Abstract
Long-term batch cultures of Escherichia coli grown in nutrient-rich medium accumulate mutations that provide a growth advantage in the stationary phase (GASP). We have examined the survivors of prolonged stationary phase to identify loci involved in conferring a growth advantage and show that a mutation in the hns gene causing reduced activity of the global regulator H-NS confers a GASP phenotype under specific conditions. The hns-66 allele bears a point mutation within the termination codon of the H-NS open reading frame, resulting in a longer protein that is partially functional. Although isolated from a long-term stationary-phase culture of the parent carrying the rpoS819 allele that results in reduced RpoS activity, the hns-66 survivor showed a growth disadvantage in the early stationary phase (24 to 48 h) when competed against the parent. The hns-66 mutant is also unstable and reverts at a high frequency in the early stationary phase by accumulating second-site suppressor mutations within the ssrA gene involved in targeting aberrant proteins for proteolysis. The mutant was more stable and showed a moderate growth advantage in combination with the rpoS819 allele when competed against a 21-day-old parent. These studies show that H-NS is a target for mutations conferring fitness gain that depends on the genetic background as well as on the stage of the stationary phase.
INTRODUCTION
The bacterial H-NS protein belongs to a family of small nucleoid-associated proteins that play an important role in the structuring of the bacterial chromosome, a function that is reminiscent of histones in higher organisms (6, 29). In addition to its role in structuring the chromosome, H-NS is also known to influence several cellular functions, such as defense against foreign DNA, virulence, general stress response, genome stability, and metabolism (1, 2, 9, 17, 26). Loss of hns function results in diverse and unrelated phenotypes, indicating that H-NS is a global regulator of gene expression. Several environmentally regulated genes are under the negative control of H-NS, including rpoS encoding another global regulator, the stationary-phase sigma factor RpoS (2, 35). RpoS plays a central role in the reprogramming of gene expression to prepare the bacterial cell to face the challenges of stationary phase such as starvation, osmotic shock, change in pH, and oxidative stress (15). Thus, RpoS and H-NS that control a large repertoire of environmentally regulated genes are key players in the physiological adaptation to the environment.
H-NS is a highly abundant protein that binds to DNA with a preference for AT-rich sequences (34). The mechanism of transcriptional repression by H-NS involves binding to high-affinity centers called nucleation sites that initiates the oligomerization of H-NS along the DNA, resulting in a higher-order nucleoprotein complex. This nucleoprotein structure leads to repression of transcription either by occluding RNA polymerase binding or by trapping RNA polymerase (5, 10).
The bgl operon of Escherichia coli, involved in the catabolism of aromatic β-glucosides such as salicin and arbutin, is one of the well-studied genetic systems regulated by H-NS. Although genetically intact, the bgl operon is phenotypically silent in wild-type E. coli strains under laboratory conditions (27). The operon consists essentially of three genes—bglG, bglF, and bglB—that encode an RNA-binding anti-terminator protein, a β-glucoside permease, and a phospho-β-glucosidase, respectively (21, 30). H-NS is one of the major factors involved in silencing the bgl promoter (16, 25, 31); both loss-of-function mutations in the hns gene and mutations that disrupt H-NS binding upstream of the bgl promoter activate the bgl operon.
Maintenance of the bgl operon in the genome without accumulating deleterious mutations, despite its silent status, is an evolutionary puzzle. One possible explanation is that expression of the operon confers a growth advantage in stationary phase, referred to as GASP (12), leading to the selection of Bgl+ mutants that harbor the activated copy of the operon. With this rationale, we had screened the bgl phenotype of 150 random survivors obtained from 10 independent L broth cultures grown for 28 days and found that five were Bgl+ (20). The growth advantage of one of the Bgl+ survivors carrying an activating point mutation within the bgl promoter has recently been shown to be associated with enhanced expression of the BglG regulator, leading to the upregulation of oppA that encodes an oligopeptide transporter, enabling the uptake of short peptides (14). The remaining four Bgl+ survivors had mutations in the hns locus (20).
The presence of hns mutants among the survivors prompted us to investigate the role of hns in the survival of E. coli during long-term stationary phase. In the present study we show that the naturally isolated hns-66 mutation, in conjunction with the rpoS819 allele, confers a strong growth disadvantage in early stationary phase. There is spontaneous appearance of suppressors of the hns-66 allele, implying the unstable nature of this mutation in early-stationary-phase conditions. Most of the suppressors carry loss-of-function mutations in the ssrA locus that encodes a transfer-messenger RNA (tmRNA) involved in rescuing ribosomes that are stalled on defective mRNA and target the aberrant protein for proteolysis (13, 19). Interestingly, the hns-66 allele in the rpoS+ background confers a modest growth advantage in the early stationary phase and, along with the rpoS819 mutation, confers a moderate growth advantage during a prolonged stationary phase. This increased fitness of the hns-66 mutant in the late stationary phase is influenced by the age of the cells and that of the medium.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in the present study are listed in Table 1 and Table 2, respectively.
Table 1.
E. coli strains used in this study
| Strain | Relevant genotype and phenotypea | Source or reference |
|---|---|---|
| AE328 | ΔlacX74 thi bglR11(bglR::IS1) tna::Tn10 Bgl+ | A. Wright |
| BGL1 | CSH26 λ (Φbgl-lacZ) Bgl− | C. Ueguchi |
| BGL819 | BGL1 rpoS819 Camr Bgl− | This study |
| BGL819-66 | BGL819 hns-66 Kanr Bgl+ | This study |
| BGL819-66ΔssrA | BGL819-66 ΔssrA Kanr Bgl− | This study |
| BGL819Δhns | BGL819 Δhns::kan Bgl+ | This study |
| BGL819ΔhnsΔssrA | BGL819 Δhns ΔssrA::kan Bgl+ | This study |
| MG1655ΔssrA | MG1655 ΔssrA::kan Kanr Bgl− | U.Varshney |
| ZK819 | ZK126 (W3110 ΔlacU169) rpoS819 Smr Bgl− | 36 |
| ZK819Tn10 | ZK819 tna::Tn10 Tetr Bgl− | 20 |
| ZK819-97T | ZK819 tna::Tn10 bglR Tetr Bgl+ | 20 |
| ZK819-66 | ZK819 hns-66 Bgl+ (the original survivor that carries additional unknown mutations) | 20 |
| ZK819-66Tn5 | ZK819-66 Tn5 Kanr Bgl+ | This study |
| ZK819-66T | ZK819 hns-66 Kanr Bgl+ (the hns-66 allele transduced into the parent strain ZK819) | This study |
| ZK819-66R | ZK819-66 ssrA Bgl− (the original Bgl− revertant of ZK819-66) | This study |
| ZK819-66ΔssrA | ZK819-66 ΔssrA::kan Kanr Bgl− | This study |
| ZK819Δhns | ZK819 Δhns::kan Bgl+ | This study |
| ZK819ΔhnsΔssrA | ZK819 Δhns ΔssrA::kan Bgl+ Kanr | This study |
| ZK819proU-lacZ | ZK819 proU610::λplacMu55 Kanr | This study |
| ZK819-66proU-lacZ | ZK819 hns-66 proU610::λplacMu55 Kanr | This study |
| ZK819-66ΔssrA proU-lacZ | ZK819 hns-66 ΔssrA proU610::λplac Mu55 Kanr | This study |
| ZK819-66rpoS+ | ZK819 hns-66 rpoS+ Camr Bgl− | This study |
| ZK819-66TrpoS+ | ZK819 hns-66 rpoS+ Kanr Camr Bgl+ | This study |
| ZK819-66hns+ | ZK819-66 hns+ Kanr (the wild-type hns allele transduced into the original survivor ZK819-66) | This study |
| ZK819-66TrssB | ZK819-66T ΔrssB::cam Kanr Bgl+ | This study |
| ZK819-66TrpoS+ rssB | ZK819-66T ΔrssB::cam rpoS+ Kanr Camr Bgl− | This study |
Camr, chloramphenicol resistance; Smr, streptomycin resistance; Kanr, kanamycin resistance; Tetr, tetracycline resistance.
