Abstract
Coxsackievirus A9 (CAV9), a member of the Picornaviridae family, uses an RGD motif in the VP1 capsid protein to bind to integrin αvβ6 during cell entry. Here we report that two CAV9 isolates can bind to the heparan sulfate/heparin class of proteoglycans (HSPG). Sequence analysis identified an arginine (R) at position 132 in VP1 in these two isolates, rather than a threonine (T) as seen in the nonbinding strains tested. We introduced a T132R substitution into the HSPG-nonbinding strain Griggs and recovered infectious virus capable of binding to immobilized heparin, unlike the parental Griggs strain. The known CAV9 structure was used to identify the location of VP1 position 132, 5 copies of which were found to cluster around the 5-fold axis of symmetry, presumably producing a region of positive charge which can interact with the negatively charged HSPG. Analysis of several enteroviruses of the same species as CAV9, Human enterovirus B (HEV-B), identified examples from 5 types in which blocking of infection by heparin was coincident with an arginine (or another basic amino acid, lysine) at a position corresponding to 132 in VP1 in CAV9. Together, these data show that membrane-associated HSPG can serve as a (co)receptor for some CAV9 and other HEV-B strains and identify symmetry-related clustering of positive charges as one mechanism by which HSPG binding can be achieved. This is a potentially powerful mechanism by which a single amino acid change could generate novel receptor binding capabilities, underscoring the plasticity of host-cell interactions in enteroviruses.
INTRODUCTION
Coxsackievirus A9 (CAV9) is a member of the genus Enterovirus (species Human enterovirus B [HEV-B]) of the family Picornaviridae and has a polyadenylated single-stranded positive-sense RNA virus of around 7.5 kb, which is enclosed in a naked icosahedral capsid (5, 30). It is an important human pathogen that can infect the central nervous system and also cause myocarditis and respiratory illness, in some cases leading to neonatal death (9, 13). CAV9 uses a functional arginine-glycine-aspartic acid (RGD) motif in the VP1 capsid protein (6) to interact with cell surface integrins αvβ3 and αvβ6 during cell entry (14, 38, 48).
Several other picornaviruses have a functional RGD motif which they use to interact with integrins. Some foot-and-mouth disease virus (FMDV) serotypes have been shown to interact with αvβ1, αvβ3, αvβ6, and αvβ8 integrins, and some human parechoviruses (HPeVs) are known to interact with αvβ1, αvβ3, and αvβ6 integrins (20, 22–24, 36, 40). Although several internalization mechanisms are known to mediate picornavirus entry, e.g., clathrin-mediated endocytosis for FMDV (26) and a caveolin-mediated mechanism for coxsackievirus B3 (CBV3) (7), recent evidence indicates that CAV9 entry is mediated not by these classical mechanisms, as was previously thought, but, rather, by a caveolin- and clathrin-independent mechanism (15). While CAV9 infection is mediated by the RGD motif, RGD-independent infection can also occur (17). However, events governing RGD-independent entry have not been identified. Molecules such as GRP78 and β2-microglobulin and/or major histocompatibility complex class I (MHC-I) have been implicated in CAV9 infection and may be involved in either RGD-dependent or -independent infection (15, 45).
In this study, we examined heparan sulfate proteoglycan (HSPG) interactions with CAV9 and other members of the HEV-B species. Several viruses are known to interact with HSPG molecules at the cell surface as a prelude to infection, and the list of such viruses continues to grow (8, 31, 50). These include some serotypes of adenovirus which can show differential preference over the various conformations of HSPG that can occur (46). Several picornaviruses, including members of different species of the genus Enterovirus, have been reported to interact with HSPG (12, 19, 28, 29, 47, 49). However, apart from an interaction between an FMDV variant and heparin studied at atomic resolution, the molecular basis of this interaction is largely unknown (10, 11).
In this report, we identify HSPG as a (co)receptor for some strains of CAV9. We have mapped the HSPG-binding domain to an R residue at position 132 in the VP1 protein of the capsid structure, which, in the folded form, clusters around the 5-fold axis of symmetry and presumably produces a positively charged area allowing interactions with HSPG. Sequence analysis combined with blocking studies using heparin identified an R or lysine (K) residue at the corresponding position in several heparin-blocked isolates from other species HEV-B types, suggesting that this clustering is a common mechanism for HSPG binding in enteroviruses. Interestingly, this finding correlates with a recent study on FMDV SAT-type viruses, in which amino acid substitutions clustering around the 5-fold axis of symmetry permitted these viruses to bind HSPG (33). Thus, a symmetry-related clustering mechanism to generate a receptor binding site is seen in two diverse members of Picornaviridae and could be relatively common in this virus family.
MATERIALS AND METHODS
Virus and cell culture.
CAV9 isolates CO62, CO79, CO85, and CO87 were described previously (6). Several CAV9 and other HEV-B strains were isolated from clinical samples at the National Institute for Health and Welfare (THL), Helsinki, Finland. GMK cells were maintained in minimal essential medium (MEM) containing 10% (vol/vol) heat-inactivated fetal calf serum (FCS), 1% (vol/vol) nonessential amino acids, and 100 μg/ml of gentamicin.
Plaque titration and viral propagation.
