106 cells) in CD8+ CD95+ T lymphocytes in tissues, but not peripheral blood mononuclear cells (PBMCs), in up to 3 of the 5 monkeys (Fig. 1E). The detection of measureable CD4 mRNA in CD8+ T lymphocyte populations could be attributed either to slight impurities (<2%) of the sorted cell populations or damage to the surface CD4 molecules during the tissue processing or lymphocyte isolation in some tissues. As the amount of CD4 mRNA detected in the CD4− T lymphocyte populations was <0.1% of that in the CD4+ T lymphocyte populations, these data support the conclusion that the sorted T lymphocyte populations were quite pure.
Memory CD4+ T lymphocyte populations in the GI tract and associated lymphoid tissue are a major source of cell-associated SIV in SIV-infected AGMs.
We next measured the amount of cell-associated SIV DNA present in the T lymphocyte subsets from each anatomic compartment. The SIV DNA copy number was quantitated as described above using the following primers and probe and was normalized to the cell number: AGMgag forward primer, 5′-GTC TAC TCC TTC CTC TAG TTC C-3′; AGMgag reverse primer, 5′-CGA TTT TGC CTC TTC AGT ATC TTT C-3′; and probe, 5′-56-FAM–CGT GTA TAC–ZEN–AAG CGA GCACGC AGC AC–3IABkFQ-3′. Not surprisingly, the memory CD4+ T lymphocytes carried the largest amount of cell-associated DNA in peripheral blood, with approximately 0.1% of the memory CD4+ T cells infected (P = 0.06 compared to all other T lymphocyte subpopulations in PBMCs). The peripheral T lymphocyte population with the next largest amount of cell-associated SIV DNA was the naïve CD4+ T lymphocytes, followed by the DN T lymphocyte population, each with approximately 1 log less cell-associated virus than that of the memory CD4+ T lymphocyte population (∼0.01% infected) (Fig. 2A). While the DN T lymphocyte population had a lower cell-associated virus load than the memory CD4+ T lymphocytes, the DN T lymphocytes were more frequent in the peripheral blood T lymphocyte population than the memory CD4+ T lymphocytes (P = 0.06) (Fig. 2D). Finally, SIV DNA was detected in the peripheral CD8+ T lymphocyte populations from only one monkey at very low levels (Fig. 2A).
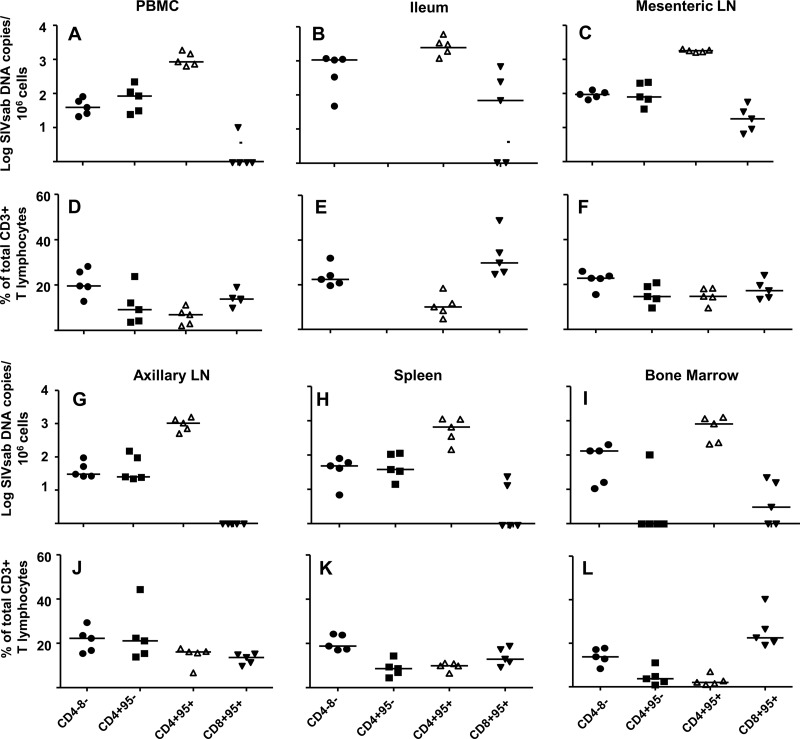
Fig 2.