Table 2.
Plasmids used in this study
| Plasmid | Genotype and description | Source or reference |
|---|---|---|
| pACDH | plac followed by multiple cloning sites, pACYC origin of replication | 33 |
| pACDH-hns66 | hns66 in pACDH | This study |
| pTRC-ssrA | E. coli ssrA cloned in pTRC99C | 33 |
| pKW23 (SsrADD) | Tetr plasmid with pACYC replicon carrying ssrADD | 18 |
| pKD3 | Template plasmid carrying the cat cassette flanked by the target for the Flp recombinase (FRT) repeats | 7 |
| pKD4 | Template plasmid carrying the kan cassette flanked by FRT repeats | 7 |
| pCP20 | flp bla cat rep101(Ts); Ampr Camr (plasmid to express the Flp recombinase used to eliminate the antibiotic cassettes) | 4 |
| pKD46 | Ampr; beta exo gam (lambda red), ori(Ts) (plasmid to express the lambda red recombinase for mediating gene replacements) | 7 |
Bacterial media and growth conditions.
E. coli cultures were grown in L broth (Hi Media) at 37°C with aeration. For determining viable counts, cultures were plated on L agar medium supplemented with 35 μg of kanamycin (Kan)/ml, 100 μg of ampicillin (Amp)/ml, 15 μg of tetracycline (Tet)/ml, or 15 μg of chloramphenicol (Cam)/ml when required. Bacterial matings and P1 transductions were performed as described by Miller (22). Competition experiments were performed as described below.
Competition assays.
For early-stationary-phase (24 to 48 h) competition experiments, monocultures of the two competing strains were grown in 5 ml of L broth at 37°C with aeration for 24 h. The 1-day-old cultures were mixed in the desired proportion reciprocally and incubated under the same conditions without the addition of fresh medium. The two populations marked with two different antibiotic resistance markers were followed by plating on L agar plates containing the respective antibiotics at different points of time. For long-term stationary-phase experiments, the parent strain was grown for 21 days at 37°C (with periodic addition of sterile-distilled water to compensate for evaporation), and the 1-day-old mutant derivatives of the parent strain were added as a minority population. The reciprocal experiment with the mutant grown for 21 days could not be carried out due to the instability of the hns-66 allele. The competing populations were monitored as described above. Since the parent population subjected to prolonged stationary phase can be genetically heterogeneous, all long-term stationary-phase experiments were carried out in six replicates. The data shown are representative of six experiments all exhibiting a uniform trend.
Construction of hns and ssrA deletion strains.
Gene knockouts were generated by the procedure of Datsenko and Wanner (7). Hybrid primers with 36-bp extensions homologous to the N terminus and C terminus of the hns locus spanning positions +37 and +445 were designed for the PCR amplification of kanamycin resistant gene flanked by FLP recognition target sites (forward primer for hns [5′-AGC GAA GCA CTT AAA ATT CTG AAC AAC ATC CGT ACT CTT GTG TAG GCT GGA GCT GCT TCG-3′] and reverse primer for hns [5′TAT TGC TTG ATC AGG AAA TCG TCG AGG GAT TTA CCT TGC CAT ATC CTC CTT A-3′]) encoded by plasmid pKD4. The PCR product was used to replace the hns locus with the Kan resistance gene (kan), a selectable marker, with the help of plasmid pKD46 expressing the phage lambda recombinase. The deletion was confirmed by PCR analysis and the Bgl+ phenotype of the hns deletion strain. A similar procedure was used for the deletion of the ssrA locus using hybrid primers with ssrA homologous 36-nucleotide extensions spanning positions +91 and +445 (forward primer for ssrA [5′-TGA TTC TGG ATT CGA CGG GAT TTG CGA AAC CCA AGG CTG TAG GCT GGA GCT GCT TCG-3′] and reverse primer for ssrA [5′-TGG AGC TGG CGG GAG TTG AAC CCG CGT CCG AAA TTC CAT ATG AAT ATC CTC CTT A-3′). Whenever necessary, the Kan resistance cassette was eliminated using the helper plasmid pCP20 expressing FLP recombinase, which leaves a scar of 80 nucleotides at the deletion site (4).
Cloning of the hns-66 allele from ZK819-66.
The DNA primers hns-66-Fp-5′-CGA GCT CGA TGA GCG AAG CAC TT-3′ and hns-66-Rp-5′-GGA ATT CCG GTG CTG ATA TAC TGG A-3′ containing SacI and EcoRI restriction sites, respectively, were designed to amplify the hns gene (without the promoter but carrying nearly 1 kb downstream covering the putative transcription terminator) from ZK819-66 using Taq-Pfu DNA polymerase (a 9:1 ratio) by PCR consisting of 30 cycles of incubation at 95°C for 1 min, 56°C for 1.5 min, and 72°C for 2 min. The PCR product was digested with SacI and EcoRI and cloned between the same sites of the vector pACDH. The authenticity of the clone was verified by DNA sequence analysis.
β-Galactosidase assay.
For measuring the β-galactosidase activity of the bgl-lacZ fusion in different mutant backgrounds, the corresponding reporter strains were grown overnight in Luria-Bertani (LB) medium at 37°C and were used to inoculate 5 ml of LB medium with 7 mM salicin as an inducer. Cells were harvested at an optical density at 600 nm (OD600) of 1.0, and β-galactosidase assays were carried out as described by Miller (22). For the proU-lacZ reporter, cells were grown in low osmolarity L broth (40 mM NaCl) at 30°C, and the β-galactosidase activity was measured as described above. The results presented are averages of at least three independent experiments.
Immunoblot analysis of in vivo H-NS66 levels.
For studies of H-NS66 levels in vivo, cells from cultures grown to mid-log phase were washed once in phosphate-buffered saline, and cell extracts were prepared by sonication. Total protein was quantified by using the Bradford dye binding assay for protein estimation with bovine serum albumin as a standard (3) Total cellular proteins were electrophoresed using SDS–15% PAGE and electroblotted onto a nitrocellulose membrane (0.45-μm pore size; Amersham) by use of a semidry blotting apparatus (1 h, 1 mA/cm2 of gel). H-NS was detected by using polyclonal anti-H-NS antibodies and peroxidase-conjugated goat anti-rabbit IgG secondary antibodies (Sigma) with the aid of a chemiluminescence detection system according to the manufacturer's protocol (Immobilon Western [Millipore]). The level of ribosome release factor (RRF), an internal loading control, was detected by using purified anti-RRF antibodies.
RESULTS
Survivors of prolonged starvation include hns mutants.