The medium was removed from confluent monolayers of GMK cells grown in 5 ml of growth medium in 25-cm2 flasks and overlaid with 1 ml of growth medium containing 500 PFU of virus. Cells were incubated for 45 to 60 min at room temperature with gentle rocking. For viral propagation, 4 ml of medium was added and the flasks were incubated at 37°C for 2 to 3 days to allow cytopathic effect (CPE) to progress. Virus was then released by freeze-thawing three times. For plaque assays, after infection cells were covered with 4 ml of warm (56°C) MEM containing 0.5% (wt/vol) carboxymethyl cellulose (CMC) and 0.5% agarose or agar. The flasks were then incubated at 37°C for 2 to 3 days until plaques were observed. Plaques were selected and propagated, or flasks were stained with crystal violet (0.1% [wt/vol]).
RNA extraction, RT-PCR, and sequencing.
RNA was isolated from 140 μl of viral stock solutions using a QIAamp viral RNA minikit (Qiagen) as per the manufacturer's instructions and eluted with 50 μl of water. Using the Superscript III one-step reverse transcription-PCR (RT-PCR) kit (Invitrogen), cDNA was generated and amplified from 5 μl of the RNA sample and 0.1 nmol of forward and reverse primers according to the manufacturer's protocol. The reactions were run in a GeneAmp PCR System 9700 (Applied Biosystems) thermal cycler. The settings included the initial cDNA synthesis step at 50°C for 30 min, followed by 35 cycles of denaturation at 94°C for 5 min, annealing between 45 and 57°C for 2 min, and elongation at 68°C for 2 min. A final extension step at 68°C for 15 min was also applied. The amplicons were purified using a QIAquick gel extraction kit (Qiagen) and sequenced commercially by Geneservice (Cambridge, United Kingdom).
Sequence analysis, graphics, and three-dimensional (3D) imaging.
CAV9 isolates CO62, CO79, CO85, and CO87 were sequenced in the capsid region and other CAV9 and HEV-B isolates in the VP1 amino acid position 132 region (601 nucleotides from VP1/3), using primers directed at conserved sequences identified from HEV-B alignments. Database searches were performed using the NCBI BLAST search facility (http://www.ncbi.nlm.nih.gov/BLAST/) (1, 35). The online tool ClustalW was used to align multiple sequences (http://www.ebi.ac.uk/Tools/clustalw/) (44). Amino acid positions of interest were plotted onto the structure of the CAV9 pentamer (16) using the program Rasmol 2.0 (39).
Plasmids and mutagenesis.
Plasmid pCAV9 contains the entire CAV9 (Griggs) sequence cloned into pBluescript and was used as the template for site-directed mutagenesis to introduce a threonine (T)-to-R change at VP1 position 132. Mutagenesis was performed by an overlap-PCR approach using Pfu DNA polymerase (NEB, United Kingdom) together with the primer pairs OL1507 (CAGTGCCACCATAGTGATGCCA)/OL1508 (TGTCCTGAGCTAGCCGTGTTCCGGGGTCT) and OL1509 (AGACCCCGGAACACGGCTAGCTCAGGACA)/OL1510 (CTACTTCCACTCACGTGTCTCAC) to generate overlapping fragments that were joined and amplified using primers OL1507 and OL1510. The product was cut at two unique restriction sites, BsiWI and BssHII, and the mutant 1,322-bp fragment was used to replace the corresponding region in pCAV9. The construct was cloned into E. coli XL1-Blue cells, and the mutant cDNA was recovered and sequenced to confirm that the desired mutation and no additional changes had been introduced.
In vitro transcription, transfection, and analysis of the mutant.
Mutated full-length CAV9 cDNA was linearized at the end of the poly(A) tract with the restriction enzyme BscI (NEB) and in vitro transcribed into infectious RNA using a T7 RNA polymerase RNA transcription kit (Invitrogen, United Kingdom). Next, GMK cells were transfected with 10 μg of RNA per flask using DEAE-dextran followed by dimethyl sulfoxide (DMSO) shock (32). After 3 days, a plaque was picked and the recovered virus propagated and sequenced.
Inhibition of infection with soluble proteoglycans.
GMK cells were grown in 25-cm2 flasks to ∼100% confluence. Virus dilutions (180 μl) were incubated with 20 μl of 1-, 2-, 5-, 10-, and 100-mg/ml heparin (Sigma, United Kingdom) or dermatan sulfate (Sigma) for 20 min. These were subsequently diluted into 1 ml of medium and added to the cell monolayers. Incubation continued for 1 h with gentle rocking at room temperature before the cells were overlaid with a 2% (wt/vol) solution of CMC-agarose. Plaques were visualized by staining after 2 to 3 days of incubation at 37°C.
Removal of viral particles from tissue culture supernatant using immobilized heparin.
Heparin-agarose and, as a control, the equivalent agarose preparation not conjugated to heparin (CL4B; Sigma) were used at a final mass of 500 mg. The slurry was washed 3 times with 400 μl of phosphate-buffered saline (PBS) and once with 400 μl of growth medium and preequilibrated with a final volume of 200 μl of growth medium. Five hundred microliters of virus suspension at 5 × 103 PFU/ml in growth medium was added to the heparin-agarose or agarose slurries. The virus-slurry mixtures were incubated for 1 h on a rolling mixer. The supernatant was separated by centrifugation at 3,000 × g for 2 min, filter sterilized through a 0.22-μm-pore disc, and used for plaque titration. The slurries were then incubated with 500 μl of 0.5 M sodium chloride and centrifuged, and the supernatant was also used for plaque titration.
Inhibition of proteoglycan sulfation with sodium chlorate.