The GI tract memory CD4+ T lymphocyte population is a major viral reservoir in SIV-infected AGMs. The numbers of SIVsab DNA copies per 106 cells in the DN, CD4+ CD95−, CD4+ CD95+, and CD8+ CD95+ T lymphocyte populations are shown in panel A for peripheral blood, panel B for ileum, panel C for mesenteric lymph nodes, panel G for axillary lymph nodes, panel H for spleen, and panel I for bone marrow. The frequencies of DN, CD4+ CD95−, CD4+ CD95+, and CD8+ CD95+ T lymphocytes in the total CD3+ T lymphocyte population in each anatomic compartment are shown in panels D to F and J to L. Horizontal lines represent medians.
Similarly, the T lymphocyte subpopulation in the ileum with the largest amount of cell-associated DNA was the memory CD4+ T lymphocyte population. In fact, the amount of SIV DNA detected in the memory CD4+ T lymphocytes isolated from the ileum was larger than that in the memory CD4+ T lymphocytes in peripheral blood (P = 0.06), indicating that the GI CD4+ T cell population is a major contributor to SIVsab replication during chronic infection (Fig. 2B). Interestingly, the median copy number of SIV DNA detected in the DN T lymphocyte population in the ileum was less than a log lower than that in the memory CD4+ T lymphocytes (Fig. 2B). Moreover, the DN T lymphocytes are more frequent in the GI T lymphocyte mucosa than that of memory CD4+ T lymphocytes (P = 0.06) (Fig. 2E). Thus, the DN T cell population is also a significant pool of cell-associated SIV in the GI tract of AGMs, and these cells may represent infected CD4+ T cells with suppressed CD4 expression. There was also a low level of SIV DNA detected in the ileal CD8+ CD95+ T lymphocyte population in three of five monkeys, which may be attributable to contamination of the sorted population by a few SIV-infected CD4+ T cells and/or damage to surface molecules during tissue processing (Fig. 2B).
In every lymphoid organ examined, CD4+ CD95+ T lymphocytes contained higher levels of cell-associated SIV DNA than all of the other T cell subpopulations in the same organ (Fig. 2C and G to I). Among the tissues sampled, only the CD4+ CD95+ T lymphocytes in the mesenteric lymph nodes had a higher cell-associated SIV DNA load than the CD4+ CD95+ T cells in peripheral blood (P = 0.06), again suggesting the GI-associated memory CD4+ T cells are a site of SIV replication in chronically infected AGMs. The median amount of SIV DNA detected in the naïve CD4+ and DN T lymphocyte populations was at least 1 log lower than that in the memory CD4+ T lymphocyte populations in all lymphoid organs. Interestingly, SIV DNA was only detected in the naïve CD4+ T lymphocyte population of bone marrow in one animal, whereas SIV DNA was detected in the bone marrow DN T lymphocyte populations of all five animals, suggesting that the DN T lymphocyte population may be a source of virus replication in the bone marrow (Fig. 2I). Finally, of the 5 animals examined, low-level SIV DNA was detected in the CD8+ CD95+ T lymphocyte population in mesenteric lymph nodes of all 5 animals, in the spleen of two monkeys, and in bone marrow of three monkeys. No SIV DNA was detected in the CD8+ T lymphocyte population in the axillary lymph nodes of any animal (Fig. 2G to I).