When 150 random survivors from 10 independent 28-day-old stationary-phase L broth cultures of the Bgl− strain ZK819 carrying the rpoS819 allele were screened for the Bgl phenotype, 5 were determined to be Bgl+ (20). One of the Bgl+ survivors, ZK819-97, carried a C-to-T transition in the CRP-cAMP binding site within the bgl promoter that resulted in the activation of the bgl operon (20). The strain ZK819-97 exhibited a growth advantage in stationary phase (GASP) phenotype when competed against the wild-type parent. The remaining four Bgl+ survivors—ZK819-66, ZK819-67, ZK819-71, and ZK819-73—carried a mutation in hns resulting in the derepression of the bgl operon although there was no selection for growth on β-glucosides (20). Nucleotide sequence analysis of the hns locus from the four hns survivors revealed an A-to-T transversion of the middle nucleotide of the stop codon TAA of the hns open reading frame (ORF), altering it to a leucine codon TTA (Fig. 1A). This is expected to result in a longer H-NS protein in the hns-66 mutant compared to the wild-type strain (Fig. 1B). Since all four hns survivors came from the same tube and harbored the identical mutation and therefore are likely to be siblings, one of these survivors, ZK819-66, was used for further studies.
Fig 1.
(A) Nucleotide sequence of the 3′ end of wild-type hns and hns-66. The A-T transversion in the stop codon at the end of the hns-66 ORF (stop codon 1) is highlighted by an arrow. The putative Rho-dependent terminator of hns is underlined by inverted arrows, after which is the next available stop codon (stop codon 2). (B) Putative sequence of the H-NS66 polypeptide with the addition of extra amino acids at the C-terminal end.
ZK819-66 showed a strong growth disadvantage in the early stationary phase.
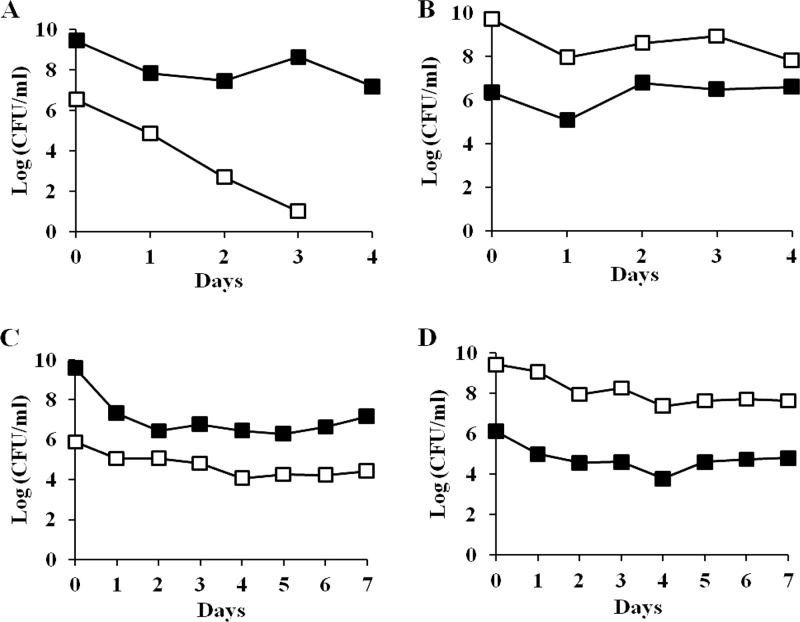
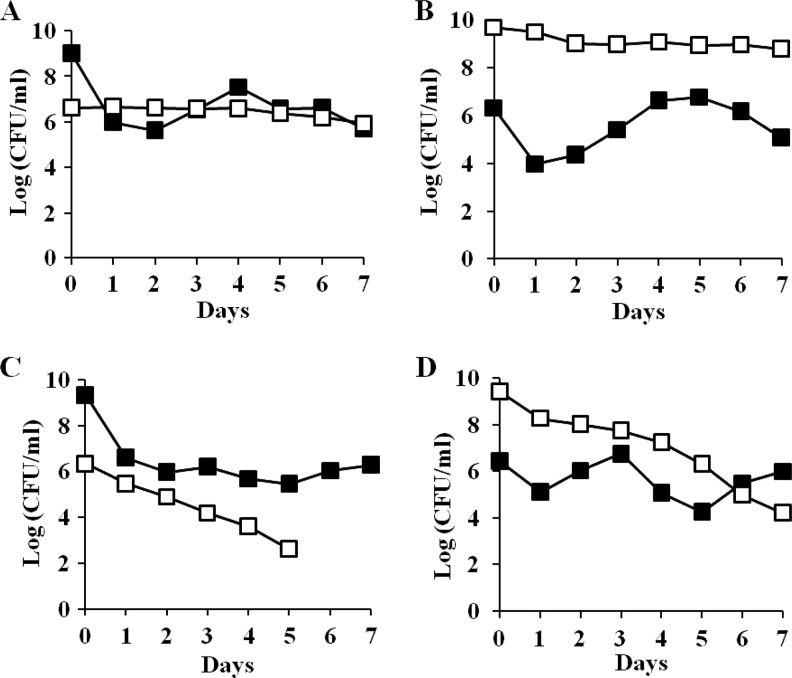
Since ZK819-66 was isolated as a survivor of prolonged starvation, it is likely to have enhanced fitness in stationary phase compared to the parent. To test this possibility, the two strains were first marked with the neutral Kanr and Tetr antibiotic markers, respectively, using Tn5 and Tn10 transposons for monitoring the growth kinetics of the two populations. One-day-old cultures of ZK819-66Tn5, grown in L broth, were competed against the parent strain ZK819Tn10, also grown for 24 h, without the addition of fresh medium. The ZK819-66Tn5 population showed a sharp decline within 48 h when present in minority, although the mutant was more stable when present in majority (Fig. 2A and B). The growth disadvantage could be rescued when the hns-66 allele was replaced by the wild-type hns allele, indicating that the sharp decline of the mutant population amounting to a “crash” is associated with the hns-66 mutation (Fig. 2C and D).
Fig 2.
(A and B) Early-stationary-phase competition between ZK819-66Tn5 (Bgl+) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (A) and 1,000:1 (B). (C and D) ZK819-66hns+ (Bgl−) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (C) and 1,000:1 (D). The mixed cultures were maintained in the original LB medium without the addition of fresh medium, and the growth of the two populations was monitored by scoring the number of CFU ml−1 (n = 3).