GMK cells were grown in medium containing 50 mM NaClO3 (Sigma, United Kingdom) for 3 days in 25-cm2 flasks to ∼100% confluence. The cells, together with mock-treated GMK cells, were used for plaque assays as described above.
Nucleotide sequence accession numbers.
The sequences obtained for the amplicons have been submitted to GenBank and given the accession numbers JN996499 to JN996516.
RESULTS
Sulfated compounds present in agar reduce infection of some CAV9 isolates.
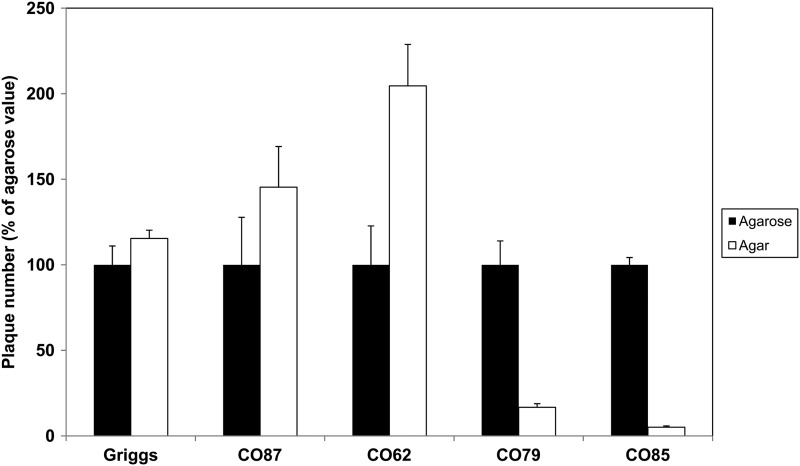
To investigate potential interactions of CAV9 with HSPG, the Griggs strain and four previously described isolates (CO62, CO79, CO85, and CO87) were studied (6). It has been known for some time that agar contains sulfated polysaccharides that can interact with some viruses and inhibit virus growth (43), while agarose lacks these sulfated polysaccharides, which gives a rapid screen for viruses that bind to HSPG (3). Through plaque titrations comparing agar and agarose overlay media, it was clear that CO79 and CO85 infections were significantly inhibited in the presence of agar, marked by a reduction in both plaque sizes (data not shown) and numbers (Fig. 1). CO62, CO87, and Griggs were not inhibited; indeed, infection appeared to be enhanced (Fig. 1).
Fig 1.
Infection by some CAV9 isolates is inhibited by agar. Plaque titration assays for several CAV9 isolates were carried out on GMK cells, using an overlay which contains either agarose or agar. The plaque number obtained using agar is expressed as a percentage of that obtained using agarose, which is set at 100%. Values shown are means from three independent experiments, and error bars indicate standard deviation.
The molecular basis of HSPG interaction.
In order to identify the molecular basis of the agar inhibition, we determined the complete sequences of the capsid-encoding P1 region of the four CAV9 isolates. From the P1 alignment, including the known Griggs sequence, 57 amino acid positions show some polymorphism, but one was particularly interesting, as it alone correlated with the observed phenotypes; i.e., it was shared by the agar-inhibited strains but differed in the noninhibited strains. At position 132 in VP1, an R residue is seen in the agar-inhibited strains CO79 and CO85, while a T residue is seen in the noninhibited strains, i.e., Griggs, CO62, and CO87 (Fig. 2; see also Fig. S1 in the supplemental material).
Fig 2.
Identification of an amino acid position which correlates with the agar inhibition phenotype. Amino acid sequences of the capsid-encoding region of CAV9 isolates CO62, CO79, CO85, CO87, and the prototype strain Griggs were aligned using ClustalW. Numbers refer to P1 position in CAV9 and are equivalent to VP1 positions 123 to 151. CO79 and CO85 (names shown in white on black background), the two isolates inhibited in the presence of agar, have an arginine (R—also shown in white on black background) at VP1 position 132, while the noninhibited viruses (names and corresponding residues all shown on a gray background) have threonine (T).
Properties of the T132R mutant virus.
To confirm the significance of this amino acid, a T-to-R change was introduced into the CAV9 Griggs strain, giving a T132R mutant. After in vitro RNA transcription, GMK cells were transfected with the newly synthesized RNA to recover mutant virus, and the presence of the mutation was confirmed. There was no appreciable effect of the mutation on virus growth (data not shown). Heparin, a highly sulfated analogue of heparan sulfate (HS), is generally used to mimic HSPG binding (34). We analyzed the ability of soluble heparin to suppress CAV9 infection (Fig. 3). Increasing concentrations of heparin were incubated with the CAV9 strains and the effects examined by plaque titration. Consistent with the agar and agarose data, CO79 and CO85 infections were inhibited by heparin treatment of the viruses prior to infection in an approximately dose-dependent manner, while a stimulatory effect was seen in Griggs, CO62, and CO87 infections. Interestingly, soluble heparin did not inhibit infection by the T132R mutant and a slight stimulatory effect was seen, although this was less than that observed for the parental strain Griggs (Fig. 3). Dermatan sulfate, the second most widely expressed and sulfated member of the glycosaminoglycan (GAG) family, showed no appreciable levels of inhibition or stimulation (data not shown).
Fig 3.