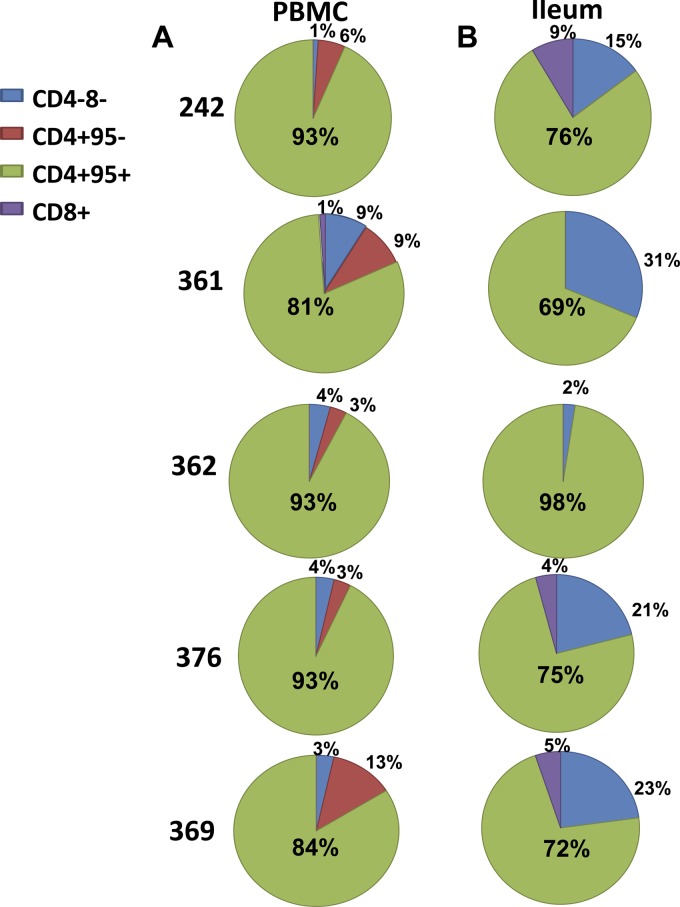
We then calculated the proportion that each T lymphocyte subpopulation contributed to the total amount of cell-associated SIV DNA in the peripheral blood (Fig. 3A) and ileum (Fig. 3B) of each SIV-infected AGM. In four of five of the SIV-infected AGMs, the memory CD4+ T lymphocyte population in the ileum made up a smaller proportion of the total number of infected T lymphocytes compared to peripheral blood; however, this comparison was not significant (P = 0.12) due to one animal (monkey 362) in which the memory CD4+ T lymphocyte population accounted for 98% of the total SIV DNA detected in the ileum. In fact, DN T lymphocytes contributed a larger proportion of the total amount of cell-associated SIV DNA in the ileum compared to those in the peripheral blood in the same four of five animals, but this comparison was again not statistically significant due to the one outlier (monkey 362).
Fig 3.
The DN T lymphocyte population is a source of cell-associated SIV in the GI tract of AGMs, comprising 2 to 31% of infected T cells in the gut. Shown are the proportions of the total number of detected SIV DNA-infected cells that are CD4+ CD95+ (green), CD4+ CD95− (red), DN (blue), or CD8+ CD95+ (purple) in peripheral blood (A) and ileum (B).
The proportion of SIV-infected memory CD4+ T lymphocytes in the GI tract predicts the plasma SIV RNA load.
As the memory CD4+ T lymphocyte population in the ileum contained a higher level of SIV DNA than that in peripheral blood, we sought to determine whether this cell-associated virus pool contributes to the chronic viremia in nonpathogenic SIV infection of AGMs. While there was a strong direct correlation between the proportion of infected memory CD4+ T lymphocytes in the periphery and the plasma SIV RNA viral load (Table 1) that trended toward significance (r = 0.9, P = 0.08), there was an even stronger direct correlation between the proportion of SIV-infected memory CD4+ T lymphocytes in the ileum and the plasma virus RNA load (r = 1.0). Moreover, this correlation was significant (P = 0.017), despite the small number of animals included in this study. Conversely, there was no correlation between the proportion of infected DN T lymphocytes in the GI tract and the plasma virus load. Thus, the memory CD4+ T lymphocytes in the GI tract appear to be a primary source of SIV viremia in this natural SIV host species, suggesting the DN T cells in the GI tract may be a latent pool of cell-associated virus. Additional analyses of the activation or proliferation status (such as expression of Ki67) of the GI DN T cell population, which was unable to be performed here due to cell number limitations, would provide insight into whether this population is a true latent reservoir of virus in SIV-infected AGMs.
Finally, as the GI tract appeared to be an important source of SIV-infected T cells, we investigated the proportion and phenotype of the CD4+ T lymphocyte population present in six chronically SIVsab9351BR-infected AGMs in the upper (duodenum) and lower (colon) GI tract 3 years following infection. Pinch biopsy samples were obtained endoscopically, and lymphocyte populations were isolated and stained with anti-CD28, anti-CD95, and anti-CCR7 monoclonal antibodies in addition to traditional T lymphocyte phenotyping molecules, as previously described (26). The T lymphocytes present in the duodenum and colon of the SIV-infected AGMs were mainly CD8+ T lymphocytes, with a lower proportion of the total GI-associated T lymphocytes consisting of DN T lymphocytes (Fig. 4A). The CD4+ T lymphocytes comprise a very minor proportion of the total GI-associated T lymphocytes, ranging from 0.9 to 2.7% in five of the six animals. One SIV-infected AGM had a higher proportion of CD4+ T lymphocytes in the gut that the others, with 21% in the duodenum and 24% in the colon. When assessing the memory phenotype of the CD4+ T lymphocyte population in the GI tract, we observed that the overwhelming majority of the GI-associated CD4+ T lymphocytes were transitional memory CD4+ T lymphocytes, and very few were fully differentiated central or effector memory T lymphocytes (P = 0.03 for both) (Fig. 4B). Thus, the GI-associated memory CD4+ T lymphocyte population is a rare population and mainly comprised of transitional memory CD4+ T lymphocytes, which may represent an evolutionary adaptation of AGMs, as these cells may be less prone to deletion than the more activated effector memory CD4+ T lymphocytes.