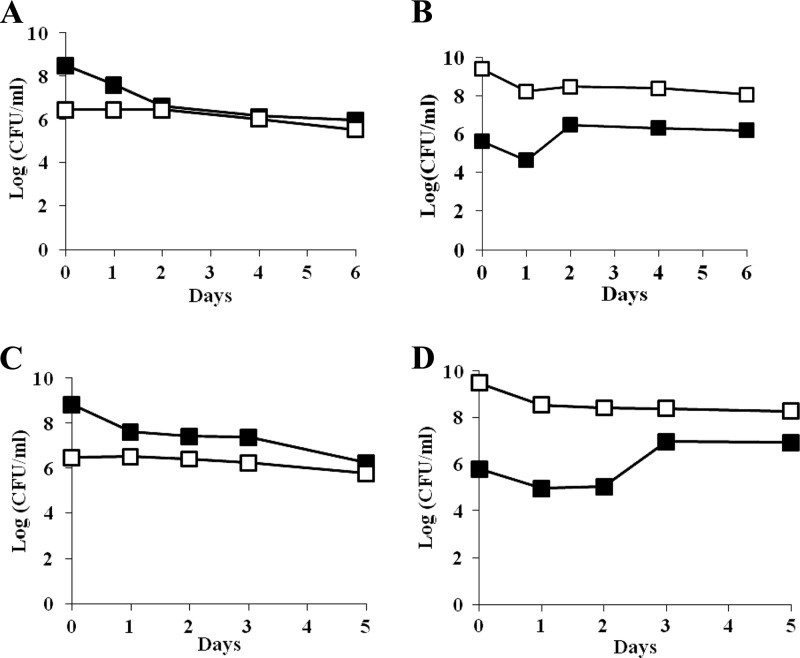
Strain ZK819-66 is likely to contain mutations in addition to the hns mutation since it is a survivor of prolonged stationary phase. To determine whether the growth disadvantage of the strain ZK819-66 is exclusively due to the mutation in hns, the hns-66 allele was transduced into the parent strain ZK819 by P1 transduction using a Tn5 transposon that is linked to hns. The transductant ZK819-66T is isogenic to the marked parent strain ZK819Tn10 except for the hns locus. When 1-day-old cultures of ZK819-66T and the parent strain ZK819Tn10 grown in L broth were mixed reciprocally in a 1:1,000 ratio, the transductant ZK819-66T also showed a steep decline in growth when present in minority (Fig. 3A and B). These observations indicate that the hns-66 allele is responsible for the growth disadvantage observed during the early stationary phase. The growth disadvantage appeared to be more drastic in the case of the transductant compared to the original survivor. This is likely to be related to the absence of the additional mutations in the transductant that are expected to be present in the survivor.
Fig 3.
(A and B) Early-stationary-phase competition between ZK819-66T (Bgl+) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (A) and 1,000:1 (B). (C and D) ZK819Δhns (Bgl+) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (C) and 1,000:1 (D). The culture conditions were similar to those described in the legend to Fig. 2 (n = 2).
To determine whether the complete loss of hns is detrimental to the cells in the stationary phase, the hns locus in the parent strain ZK819 was deleted. The resultant strain ZK819Δhns also showed a drastic growth disadvantage during early stationary phase (Fig. 3C and D), indicating that optimal expression of hns is necessary for survival during early stationary phase, at least in the genetic background tested.
The hns-66 mutant undergoes rapid reversion from Bgl+ to Bgl−.
Since the hns-66 mutant ZK819-66 showed a strong growth disadvantage in early stationary phase when competed against the hns+ parent, we tested the stability of the hns-66 allele. Spontaneous Bgl− revertants started appearing at a high frequency when ZK819-66 (Bgl+) was grown for more than 48 h. When two other Bgl+ strains—AE328 and ZK819-97T—that carry activating mutations within the bgl regulatory region were used as controls, no reversion was detected, indicating that the strains are stable and the reversion is specific to the hns-66 allele. Transduction of the hns locus from ZK819-66R, one of the Bgl− revertants, into the parent strain ZK819 resulted in the derepression of the bgl operon, indicating that the revertant carried the original hns-66 allele. This was confirmed by DNA sequence analysis of the hns locus from the revertant. The reversion of the hns-66 mutant to the Bgl− state is therefore due to a second site suppressor mutation.
Mapping of the suppressor locus.
The suppressor mutation present in the Bgl− revertant ZK819-66R was located between 50 and 70 min of the E. coli chromosome by conjugation using Hfr mapping strains (32). The suppressor locus was mapped to the 59- to 60-min region of the chromosome by P1 transduction using mapping strains carrying transposons linked to the 50- to 70-min interval (32) and the Bgl− revertant strain as the recipient. Transduction using P1 lysate raised on the strain MG1655ssrA::kan and the original Bgl+ survivor ZK819-66 as the recipient resulted in 100% cotransduction of the kanamycin resistance and Bgl− phenotype, indicating that the suppressor mutation is located within the ssrA locus (59.35 min) involved in the turnover of aberrant proteins by directing them for proteolysis, in the process also releasing stalled ribosomes. In a reciprocal experiment, when the Bgl− revertant ZK819-66R was transformed with the plasmid pTRC-ssrA carrying the wild-type ssrA allele, the transformants showed a Bgl+ phenotype. Therefore, the loss of ssrA function can suppress the hns-66 mutation. The Bgl− phenotype of 10 of 12 independently isolated revertants could be rescued by the plasmid carrying wild-type ssrA, indicating that ssrA mutations constitute the major class of mutations resulting in the suppression of the hns-66 allele. Southern analysis of the ssrA locus in ZK819-66R indicated genetic rearrangements (data not shown).
Suppression mediated by ssrA is allele specific.
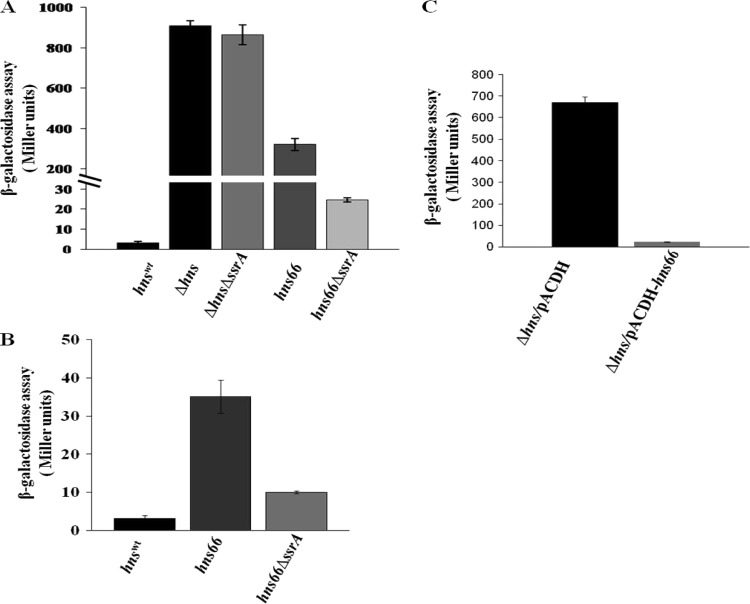
Although loss of ssrA function could suppress the Bgl+ phenotype of the hns-66 mutant, a strain carrying a deletion of hns remained Bgl+ when the ssrA locus was deleted, indicating the allele specificity of the suppression. To confirm this further, the functional status of H-NS66 was tested by measuring transcription from the wild-type bgl promoter using a chromosomal bgl-lacZ transcriptional fusion in different genetic backgrounds (Fig. 4A). H-NS66 is partially functional since it resulted in only one-third of the derepression seen in a Δhns strain in the ssrA+ background. In the ssrA mutant background the bgl promoter activity was brought down to almost basal level in the hns-66 strain, whereas in the Δhns background it remained fully derepressed (Fig. 4A). Mutation in ssrA also partially suppressed the derepression of another H-NS-sensitive promoter PproU, seen in the hns-66 background, as observed using a proU-lacZ transcriptional fusion (Fig. 4B).
Fig 4.