Heparin inhibits plaque formation of some CAV9 isolates in a dose-dependent manner. CAV9 isolates and the T132R mutant were treated with increasing concentrations of heparin and then subjected to plaque assay on GMK cells. In each case, the number of plaques obtained was expressed as a percentage of the number obtained in a no-heparin control (0 mg/ml). Values are means from three independent experiments, and error bars indicate standard deviations.
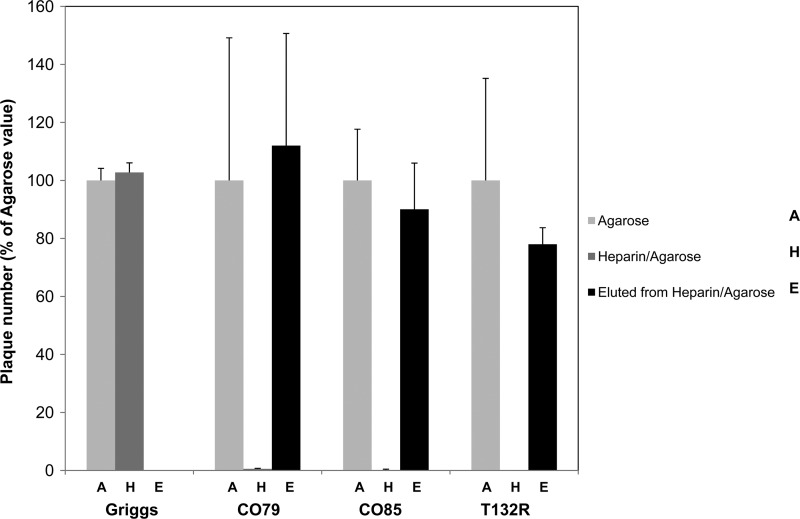
Next, we examined whether CAV9 strains could bind to immobilized heparin. Heparin-agarose and agarose (control) slurries were incubated with virus cultures. The supernatants were recovered from the slurry by centrifugation and filtered sterilized prior to plaque titration. Infectivity of the T132R mutant, CO79, and CO85 supernatants was almost completely lost when heparin-agarose was used compared to agarose, while the Griggs sample retained full infectivity (Fig. 4). To confirm that the loss of activity was due to virus binding to immobilized heparin, this was treated with 0.5 M NaCl and the eluate was subjected to plaque assay. This released most of the infectivity from the T132R mutant, CO79, and CO85 slurries, demonstrating that virus particle binding to immobilized heparin had occurred and providing definitive evidence that the T132R change permitted heparin binding (Fig. 4).
Fig 4.
Some CAV9 isolates and the T132R mutant can bind to immobilized heparin. Viruses were incubated with agarose or heparin-agarose beads for 1 h and the supernatants subjected to plaque assays on GMK cells. Any bound viruses were eluted using 0.5 M NaCl2 and again analyzed by plaque assays. The results are the means of three experiments, and error bars indicate standard deviations.
Growing GMK cells in the presence of sodium chlorate reduced infection by HSPG-interacting strains.
Further evidence for the involvement of HSPG in virus binding was demonstrated using sodium chlorate. Sodium chlorate was previously shown to inhibit human rhinovirus A54 (HRVA54) infection in RD cells (28) by blocking a crucial stage in HSPG biosynthesis. GMK cells were grown for 3 days in media with and without 50 mM sodium chlorate before they were infected with virus. The results show that the presence of chlorate reduced infection by isolates CO79 and CO85, compared to the non-chlorate-treated control (Fig. 5). Chlorate did not inhibit T132R mutant infection of cells, and a slight increase in Griggs infectivity was observed. Total inhibition of infection by CO79 and CO85 was not observed, possibly due to chlorate not completely blocking sulfation of GAG moieties or to use of other receptors.
Fig 5.
GMK cells grown in the presence of sodium chlorate show reduced susceptibility to some CAV9 isolates. Cells were grown for 3 days in medium containing 50 mM sodium chlorate. They were then used for plaque assays using CAV9 isolates and the T132R mutant. In each case, the number of plaques obtained was expressed as a percentage of the number obtained in mock-treated cells. Values are means from three independent experiments, and error bars indicate standard deviations.
Assessing the HSPG-binding phenotype in other HEV-B species members.
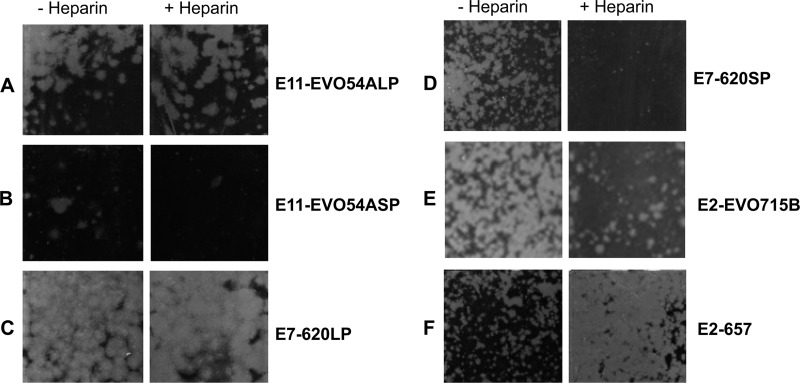
Several HEV-B species isolates were screened for HSPG interactions using the soluble heparin assay (typical data are shown in Fig. 6), and a 601-nucleotide fragment including the CAV9 VP1 position 132 equivalent was sequenced in both orientations following RT-PCR (Fig. 7). These samples included a further 5 CAV9 isolates, all of which were found to have the T132 genotype and were not inhibited by heparin. Of the two echovirus 2 (E2) isolates examined (E2-657 and E2-EVO715B), E2-EVO715B had the basic residue K at the position equivalent to CAV9 VP1 position 132 and was blocked by soluble heparin, while E2-657 was not blocked and had glutamine (Q) at position 132. The E5 isolate tested (E5-EVO59B) was also inhibited by soluble heparin, and it, too, had K132. One of the E11 isolates (E11-EVO427B) had Q132 and was not blocked by heparin. The other E11 isolate (E11-EV054A) and the E7 isolate (E7-620) both contained a mixture of small and large plaque viruses. These were plaque purified and analyzed separately. In each case, the small but not the large plaque variant was blocked by heparin. The sequences were consistent with the pattern observed for the other isolates, as the blocked variants both had amino acid R132. The nonblocked E7-620 had S132, and the nonblocked E11-EVO54A had Q132. In contrast, both E6 isolates analyzed (E6-EVO511B and E6-EVO415B) had R132, but neither was inhibited by soluble heparin.