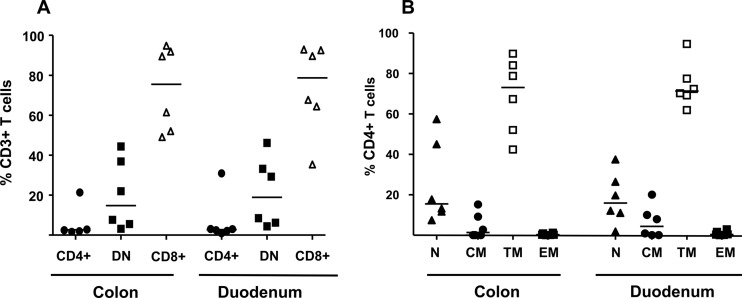
Fig 4.
CD4+ T lymphocyte subpopulations in the gastrointestinal tract of SIV-infected AGMs. The gastrointestinal T lymphocyte population of SIV-infected AGMs comprises low levels of CD4+ T lymphocytes (colon median, 2.7%, and range, 1.6 to 24.4%; duodenum median, 2.7%, and range, 0.9 to 30.9%), a substantial proportion of DN T lymphocytes (colon median, 14.8%, and range, 3.2 to 44.4%; duodenum median, 18.9, and range, 4.4 to 46.2%), and a large proportion of CD8+ T lymphocytes (colon median, 75.5%, and range, 49.1 to 94.8%; duodenum median, 78.7%, and range, 35.4 to 92.9%) (A). The CD4+ T lymphocyte population in the colon and the duodenum of SIV-infected AGMs is mainly the transitional memory (TM) phenotype (CD95+ CD28+ CCR7−: colon median, 73%, and range, 42.4 to 89.8%; duodenum median, 71.3%, and range, 61.9 to 94.6%), with lower proportions of naive (N) (CD95− CD28+: colon median, 15.5%, and range, 7.4 to 57.4%; duodenum median, 15.9%, and range, 1.8 to 37.5%) and central memory (CM) (CD95+ CD28+ CCR7+: colon median, 1.3%, and range, 0 to 15.2%; duodenum median, 4.4%, and range, 0 to 20.1%) CD4+ T lymphocytes and an extremely low proportion of effector memory (EM) (CD95+ CD28−: colon median, 0.2%, and range, 0 to 1.3%; duodenum median, 0.4%, and range, 0 to 2.9%) CD4+ T lymphocytes (B). Horizontal lines indicate medians.
Our finding that the CD4+ T lymphocyte population in the GI tract is a major source of cell-associated virus in SIV infection in this natural SIV host species is similar to the findings in pathogenic SIV and HIV infections, suggesting that the anatomic site of virus replication does account for the differences in pathogenicity in natural and aberrant primate hosts. Perhaps then, the difference in the course of SIV infection in natural and aberrant hosts is mainly due to the ability of natural SIV host species to maintain an intact GI epithelial barrier throughout infection (3, 4). Our results are in concert with the observation that there is an acute loss of CD4+ T lymphocyte population when the infection is first established in these species, potentially due to direct infection (8, 21, 23). This GI-associated CD4+ T cell population also appears to be an active site of SIV replication supporting chronic viremia, despite comprising only a very small proportion of the total T lymphocytes in the GI tract. The intact intestinal immune barrier in AGMs is thus likely maintained through mechanisms other than modulation of the susceptibility of CD4+ target cells to SIV infection. Our findings are consistent with the hypothesis that the AGM mucosal immune system may have evolved to be less dependent on CD4+ T lymphocytes, with non-CD4-expressing cells compensating for the low total numbers and high SIV-infected proportion of CD4+ T cells (10, 13, 25). Our study is also consistent with the large body of work suggesting that dampening of chronic immune activation is the key to maintaining nonpathogenic SIV infection in these hosts (6, 16, 19, 23, 24), as the major SIV-infected cell populations do not appear to be distinct from that of pathogenic HIV/SIV infection.