(A and B) Effect of ssrA deletion on bgl (A) and proU (B) promoter activity in the presence of different hns alleles. (A) Strains carrying a bglR0-lacZ fusion (where bglR0 is the silent allele of bglR) in the genetic backgrounds indicated were grown at 37°C to an OD600 of 1.0 in L broth with 7 mM salicin as an inducer. The activity of the chromosomal bglR0-lacZ fusion was measured as described by Miller (22). (B) Strains carrying a proU-lacZ fusion in the genetic backgrounds indicated were grown at 30°C in low osmolarity L broth to an OD600 of 1.0, and the β-galactosidase activity of the proU-lacZ fusion was measured as described above (n = 3). (C) Overexpression of H-NS66 from a plasmid can repress the bgl promoter. The Δhns strain carrying a bglR0-lacZ fusion was transformed with pACDH (vector control) (black bar) and pACDH-hns66 (gray bar) and grown in L broth supplemented with 15 μg of tetracycline/ml and 7 mM salicin to an OD of 1. The β-galactosidase activity was measured as described for panel A. Error bars represent the standard deviation (n = 3).
SsrA is involved in the turnover of the H-NS66 protein.
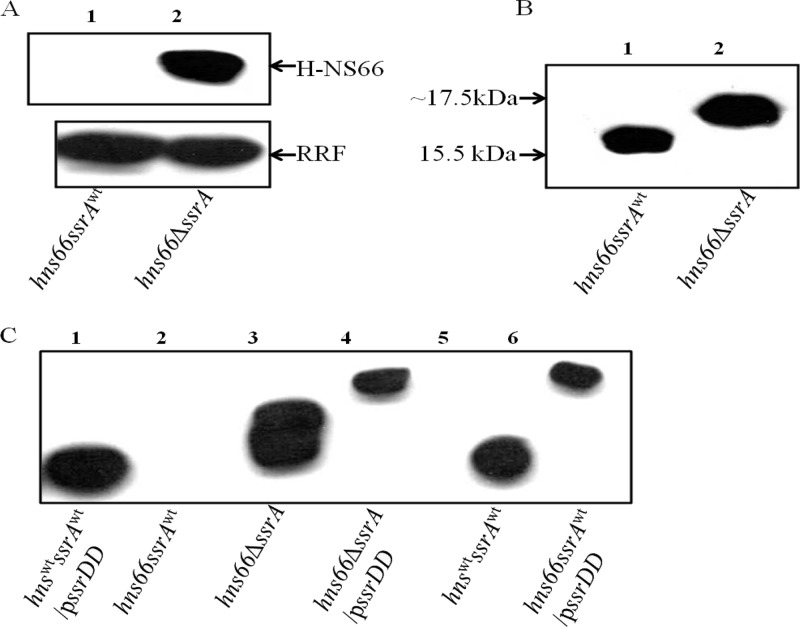
The results described above suggest that the absence of SsrA leads to the stabilization of the aberrant but partially active H-NS66 protein resulting in suppression. In such a situation, overexpression of the H-NS66 protein should result in suppression of the Bgl+ phenotype even in an ssrA+ strain. Suppression of the Bgl+ phenotype was observed when H-NS66 was overexpressed using a plasmid construct in the Δhns mutant carrying the wild-type ssrA allele (Fig. 4C). This was further confirmed by observing the levels of H-NS66 in the original hns-66 mutant using anti-H-NS polyclonal antibodies. H-NS66 accumulated in the suppressor background but was not detectable in the original mutant (Fig. 5A). Even at equal loadings of total protein, there was no detectable H-NS66 protein in the original mutant (Fig. 5A, top panel, lane 1), whereas H-NS66 was detected in the revertant (Fig. 5A, top panel, lane 2). Anti-RRF antibodies were used to detect the RRF as an internal loading control (Fig. 5A, bottom panel). The size of the H-NS66 protein was also larger than the wild-type protein (Fig. 5B).
Fig 5.
Immunoblot analysis of H-NS66 in the presence or absence of ssrA. (A) Total cell extracts of the strains ZK819-66 (lane 1) and ZK819-66ΔssrA (lane 2) were probed using polyclonal anti-H-NS antibody. Anti-RRF antibody was used to detect the loading control RRF. (B) Wild-type H-NS in cell extracts of ZK819 (lane 1) and H-NS66 in ZK819-66ΔssrA (lane 2). (C) H-NS and HNS66 in total cell extracts from different strain with or without plasmid-encoded SsrADD. Wild-type H-NS was detected in the presence or absence of SsrADD (lanes 1 and 5). H-NS66 was detected in the presence or absence of SsrA (lanes 2 and 3). H-NS66 was detected in the presence of SsrADD (lanes 4 and 6) (n = 2).
SsrA facilitates the release of aberrant polypeptides from ribosomes that are stalled on defective mRNA, typically truncated mRNA that lack a stop codon. SsrA aids in the addition of an 11-amino acids tag at the C terminus of the aberrant polypeptide which is recognized by ATP-dependent proteases and is targeted for degradation (13). In the case of the hns-66 allele, the point mutation within the stop codon is predicted to allow translation past the normal C-terminal into the putative transcriptional terminator of the hns gene. To test whether H-NS66 is a substrate that is tagged by SsrA, we used a modified ssrA allele referred to as ssrADD. The ssrADD allele facilitates the addition of a tag, but the tagged protein is not targeted for degradation since the tag is not recognized by the proteases (18). The H-NS66 polypeptide is longer than wild-type H-NS and addition of the 11-amino-acid tag by SsrA is expected to increase the size of the polypeptide further.
To compare the sizes of H-NS and H-NS66 in the presence or absence of SsrADD, immunoblot analysis of H-NS from the wild-type strain, the hns-66 mutant, and the suppressor strain, all three transformed with a plasmid encoding ssrADD, was carried out using anti-H-NS antibodies (Fig. 5C). Wild-type H-NS is unaltered in the presence or absence of pssrADD (Fig. 5C, lanes 1 and 5). The size of H-NS66, which is already larger than the wild-type H-NS, increased further in the presence of pssrADD (Fig. 5C, lanes 3, 4, and 6). Although H-NS66 in the original mutant was not detectable in the presence of wild-type ssrA (Fig. 5C, lane 2), the presence of SsrADD resulted in the accumulation the tagged H-NS66 (Fig. 5C, lane 6). These results indicate that H-NS66 is a substrate for SsrA-mediated degradation.
Suppression by ssrA rescues the hns-66-mediated growth disadvantage in the early stationary phase.