Fig 6.
Infection with several members of the species HEV-B can be blocked with heparin. Plaque assays were performed on 5 CAV9 and 10 echovirus isolates or variants after treating virus dilutions with 100 mg/ml of heparin or mock treating. Results are shown for large plaque (LP) variants of E11-EVO419B (A) and E7-620 (C), representing nonblocked viruses, and small plaque (SP) variants of E11-EVO419B (B) and E7-620 (D), representing blocked viruses. Two E2 strains, E2-EVO715B (E) (partially blocked) and E2-657 (F) (enhanced infection), are also shown.
Fig 7.
HEV-B isolates are polymorphic at the position equivalent to VP1 position 132. The amino acid sequences of part of VP1, including the residue equivalent to position 132 in CAV9, were determined for the viruses tested for heparin blocking and aligned using ClustalW. Numbers refer to P1 position in CAV9 and are equivalent to VP1 positions 127 to 151. Several (names shown in white on a black background) were blocked by heparin and had a basic amino acid at this position (highlighted in white on a black background). Isolates that were not blocked by heparin did not have a basic residue at the corresponding position (on a gray background) apart from the E6 isolates analyzed, which were not blocked by soluble heparin but did have a basic residue. Perfectly conserved residues are indicated by an asterisk (*), and conservation of residues with strongly and weakly similar properties by two dots (:) or by one dot (.), respectively.
Symmetry-related clustering at VP1 position 132.
To understand how the T-to-R VP1 position 132 change may exert its effect, we looked at the 3D structure of the CAV9 Griggs strain (16) using the Rasmol program (39). It was observed that VP1 position 132, which is located in the VP1 DE loop, is a highly surface-exposed position in the 3D structure. Moreover, 5 copies of this amino acid come together to form a tight cluster around the 5-fold axis of symmetry (Fig. 8). The implications of this are that the T-to-R change forms a cluster of positive charge around the 5-fold axis which would permit HSPG binding at the cell surface.
Fig 8.
Five copies of VP1 position 132 cluster at the 5-fold axis. The location of VP1 position 132 (shown in spacefill) was identified in the three-dimensional structure of CAV9 using Rasmol (16, 39). Two projections are shown to illustrate the clustering (A, top view of the CAV9 pentamer) and the surface exposure (B, side view).
DISCUSSION
In this report, we demonstrate a molecular interaction of some CAV9 isolates with HSPG and identify a critical amino acid, that at position 132 in VP1, involved in defining this phenotype. HSPG usage was demonstrated by reduction of plaque formation in the presence of agar, which is known to contain sulfated polysaccharides; inhibition of viral binding using soluble heparin; removal of virus particles from cell culture supernatant using immobilized heparin, followed by elution; and a reduction of virus infectivity in cells grown in the presence of the sulfation inhibitor sodium chlorate. Introducing a mutation (T132R) at this position in CAV9 Griggs, which does not bind HSPG, conferred the ability to bind to immobilized heparin.
HSPG-interacting regions must be exposed on the surface of the potential ligand in order to bind and are made up of multiple positively charged amino acids that interact with the negatively charged HSPG. They can be composed of linear motifs of the form BBXB and BBBXXB, where X is a neutral or hydrophobic amino acid and B is any one of the basic amino acids (4). However, they are often formed from clusters of basic residues that tend to be far apart in the protein sequence but are brought together in the folded structure. One example is seen in FMDV, in which an HSPG-binding site studied by X-ray crystallography is made up of residues from both VP1 and VP3 (10, 11). Analysis of the capsid region of two HSPG-interacting CAV9 isolates did not reveal a linear domain; rather, a single VP1 amino acid difference (R at position 132) from nonbinding strains is shared by these isolates and is a key factor in binding to HSPG (Fig. 2). VP1 position 132 is unique, as copies of no other amino acid in a surface-exposed position are clustered by symmetry so tightly around the 5-fold axis (Fig. 8). Thus, a change at no other single position in the P1 region could generate such a concentration of additional positive charges. The adjacent amino acid, at position 131, has a similarly exposed location but clusters slightly more loosely, as it is farther away from the 5-fold axis. The unique location of R132 strongly supports the idea that clustering of this amino acid enables HSPG binding. This means that the virus particle will have 12 HSPG-binding sites, each located at a vertex of the icosahedron. It remains possible that other residues contribute to HSPG binding, such as a histidine at VP1 position 230, which is the only other nearby positively charged amino acid in any of the CAV9 isolates. In confirmation of a role in HSPG binding, substitution of R132 for the naturally occurring T in the nonbinding Griggs strain at this position gave a virus capable of binding to immobilized heparin (Fig. 4). Database sequences suggest that a basic residue at position 132 in VP1 is rare in CAV9 isolates, as there are no other examples with R and only a few with K at this position. Accordingly, none of the 15 CAV9 isolates we studied was inhibited by heparin, and all 5 chosen for sequencing had a T at this position (Fig. 7).