ACKNOWLEDGMENTS
This work was supported by the Center for HIV/AIDS Vaccine Immunology (CHAVI) Early Career NHP Investigator Award (AI067854) (S.R.P.) and NIH grants K08AI087992 (S.R.P.) and AI065335 (J.E.S.).
Footnotes
Published ahead of print 15 August 2012
REFERENCES
- 1. Beaumier CM, et al. 2009. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat. Med. 15:879–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brenchley JM, et al. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 78:1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brenchley JM, et al. 2008. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112:2826–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brenchley JM, et al. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12:1365–1371 [DOI] [PubMed] [Google Scholar]
- 5. Brenchley JM, et al. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chakrabarti LA, et al. 2000. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74:1209–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Estes JD, et al. 2010. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 6:e1001052 doi:10.1371/journal.ppat.1001052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gordon SN, et al. 2007. Severe depletion of mucosal CD4+ T cells in AIDS-free simian immunodeficiency virus-infected sooty mangabeys. J. Immunol. 179:3026–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klatt NR, et al. 2008. Availability of activated CD4+ T cells dictates the level of viremia in naturally SIV-infected sooty mangabeys. J. Clin. Invest. 118:2039–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Milush JM, et al. 2011. Lack of clinical AIDS in SIV-infected sooty mangabeys with significant CD4+ T cell loss is associated with double-negative T cells. J. Clin. Invest. 121:1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Milush JM, et al. 2007. Virally induced CD4+ T cell depletion is not sufficient to induce AIDS in a natural host. J. Immunol. 179:3047–3056 [DOI] [PubMed] [Google Scholar]
- 12. Murayama Y, et al. 1997. CD4 and CD8 expressions in African green monkey helper T lymphocytes: implication for resistance to SIV infection. Int. Immunol. 9:843–851 [DOI] [PubMed] [Google Scholar]
- 13. Murayama Y, Mukai R, Inoue-Murayama M, Yoshikawa Y. 1999. An African green monkey lacking peripheral CD4 lymphocytes that retains helper T cell activity and coexists with SIVagm. Clin. Exp. Immunol. 117:504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. National Research Council 1996. Guide for the care and use of laboratory animals. National Research Council, National Academy Press, Washington, DC [Google Scholar]
- 15. Paiardini M, et al. 2011. Low levels of SIV infection in sooty mangabey central memory CD(4)(+) T cells are associated with limited CCR5 expression. Nat. Med. 17:830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pandrea I, et al. 2006. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J. Virol. 80:4858–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pandrea I, et al. 2007. Paucity of CD4+CCR5+ T cells is a typical feature of natural SIV hosts. Blood 109:1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pandrea I, et al. 2008. Cutting edge: experimentally induced immune activation in natural hosts of simian immunodeficiency virus induces significant increases in viral replication and CD4+ T cell depletion. J. Immunol. 181:6687–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pandrea I, et al. 2003. High levels of SIVmnd-1 replication in chronically infected Mandrillus sphinx. Virology 317:119–127 [DOI] [PubMed] [Google Scholar]
- 20. Pandrea I, et al. 2008. Simian immunodeficiency virus SIVagm dynamics in African green monkeys. J. Virol. 82:3713–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pandrea IV, et al. 2007. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J. Immunol. 179:3035–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Picker LJ, et al. 2004. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J. Exp. Med. 200:1299–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Silvestri G, et al. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441–452 [DOI] [PubMed] [Google Scholar]
- 24. Sumpter B, et al. 2007. Correlates of preserved CD4(+) T cell homeostasis during natural, nonpathogenic simian immunodeficiency virus infection of sooty mangabeys: implications for AIDS pathogenesis. J. Immunol. 178:1680–1691 [DOI] [PubMed] [Google Scholar]
- 25. Vinton C, et al. 2011. CD4-like immunological function by CD4− T cells in multiple natural hosts of simian immunodeficiency virus. J. Virol. 85:8702–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilks AB, et al. 2011. High cell-free virus load and robust autologous humoral immune responses in breast milk of simian immunodeficiency virus-infected African green monkeys. J. Virol. 85:9517–9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zahn RC, et al. 2008. Simian immunodeficiency virus (SIV)-specific CD8+ T-cell responses in vervet African green monkeys chronically infected with SIVagm. J. Virol. 82:11577–11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zahn RC, et al. 2010. Suppression of adaptive immune responses during primary SIV infection of sabaeus African green monkeys delays partial containment of viremia but does not induce disease. Blood 115:3070–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]