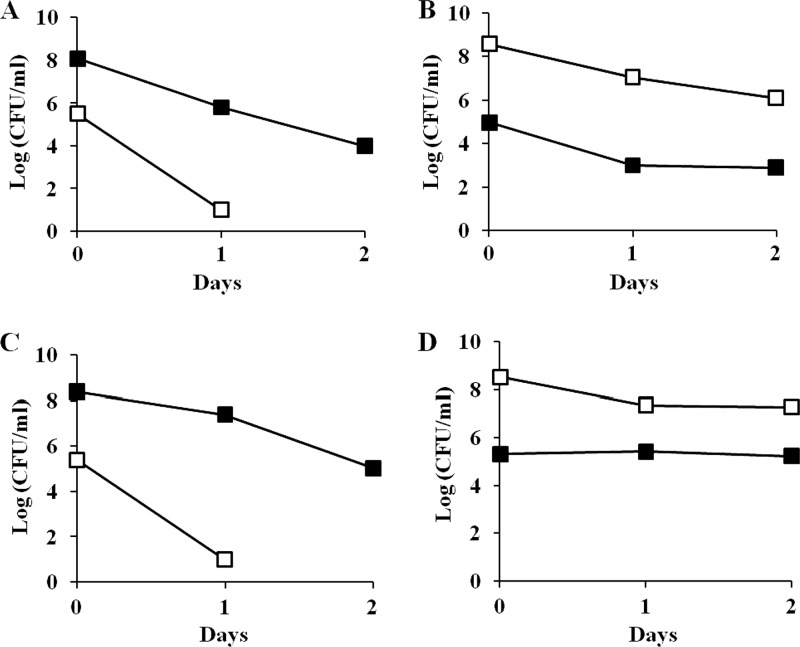
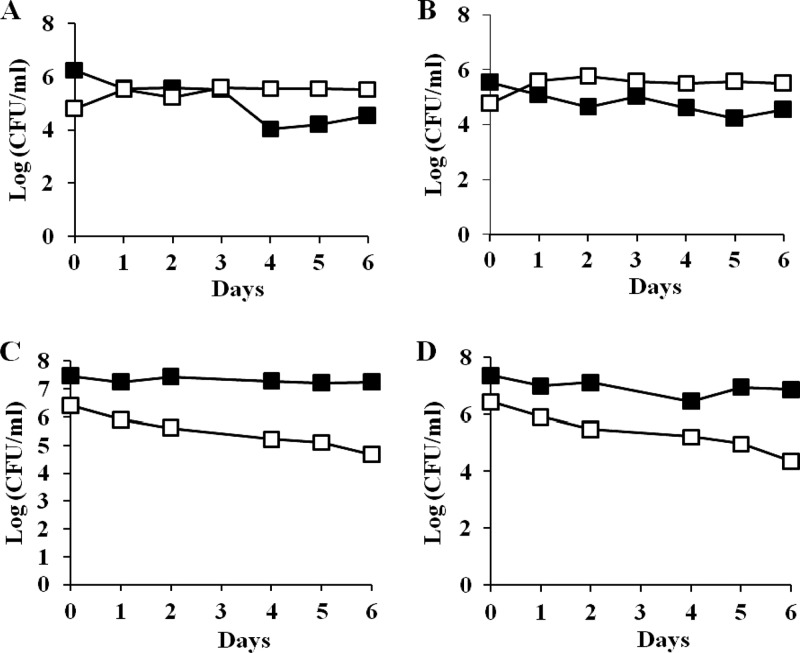
Since mutation in ssrA restores the function of H-NS66, it is conceivable that it will also rescue the hns-66-mediated growth disadvantage. To test this possibility, the original survivor carrying a deletion of the ssrA locus, ZK819-66ΔssrA (Bgl−), was competed against the parent strain. The ssrA deletion helped to overcome the growth disadvantage seen in the hns-66 mutant (Fig. 6, upper panel). As expected, the ssrA mutation could not rescue the Δhns strain from crash since the ZK819ΔhnsΔssrA double mutant continued to exhibit a growth disadvantage when competed against the parent similar to the Δhns strain (Fig. 6, lower panel).
Fig 6.
(A and B) Early-stationary-phase competition between ZK819-66ΔssrA (Bgl−) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (A) and 1,000:1 (B). (C and D) ZK819ΔhnsΔssrA (Bgl+) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (C) and 1,000:1 (D). The culture conditions were similar to those described in the legend to Fig. 2 (n = 2).
The status of rpoS influences the phenotype of the hns-66 mutation.
The strains used in the competition experiments described above also carry the rpoS819 allele that leads to reduced activity of RpoS. The rpoS819 allele was isolated as the first GASP mutation that by itself can confer growth advantage in the stationary phase. The interplay between the two global regulators RpoS and H-NS has been well established. H-NS represses a large number of genes that constitute the rpoS regulon in the exponential phase (2). To test the possible involvement of the rpoS819 allele in the phenotype of the hns-66 mutant, the rpoS819 allele was replaced by the fully functional rpoS+ allele in the original survivor ZK819-66. This resulted in the suppression of the Bgl+ phenotype of the hns-66 allele in the original survivor but not in the transductant, suggesting the presence of additional mutation(s) in the original survivor likely to be involved in the suppression by rpoS+. Interestingly, the rpoS+ allele could suppress the hns-66 mutation in the transductant in the presence of a mutation in rssB that is known to influence the stability of RpoS (24, 28). The functional status of rpoS was irrelevant in the absence of SsrA activity.
Since the hns-66-mediated growth disadvantage was rescued when the activity of H-NS66 was restored by the loss of ssrA function, it is likely that introduction of wild-type rpoS allele may show a similar rescue since rpoS+ can suppress the hns-66 mutation in the original survivor. When the strain ZK819-66rpoS+ was competed against the parent strain ZK819Tn10, the growth disadvantage displayed by the original survivor was lost (Fig. 7, upper panel); the rpoS+ strain in fact showed a modest growth advantage in the early stationary phase when in minority. Interestingly, although wild-type rpoS could not suppress the hns-66 mutation in the transductant, it could rescue the hns-66-mediated growth disadvantage of the transductant. ZK819-66TrpoS+, when competed against the parent strain, failed to show the growth disadvantage seen in the case of ZK819-66T and also showed a modest GASP phenotype when in minority (Fig. 7, lower panel). This rescue was not observed in a strain carrying a deletion of hns. Strain ZK819Δhns rpoS+ continued to show a growth disadvantage in the stationary phase when competed against the parent similar to the strain ZK819Δhns (data not shown). These results indicate that the combined presence of the rpoS819 and hns-66 mutations, both resulting in reduced function of the respective regulatory protein, is detrimental in the early stationary phase.
Fig 7.
(A and B) Early-stationary-phase competition between ZK819-66 rpoS+ (Bgl−) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (A) and 1,000:1 (B). (C and D) ZK819-66TrpoS+ (Bgl−) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (C) and 1,000:1 (D). The culture conditions were similar to those described in the legend to Fig. 2 (n = 2).
The original survivor ZK819-66 exhibits a moderate GASP phenotype when competed against the parent during long-term stationary phase.
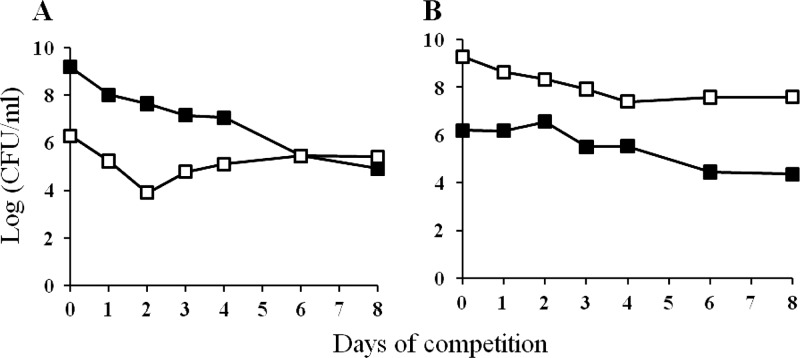
The experiments described above indicate that the original hns-66 survivor has a lower fitness in early stationary phase compared to the parent in the presence of the rpoS819 mutation. These competition experiments described thus far were performed within the first 7 days of entry into stationary phase. Since the hns-66 survivor was isolated from a 28-day-old culture, it is conceivable that this survivor arose and was selected during long-term stationary phase because it has greater fitness compared to the parent under these conditions. To test this possibility, 1-day-old cultures of ZK819-66Tn5 were competed against the parent grown for 21 days. Six replicates of the parent culture were used in each experiment to compensate for the possible genetic diversity arising during prolonged stationary phase. ZK819-66Tn5 not only avoided the crash but displayed a modest growth advantage during long-term stationary phase compared to the parent (Fig. 8, upper panel). The Bgl+ phenotype of ZK819-66Tn5 was also stable, indicating that hns-66 did not undergo suppression. The original survivor with wild-type hns could avoid the crash but failed to exhibit any growth advantage per se (data not shown), confirming the role of the hns-66 allele in the GASP phenotype.