The fact that a single substitution permitted HSPG usage by some CAV9 isolates in our study prompted further examination of other HEV-B species members. Analysis of database sequences shows that there is extensive polymorphism at the equivalent position in other HEV-B members (data not shown). In 11 types, a basic residue occurs at this position in at least some isolates, suggesting the possibility of HSPG binding, although without the 3D structures of many of these types, it is not possible to tell whether folding generates a domain similar to that in CAV9. Possible interactions with HSPG were, however, addressed directly by sequence analysis and heparin-blocking studies (Fig. 6 and 7). Isolates from 6 HEV-B types were analyzed. In 5 of these types, isolates with a basic residue at position 132 were inhibited by heparin, while isolates with a nonbasic residue were not blocked. The exception was E6, where two isolates studied had R132 but were not inhibited by heparin. Similarly, they were not able to bind to immobilized heparin (data not shown).
It is possible that HSPG usage in enteroviruses arises from adaptation to the cells in which isolates are propagated, as seen in FMDV (21). Differences in initial culture protocols may be responsible for the HSPG usage or nonusage in the CAV9 isolates, particularly as these were isolated over a period of 25 years (6). In two of the echovirus isolates examined in this study, HSPG-binding and nonbinding variants were recovered, which may add weight to this conclusion. However, when these variants were isolated by plaque purification, their phenotypes appeared to be stable upon further passage, implying that tissue culture did not impose a severe selection. It may therefore be that the variants were both present in the original infection, raising the possibility that populations with different receptor tropisms are present in enterovirus infections. The correlation between a basic residue at position 132 and HSPG binding suggests that receptor or coreceptor usage in these viruses is at least partially predictable from the sequence alone. This opens up the possibility of future studies on the significance of HSPG usage in enterovirus infections, particularly if this affects tropism or pathogenesis, by simply recovering and directly sequencing viruses from different sites of infection. Interestingly, attachment and subsequent infection by a highly neurovirulent strain of Theiler's virus are mediated by HSPG, which contrasts with infection by a strain with low neurovirulence, whose attachment is mediated by sialic acid moieties (37, 41). There is evidence which suggests that an increase in virulence due to HSPG usage is seen in other picornaviruses such as FMDV (33).
Prediction of HSPG binding from sequence alone is complicated by our results on E6. Over 100 E6 sequences are available in the database, and almost universally, isolates have a basic residue (nearly always an R) at the position equivalent to 132 in CAV9. Moreover, previous work by Goodfellow et al. (12) identified several HSPG-binding E6 strains. It was therefore surprising that neither E6 strain in our study binds HSPG, despite having an R residue at the apparently key position. One possibility is that the precise structure is critical, and there may be a sufficient difference in the location of the 132-equivalent residue in E6 to prevent clustering of positive charges and HSPG utilization. In this regard, it is interesting that a phylogenetic tree based on VP1 sequences indicates two broad clusters of enterovirus types (see Fig. S2 in the supplemental material). Most (i.e., 9) of the 11 types, where some isolates have a basic residue at the 132-equivalent position, are in the same group as CAV9, including 4 (E7, E11, E19, and enterovirus 85) that group more tightly with CAV9. E6 is in the other group, related more closely to coxsackie B viruses, which could imply significant structural or functional differences. It is also possible that other local charges may interfere with potential HSPG binding in our E6 isolates. HSPG binding in isolates studied by Goodfellow et al. (12) could, then, be due to differences in these local charges or due to an entirely different mechanism. These isolates would clearly repay further study. It is interesting that in contrast to the discordance seen in E6 results, the recent observation that E5 Noyce binds to HSPG is consistent with our predictions, as this strain has a K residue at the 132-equivalent position (19). Preincubation of the CAV9 strains with heparin prior to cell adsorption reduced virus infection dramatically in the case of CO79 and CO85 but not Griggs, CO62, or CO87, with which, in fact, infection was stimulated. The engineered T132R mutant was also not inhibited by heparin (Fig. 3). All the isolates have the RGD motif (6) used to interact with integrin αvβ6 (48), and all can be blocked by an antibody to this integrin (data not shown), suggesting that the interaction is critical. Thus, HSPG does not appear to be an alternative receptor, and it might be expected that HSPG binding would be advantageous rather than necessary for infection. It may function by binding the virus on the cell surface, facilitating a more specific interaction with the integrin. The sensitivity to competitive inhibition with soluble heparin and substantial reduction in infection of chlorate-treated cells suggest that this advantage is highly significant for these isolates. Alternatively, binding to heparin interferes with binding to the integrin receptor or other molecules that are involved in entry. However, if interference was occurring, or there was a nonspecific effect due to aggregation of virus particles, it would be expected that the acquisition of heparin-binding ability by the T132R mutant would also lead to inhibition of infection. Overall, it is likely that HSPG binding is advantageous for CO79 and CO85, and so blocking this reduces infectivity. It has recently been observed that acquisition of HSPG binding allows an HRVA8 variant to enter cells by a distinct mechanism (29), and it would be interesting to explore whether the different effects of heparin on the CAV9 strains are mediated by alternative entry pathways.
