Abstract
Escherichia coli O157:H7 is a gastrointestinal pathogen that has become a serious public health concern, as it is associated with outbreaks and severe diseases such as hemolytic-uremic syndrome. The molecular basis of its greater virulence than that of other serotypes is not completely known. OI-1 is a putative fimbria-encoding genomic island that is found almost exclusively in O157:H7 Shiga toxin-producing E. coli strains and may be associated with the enhanced pathogenesis of these strains. In this study, we identified and characterized a novel repressor of flagellar synthesis encoded by OI-1. We showed that deletion of Z0021 increased the motility of E. coli O157:H7, which correlated with an increase in flagellin production and enhanced assembly of flagella on the cell surface. In contrast, overexpression of Z0021 inhibited motility. We demonstrated that Z0021 exerted its regulatory effects downstream of the transcription and translation of flhDC but prior to the activation of class II/III promoters. Furthermore, the master regulator of flagellar synthesis, FlhD4C2, was shown to be a high-copy suppressor of the nonmotile phenotype associated with elevated levels of Z0021—a finding consistent with Z0021-FlhD4C2 being a potential regulatory complex. This work provides insight into the mechanism by which Z0021, which we have named fmrA, represses flagellar synthesis and is the first report of a fimbrial-operon-encoded inhibitor of motility in E. coli O157:H7.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) is a zoonotic food-borne and waterborne pathogen that has been implicated in outbreaks worldwide and can cause hemorrhagic colitis and the potentially fatal hemolytic-uremic syndrome (7, 14, 16, 17, 24). The O157:H7 serotype of enterohemorrhagic STEC (EHEC), classified as a seropathotype A strain, is one of the most prevalent and is recognized for its high virulence in human populations (10, 13, 24). A genomic comparison of E. coli O157:H7 with serotype O26:H11, which is less frequently associated with human outbreaks and disease, revealed the presence of four fimbria-encoding genomic islands unique to seropathotype A strains: O-island 1 (OI-1), OI-47, OI-141, and OI-154 (30). Although the mechanisms underlying the unique pathogenesis of E. coli O157:H7 compared to that of other STEC strains are not fully known, these four OIs may be associated with the ability of O157:H7 to colonize humans and cause disease more readily than other serotypes do (30).
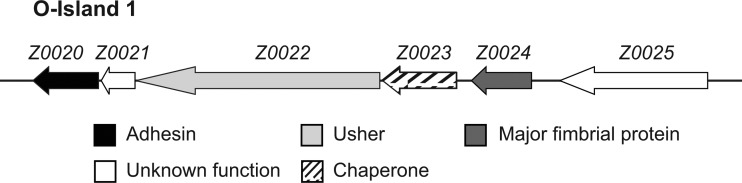
Fimbria-mediated adherence of E. coli O157:H7 to intestinal epithelial cells is an important and early step in the colonization process. The sequencing of two E. coli O157:H7 outbreak strains revealed the presence of at least 16 fimbrial operons, many of the chaperone/usher class (12, 26). Of the four fimbria-encoding OIs that are present in seropathotype A strains, two have been subjects of investigation. OI-141 and OI-154 encode long polar fimbriae (LPF), which were first described in Salmonella enterica serovar Typhimurium and show similarity to type I fimbriae (2). Torres and colleagues showed that the LPF1 cluster of E. coli O157:H7 located in OI-141 increases fimbrial expression and adherence to tissue culture cells when introduced into a nonfimbriated E. coli K-12 strain (35). Furthermore, the LPF1 cluster has a demonstrated role in adherence to epithelial cells and microcolony formation (35). The second lpf gene cluster of E. coli O157:H7, located in OI-154, is implicated in the initial stages of adhesion, and a similar region in E. coli O113:H21 has been shown to mediate adherence to epithelial cells (9, 36). In contrast to OI-141 and OI-154, little is known about OI-1 and OI-47. The major fimbrial proteins encoded by OI-47 are also related to Salmonella LPF, and this OI is predicted to contain additional putative virulence genes (30). OI-1 is distinct from these three LPF-related clusters and is predicted to encode type 1-like fimbriae. The putative fimbrial genes in OI-1 are present in an order highly conserved with respect to that of the E. coli type 1 fimbrial locus (30) (Fig. 1).
Fig 1.
Genetic organization of OI-1 in E. coli O157:H7 strain EDL933. Arrows represent open reading frames, and the putative functions of the open reading frames based on BLAST similarity are shown.
In addition to fimbriae, flagella also play an important role in E. coli O157:H7 pathogenesis, as they enable the bacteria to breach the intestinal mucus layer to access the intestinal epithelium. Flagellar synthesis is a tightly regulated and highly energetic three-tier process (21). The early genes flhDC encode the master regulator FlhD4C2, which is required for the transcription of the middle genes from class II promoters (5). The middle genes encode the structural and assembly proteins required for the synthesis of the hook-basal body, the alternative sigma factor FliA (σ28), and its anti-sigma factor FlgM (anti-σ28). Once the hook-basal body has been assembled, FlgM is secreted, thereby allowing FliA to activate the transcription of the late genes that encode flagellin and the motor and chemotaxis proteins (4). Given that adherence and motility represent antagonistic functions, bacteria use mechanisms to reciprocally regulate these processes. For instance, the BvgAS two-component signal transduction system in Bordetella pertussis activates adhesin genes while repressing those involved in motility (1). There are also proteins encoded within fimbrial operons that have been shown to regulate motility. In Proteus mirabilis, MrpJ, which is encoded by the MR/P fimbrial gene cluster, inhibits swimming and swarming motility by repressing the transcription of flhDC (19). The functional homologue of MrpJ in uropathogenic E. coli, PapX, has been shown to repress motility by binding directly to the flhDC promoter (27, 31). To date, there have been no fimbrial-operon-encoded regulators of motility identified in E. coli O157:H7.