Fig 8.
(A and B) Long-term stationary-phase competition between ZK819-66Tn5 (Bgl+) (□) and ZK819Tn10 (Bgl−) (■). A 1-day-old culture of ZK819-66Tn5 (Bgl+) was mixed with a 21-day-old culture of the parent ZK819Tn10 in the ratio of 1:10 (n = 6). (C and D) ZK819-66T (Bgl+) (□) and ZK819Tn10 (Bgl−) (■). A 1-day-old culture of ZK819-66T (Bgl+) was mixed with a 21-day-old parent ZK819Tn10 in the ratio of 1:10. The data shown are representative of six replicates, all showing a similar trend (n = 6).
To determine whether the hns-66 mutation alone is responsible for the GASP phenotype observed in the long-term stationary phase, the 1-day-old transductant strain ZK819-66T was competed against the 3-week-old parent. The transductant survived the crash but did not exhibit a GASP phenotype shown by the original survivor in long-term stationary phase (Fig. 8, lower panel).
The difference in the phenotypes of the two strains is likely to be related to the additional mutations that are expected to be present in the original survivor that are absent in the transductant. As a result, the original survivor is likely to be genetically closer to the 21-day-old parent competing against it. To compensate for the absence of the possible additional mutations in the transductant, it was competed against the 1-day-old parent in medium derived from a 21-day-old culture. The transductant, which is genetically identical to the parent except for the hns locus, showed a distinct growth advantage under these conditions when in minority, suggesting that the hns-66 allele enables the utilization of limited nutrients present in the 21-day-old medium. This is consistent with the observation that the GASP phenotype of the transductant is lost when in majority (Fig. 9). Therefore, the hns-66 allele is sufficient to provide a growth advantage in late stationary phase to the transductant that does not carry additional mutations. It is also interesting that the GASP phenotype is displayed by both the original survivor and the transductant in the presence of the rpoS819 allele that interfered with the growth advantage during the early stationary phase.
Fig 9.
(A and B) Competition between ZK819-66T (Bgl+) (□) and ZK819Tn10 (Bgl−) (■). One-day-old cultures of the two strains were mixed in ratios of 1:1,000 (A) and 1,000:1 (B) in spent medium derived from a 3-week-old parent culture (n = 2).
DISCUSSION
In this study we have characterized the hns-66 mutation present in a survivor of prolonged starvation. The parent strain ZK819 used in this experiment carries the rpoS819 mutation, which itself was originally isolated from survivors of prolonged stationary phase (36). The presence of the hns-66 allele in the survivors suggested the possibility that it is also under selection and, like the rpoS819 allele, confers a growth advantage in the stationary phase. The investigations described here were primarily aimed at addressing this possibility.
The hns-66 allele present in ZK819-66 results in reduced activity of H-NS, which is strikingly similar to the nature of rpoS819 allele. Surprisingly, the original survivor ZK819-66 and the transductant strain ZK819-66T showed a strong growth disadvantage when competed against the parent in early stationary phase. This was further supported by the observation that the hns-66 allele was unstable and was suppressed by second-site mutations. One such suppressor mutation was identified within the ssrA locus that encodes for a tmRNA, a hybrid tRNA-mRNA entity that is a main player in the trans-translation process that rescues stalled ribosomes from mRNA that lack translation termination signals (19). The trans-translation complex shifts the translation machinery from the defective mRNA to the mRNA segment of the tmRNA. Resumption of translation results in the addition of an 11-amino-acid tag that targets the defective polypeptide to proteases such as ClpAP/ClpXP (13).
In the original hns-66 mutant with functional SsrA, we failed to detect H-NS66 in immunoblots. However, in the ssrA mutant a significant level of the longer H-NS66 protein was detectable, indicating the stabilization of H-NS66 in the absence of SsrA function. This is consistent with the observation that overexpression of H-NS66 from a plasmid led to the suppression of the hns-66 phenotype. The presence of a protein larger than H-NS66 in strains carrying the ssrADD allele further confirmed that H-NS66 is modified by SsrA. The ssrA mutation also reversed the growth disadvantage of the hns-66 mutant observed in stationary-phase competition experiments, indicating that in the rpoS819 background, there is selection for functional hns in the early stationary phase, the loss of which results in a significant growth disadvantage.
RpoS and H-NS play very important roles in the homeostatic control of the genes of the rpoS regulon due to their opposing actions. It has been reported that strains carrying loss-of-function mutations in hns spontaneously undergo selection for mutations in the rpoS gene (2), and conversely rpoS strains accumulate hns mutations (23) specifically in the stationary phase (8). The complete reversal of the GASP phenotype conferred by the rpoS819 allele in the presence of the hns-66 allele prompted us to study the interrelation between these two independently isolated mutant alleles from long-term stationary-phase survivors and their role in survival during the stationary phase. Our studies have shown that wild-type rpoS could suppress the Bgl+ phenotype of the hns-66 mutation in the original survivor but not in the hns-66 transductant, suggesting the involvement of an additional mutation(s) in the original survivor. However, wild-type rpoS in conjunction with a mutation in rssB could suppress the hns-66 mutation in the transductant. Since rssB is known to be involved in the regulation of RpoS levels (24, 28), the additional mutation(s) in the original survivor might be similarly affecting the level of RpoS. The mechanism by which rpoS+ suppresses the hns-66 allele is at present unclear.
Wild-type rpoS could not only rescue the crash associated with the original survivor but also confer a modest growth advantage in the early stationary phase compared to the parent. Therefore, the hns-66 allele can contribute to fitness during early stationary phase if the rpoS819 allele is replaced by the wild-type rpoS+ allele, indicating that the combined presence of the two mutant alleles leads to growth inhibition. Interestingly, although wild-type rpoS by itself could not suppress the Bgl+ phenotype of the hns-66 allele in the transductant, it could rescue the crash of the transductant in the early stationary phase.
In the light of these results, the presence of the hns-66 mutation in survivors of prolonged stationary phase was puzzling since it is unlikely that the hns-66 mutation would have been selected during early stationary phase because of the reduced fitness of the mutant. One possibility is that since culture conditions during extended stationary phase are constantly changing, the selective advantage of the hns-66 allele is not in the early stationary phase but in the long-term stationary phase. This is consistent with our observation that when the 1-day-old original hns-66 survivor was competed against the 21-day-old parent, it not only survived the crash but also displayed a modest growth advantage in the presence of rpoS819. The stability of the hns-66 allele under prolonged stationary phase also suggests selection for the mutant allele. Replacing the hns-66 allele by hns+ in the original mutant abolished the GASP phenotype during long-term stationary phase, confirming the role of the hns-66 allele in the growth advantage. The Δhns mutation retained the growth disadvantage phenotype in a prolonged stationary phase, indicating that the complete loss of hns function is detrimental.