Rather than blocking infection, treatment of the non-HSPG-binding strains with heparin enhanced infectivity (Fig. 3). Low concentrations of heparin enhanced infection of an HSPG-binding HRVA89 mutant (47), while higher concentrations inhibited infection, and similar results have been observed for murine leukemia virus PVC-211 (25). In these cases, enhanced binding and/or entry by bridging of heparin on the cell surface and bound to the virus, or alteration of pH of intracellular compartments, has been suggested. In the case of CAV9, it seems likely that the effect is not due to alterations by the soluble heparin itself, as enhanced infectivity growth was also seen in cells treated with sodium chlorate in the absence of heparin (Fig. 5). One possibility is that the apparently nonbinding strains in fact bind heparin very weakly, and this interferes with productive interactions at the cell surface. Blocking with soluble heparin prior to cell surface interactions or reducing the level of interacting molecules by chlorate treatment would both be expected to enhance infection as observed.
The interactions between HSPG, its cellular ligands, and viruses have been shown to be mostly ionic. Some viruses which bind HSPG at the cell surface do so as a result of cell culture adaptation, and in some instances, the mutant virus is attenuated when introduced back into the natural host (2, 18, 42, 47). Recently, in a similar study where FMD SAT 1 and SAT 2 viruses were adapted to BHK-21 cells, Maree et al. (33). showed that the adaptation resulted in several mutations that occurred throughout the capsid proteins. Of particular interest were mutations that occurred at positions 110 and 112 in the VP1 protein which, when plotted onto the 3D image of the FMDV pentamer, clustered around the 5-fold axis of symmetry. The findings of Maree et al. (33), together with work on adenovirus 2 in which multiple amino acids formed a functional heparin binding domain (HBD) by clustering around the 3-fold axis of symmetry in the folded structure, highlight the fact that uncommon HBDs exist (27). Here we show that in a number of viruses of HEV-B species, a similar symmetry-related clustering of copies of one basic amino acid can allow HSPG binding. Thus, a single amino acid change can lead to the acquisition of the ability to bind to a cell surface molecule, which emphasizes the potential flexibility of receptor usage in picornaviruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank the University of Essex for financial support.
We also thank Jane Stanway for proofreading the manuscript.
Footnotes
Published ahead of print 1 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Bernard KA, Klimstra WB, Johnston RE. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93–103 [DOI] [PubMed] [Google Scholar]
- 3. Byrnes AP, Griffin DE. 2000. Large-plaque mutants of Sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J. Virol. 74:644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cardin AD, Weintraub HJ. 1989. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 9:21–32 [DOI] [PubMed] [Google Scholar]
- 5. Chang KH, Auvinen P, Hyypiä T, Stanway G. 1989. The nucleotide sequence of coxsackievirus A9; implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269–3280 [DOI] [PubMed] [Google Scholar]
- 6. Chang KH, Day C, Walker J, Hyypiä T, Stanway G. 1992. The nucleotide sequences of wild-type coxsackievirus A9 strains imply that an RGD motif in VP1 is functionally significant. J. Gen. Virol. 73:621–626 [DOI] [PubMed] [Google Scholar]
- 7. Chung SK, et al. 2005. Internalization and trafficking mechanisms of coxsackievirus B3 in HeLa cells. Virology 333:31–40 [DOI] [PubMed] [Google Scholar]
- 8. Dasgupta J, et al. 2011. Structural basis of oligosaccharide receptor recognition by human papillomavirus. J. Biol. Chem. 286:2617–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eisenhut M, et al. 2000. Fatal coxsackie A9 virus infection during an outbreak in a neonatal unit. J. Infect. 40:297–298 [DOI] [PubMed] [Google Scholar]
- 10. Fry EE, et al. 1999. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 18:543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fry EE, et al. 2005. Structure of foot-and-mouth disease virus serotype A10 61 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation. J. Gen. Virol. 86:1909–1920 [DOI] [PubMed] [Google Scholar]
- 12. Goodfellow IG, Sioofy AB, Powell RM, Evans DJ. 2001. Echoviruses bind heparan sulfate at the cell surface. J. Virol. 75:4918–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grist NR, Reid D. 1988. General pathogenicity and epidemiology, p 221–239 In Berdinelli M, Friedman H. (ed), Coxsackieviruses: a general update. Plenum Press, New York, NY [Google Scholar]
- 14. Heikkilä O, Susi P, Stanway G, Hyypiä T. 2009. Integrin alphaVbeta6 is a high-affinity receptor for coxsackievirus A9. J. Gen. Virol. 90:197–204 [DOI] [PubMed] [Google Scholar]
- 15. Heikkilä O, et al. 2010. Internalization of coxsackievirus A9 is mediated by β2-microglobulin, dynamin, and Arf6 but not by caveolin-1 or clathrin. J. Virol. 84:3666–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hendry E, et al. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527–1538 [DOI] [PubMed] [Google Scholar]
- 17. Hughes PJ, Horsnell C, Hyypiä T, Stanway G. 1995. The coxsackievirus A9 RGD motif is not essential for virus viability. J. Virol. 69:8035–8040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hulst MM, van Gennip HG, Moormann RJ. 2000. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein E(rns). J. Virol. 74:9553–9561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Israelsson S, et al. 2010. Studies of echovirus 5 interactions with the cell surface: heparan sulfate mediates attachment to the host cell. Virus Res. 151:170–176 [DOI] [PubMed] [Google Scholar]
- 20. Jackson T, et al. 2004. Integrin αvβ8 functions as a receptor for foot-and-mouth disease virus: role of the β-chain cytodomain in integrin-mediated infection. J. Virol. 78:4533–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jackson T, et al. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 70:5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jackson T, Mould AP, Sheppard D, King AM. 2002. Integrin αvβ1 is a receptor for foot-and-mouth disease virus. J. Virol. 76:935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson T, et al. 1997. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth disease viruses to the purified integrin alpha (v) beta3 in vitro. J. Virol. 71:8357–8361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jackson T, Sheppard D, Denyer M, Blakemore W, King AM. 2000. The epithelial integrin αvβ6 is a receptor for foot-and-mouth disease virus. J. Virol. 74:4949–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jinno-Oue A, Oue M, Ruscetti SK. 2001. A unique heparin-binding domain in the envelope protein of the neuropathogenic PVC-211 murine leukemia virus may contribute to its brain capillary endothelial cell tropism. J. Virol. 75:12439–12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johns HL, Berryman S, Monaghan P, Belsham GJ, Jackson T. 2009. A dominant-negative mutant of rab5 inhibits infection of cells by foot-and-mouth disease virus: implications for virus entry. J. Virol. 83:6247–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kern A, et al. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khan AG, Pichler J, Rosemann A, Blaas D. 2007. Human rhinovirus type 54 infection via heparan sulfate is less efficient and strictly dependent on low endosomal pH. J. Virol. 81:4625–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khan AG, et al. 2011. Entry of a heparan sulphate-binding HRV8 variant strictly depends on dynamin but not on clathrin, caveolin, and flotillin. Virology 412:55–67 [DOI] [PubMed] [Google Scholar]
- 30. Knowles NJ, et al. 2011. Picornaviridae, p 855–880 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA [Google Scholar]
- 31. Kureishy N, Faruque D, Porter CD. 2006. Primary attachment of murine leukaemia virus vector mediated by particle-associated heparan sulfate proteoglycan. Biochem. J. 400:421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lopata MA, Cleveland DW, Sollner-Webb B. 1984. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 12:5707–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maree FF, Blignaut B, de Beer TA, Visser N, Rieder EA. 2010. Mapping of amino acid residues responsible for adhesion of cell culture-adapted foot-and-mouth disease SAT type viruses. Virus Res. 153:82–91 [DOI] [PubMed] [Google Scholar]
- 34. Mulloy B, Forster MJ. 2000. Conformation and dynamics of heparin and heparan sulfate. Glycobiology 10:1147–1156 [DOI] [PubMed] [Google Scholar]
- 35. Pearson WR, Lipman DJ. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U. S. A. 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pulli T, Koivunen E, Hyypiä T. 1997. Cell-surface interactions of echovirus 22. J. Biol. Chem. 272:21176–21180 [DOI] [PubMed] [Google Scholar]
- 37. Reddi HV, Lipton HL. 2002. Heparan sulfate mediates infection of high-neurovirulence Theiler's viruses. J. Virol. 76:8400–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roivainen M, et al. 1991. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J. Virol. 65:4735–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sayle RA, Milner-White EJ. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 40. Seitsonen J, et al. 2010. Interaction of alphaVbeta3 and alphaVbeta6 integrins with human parechovirus 1. J. Virol. 84:8509–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shah AH, Lipton HL. 2002. Low-neurovirulence Theiler's viruses use sialic acid moieties on N-linked oligosaccharide structures for attachment. Virology 304:443–450 [DOI] [PubMed] [Google Scholar]
- 42. Smit JM, et al. 2002. Adaptation of alphaviruses to heparan sulfate: interaction of Sindbis and Semliki Forest viruses with liposomes containing lipid-conjugated heparin. J. Virol. 76:10128–10137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takemoto KK, Liebhaber H. 1961. Virus-polysaccharide interactions. I. An agar polysaccharide determining plaque morphology of EMC virus. Virology 14:456–462 [DOI] [PubMed] [Google Scholar]
- 44. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Triantafilou K, Fradelizi D, Wilson K, Triantafilou M. 2002. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J. Virol. 76:633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tuve S, et al. 2008. Role of cellular heparan sulfate proteoglycans in infection of human adenovirus serotype 3 and 35. PLoS Pathog. 4:e1000189 doi:10.1371/journal.ppat.1000189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vlasak M, Goesler I, Blaas D. 2005. Human rhinovirus type 89 variants use heparan sulfate proteoglycan for cell attachment. J. Virol. 79:5963–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Williams ÇH, et al. 2004. Integrin αvβ6 is an RGD-dependent receptor for coxsackievirus A9. J. Virol. 78:6967–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zautner AE, Jahn B, Hammerschmidt E, Wutzler P, Schmidtke M. 2006. N- and 6-O-sulfated heparan sulfates mediate internalization of coxsackievirus B3 variant PD into CHO-K1 cells. J. Virol. 80:6629–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Z, et al. 2010. Heparin sulphate D-glucosaminyl 3-O-sulfotransferase 3B1 plays a role in HBV replication. Virology 406:280–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.