Interestingly, the hns-66 transductant survived the crash but failed to show any growth advantage in extended stationary phase when competed against 21-day-old parent cells. The GASP phenotype could be recovered if the parent culture used is also 1 day old, thereby genetically identical to the transductant except for the presence of the hns-66 allele, in medium derived from a 21-day-old culture. This observation indicates that the hns-66 allele in conjunction with the rpoS819 allele is sufficient for the GASP phenotype during long-term stationary phase. The combined presence of the rpoS819 and hns-66 alleles is advantageous during prolonged stationary phase but detrimental in the early stationary phase. Either mutation can provide a growth advantage individually in the early stationary phase. The advantage provided by the hns-66 allele is most likely to be related to the increased ability of the mutant to scavenge a specific nutrient available in low concentrations since the GASP phenotype is observed only when the mutant is in minority.
The results described above have raised several interesting questions regarding the chronological appearance of GASP mutations. Did the additional mutations arise before the appearance of the hns-66 mutation? If so, was there any fitness advantage associated with the additional mutations by themselves at any point of the stationary phase? As stated above, it is unlikely that the hns-66 allele arose in the absence of the additional mutations because of the growth disadvantage shown during the early stationary phase. It is likely that the alterations in the genetic composition of the parent as well as the growth conditions enabled the selection of the hns-66 allele during the late stationary phase. Identification and characterization of the additional mutations will shed light on the nature of the possible genetic interaction between them and the hns-66 mutation.
Our studies have added another global regulator hns to the list of pleiotropic regulators, mutations which confer a selective advantage under prolonged nutrient starvation conditions. These results are consistent with the hypothesis that mutations in global regulatory genes are likely to have a more profound effect on survival due to their impact on the expression of a large number of genes (39). The rpoS819 and lrp-1141 mutations that are known to enhance the ability to scavenge nutrients such as amino acids released from the dying cells (36, 38) also fall under the category of global regulators of transcription in E. coli (11, 37). A rapidly evolving population probably favors altered gene expression instead of the gain of a new function that might involve more than one step and hence be costly in terms of time, consistent with the nature of the rpoS819 and hns-66 mutations.
ACKNOWLEDGMENTS
We thank C. Ueguchi for anti-H-NS antibodies, U. Varshney for anti-RRF antibodies and the plasmid pACDH-ssrAwt, and R. T. Sauer for the plasmid pKW23 (ssrADD). We also thank the two anonymous referees for their constructive suggestions that have helped to improve the manuscript considerably.
This study was supported by a grant to S.M. from the Department of Science and Technology, Government of India.
Footnotes
Published ahead of print 27 July 2012
REFERENCES
- 1. Atlung T, Ingmer H. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7–17 [DOI] [PubMed] [Google Scholar]
- 2. Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. 1995. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σS and many σS-dependent genes in Escherichia coli. J. Bacteriol. 177:3455–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bradford M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 72:248. [DOI] [PubMed] [Google Scholar]
- 4. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 5. Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. 2002. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277:2146–2150 [DOI] [PubMed] [Google Scholar]
- 6. Dame RT, Noom MC, Wuite GJL. 2006. Bacterial chromatin organization by H-NS protein unraveled using dual DNA manipulation. Nature 444:387–390 [DOI] [PubMed] [Google Scholar]
- 7. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desai SK, Mahadevan S. 2006. Accumulation of hns mutations specifically in stationary phase in an Escherichia coli strain carrying an impaired rpoS locus. J. Genet. 85:221–224 [DOI] [PubMed] [Google Scholar]
- 9. Erol I, et al. 2006. H-NS controls metabolism and stress tolerance in Escherichia coli O157: H7 that influence mouse passage. BMC Microbiol. 12:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Esposito D, et al. 2002. H-NS oligomerization domain structure reveals the mechanism for high-order self-association of the intact protein. J. Mol. Biol. 324:841–850 [DOI] [PubMed] [Google Scholar]
- 11. Farrell MJ, Finkel SE. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 185:7044–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nature Rev. Microbiol. 4:113–120 [DOI] [PubMed] [Google Scholar]
- 13. Gottesman S, Roche E, Zhou Y, Sauer RT. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harwani D, Zangoui P, Mahadevan S. 2012. The β-Glucoside (bgl) operon of Escherichia coli is involved in the regulation of oppA, encoding an oligopeptide transporter. J. Bacteriol. 194:90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hennge-Aronis R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in Escherichia coli. Cell 72:165–168 [DOI] [PubMed] [Google Scholar]
- 16. Higgins CF, et al. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in Salmonella typhimurium and Escherichia coli. Cell 52:569–584 [DOI] [PubMed] [Google Scholar]
- 17. Hommais F, et al. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20–36 [DOI] [PubMed] [Google Scholar]
- 18. Karzai AW, Susskind MM, Sauer RT. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18:3793–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keiler KC, Waller PRH, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993 [DOI] [PubMed] [Google Scholar]
- 20. Madan R, Kolter R, Mahadevan S. 2005. Mutations that activate the silent bgl operon of Escherichia coli confers a growth advantage in stationary phase. J. Bacteriol. 187:7912–7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mahadevan S, Reynolds AE, Wright A. 1987. Positive and negative regulation of the bgl operon in Escherichia coli. J. Bacteriol. 169:2570–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23. Moorthy S, Mahadevan S. 2002. Differential spectrum of mutations that activate the Escherichia coli bgl operon in an rpoS genetic background. J. Bacteriol. 184:4033–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muffler A, Fischer D, Altuvia S, Storz GHengge-aronis R. 1996. The response regulator RssB controls stability of the σS subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333–1339 [PMC free article] [PubMed] [Google Scholar]
- 25. Mukerji M, Mahadevan S. 1997. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol. Microbiol. 24:617–627 [DOI] [PubMed] [Google Scholar]
- 26. Navarre WW, McClelland M, Libby SJ, Fang FC. 2007. Silencing of xenogeneic DNA by H-NS: facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21:1456–1471 [DOI] [PubMed] [Google Scholar]
- 27. Prasad I, Schaefler S. 1974. Regulation of the β-glucoside system in Escherichia coli K-12. J. Bacteriol. 120:638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pratt LA, Silhavy TJ. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. U. S. A. 93:2488–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rimsky S, Travers A. 2011. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr. Opin. Microbiol. 14:136–141 [DOI] [PubMed] [Google Scholar]
- 30. Schnetz K, Toloczyki C, Rak B. 1987. β-Glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis Genes J. Bacteriol. 169:2579–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schnetz K, Wang JC. 1996. Silencing of the Escherichia coli bgl promoter: effects of template supercoiling and cell extracts on promoter activity in vitro. Nucleic Acids Res. 24:2422–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singer M, et al. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh NS, Varshney U. 2004. A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Res. 32:6028–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamada H, Muramatsu S, Mizino T. 1990. An Escherichia coli protein that preferentially binds to sharply curved DNA. J. Biochem. 108:420–425 [DOI] [PubMed] [Google Scholar]
- 35. Yamashino T, Ueguchi C, Mizuno T. 1995. Quantitative control of the stationary phase-specific sigma factor, σS, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 14:594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zambrano MM, Siegele DA, Almiron M, Tormo A, Kolter R. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757–1760 [DOI] [PubMed] [Google Scholar]
- 37. Zinser ER, Kolter R. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zinser ER, Kolter R. 2000. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J. Bacteriol. 182:4361–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zinser ER, Kolter R. 2004. Escherichia coli evolution during stationary phase. Res. Microbiol. 155:328–336 [DOI] [PubMed] [Google Scholar]