We turned our attention to the OI-1 genomic island as a potential source of virulence determinants associated with O157:H7 EHEC. In this work, we identified and characterized a novel repressor of motility in E. coli O157:H7, Z0021, encoded by OI-1. We show that deletion of Z0021 leads to greater swimming motility and flagellar production in E. coli O157:H7 than in the parental strain while overexpression of Z0021 inhibits motility. Using a transcriptional reporter system, we demonstrate that Z0021 regulates motility prior to middle and late gene transcription but downstream of the master regulator, FlhD4C2. In addition, we show that FlhD4C2 is a high-copy suppressor of the nonmotile phenotype associated with the overexpression of Z0021. In keeping with this functional role, we propose that Z0021 be renamed fmrA (fimbrial-operon-encoded motility regulator A). This paper reports on a novel fimbrial-operon-encoded regulator of flagellar synthesis in E. coli O157:H7 and provides insight into the mechanism by which Z0021/FmrA represses motility.
MATERIALS AND METHODS
General methods.
Table 1 lists the strains, plasmids, and oligonucleotides used in this work. E. coli strains were grown in Luria broth (LB), and when necessary, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 34 μg/ml. Gel extraction and plasmid miniprep kits were purchased from Qiagen (Mississauga, Ontario, Canada). Vent polymerase was obtained from New England BioLabs (Beverly, MA), and restriction enzymes were purchased from Fermentas (Burlington, Ontario, Canada).
Table 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| BKC15-1 | EHEC serotype O157:H7, wild-type strain EDL933 | 26 |
| BKC19-79 | EDL933 with deletion of OI-1 (ΔOI-1::kan) | This study |
| BKC19-81 | EDL933 ΔZ0020; Kanr | This study |
| BKC20-1 | EDL933 ΔZ0021; Kanr | This study |
| BKC15-3 | EDL933 ΔZ0021; kan cassette excised | This study |
| BKC20-7 | EDL933 ΔZ0022 ΔZ0023 ΔZ0024 ΔZ0025; Kanr | This study |
| BKC30-74 | EDL93/pCS26-flhD | This study |
| BKC30-75 | EDL93/pCS26-fliE | This study |
| BKC30-76 | EDL93/pCS26-fliL | This study |
| BKC30-77 | EDL93/pCS26-flhB | This study |
| BKC30-78 | EDL93/pCS26-flgB | This study |
| BKC30-79 | EDL93/pCS26-flgA | This study |
| BKC30-80 | EDL93/pCS26-fliF | This study |
| BKC30-81 | EDL93/pCS26-fliA | This study |
| BKC31-1 | EDL93/pCS26-fliC | This study |
| BKC31-2 | EDL93/pCS26-motA | This study |
| BKC31-3 | EDL933 ΔZ0021 (BKC15-3)/pCS26-flhD | This study |
| BKC31-4 | EDL933 ΔZ0021 (BKC15-3)/pCS26-fliE | This study |
| BKC31-5 | EDL933 ΔZ0021 (BKC15-3)/pCS26-fliL | This study |
| BKC31-6 | EDL933 ΔZ0021 (BKC15-3)/pCS26-flhB | This study |
| BKC31-7 | EDL933 ΔZ0021 (BKC15-3)/pCS26-flgB | This study |
| BKC31-8 | EDL933 ΔZ0021 (BKC15-3)/pCS26-flgA | This study |
| BKC31-9 | EDL933 ΔZ0021 (BKC15-3)/pCS26-fliF | This study |
| BKC31-10 | EDL933 ΔZ0021 (BKC15-3)/pCS26-fliA | This study |
| BKC31-11 | EDL933 ΔZ0021 (BKC15-3)/pCS26-fliC | This study |
| BKC31-12 | EDL933 ΔZ0021 (BKC15-3)/pCS26-motA | This study |
| BKC16-15 | EDL933 flhD::flhD-HA | This study |
| BKC16-16 | EDL933 flhC::flhC-HA | This study |
| BKC31-14 | EDL933 ΔZ0021 flhC::flhC-HA | This study |
| BKC31-15 | EDL933/pFLAG-CTC | This study |
| BKC31-16 | EDL933/pFLAG-Z0021 | This study |
| BKC31-17 | EDL933/pFLAG-Z0021-araC-PBAD | This study |
| BKC31-18 | EDL933/pFLAG-Z0021-flhDC | This study |
| BKC1-14 | DH5α (hsdR recA lacZYA ϕ80dlacZΔM15) | Lab collection |
| Plasmids | ||
| pKD4 | oriRγ; Kanr cassette flanked by FRT sites | 8 |
| pSU315 | HA epitope sequence with Kanr cassette flanked by FRT sites | 37 |
| pKD46 | RepA1019(Ts), λ, γ, β, and exo expressed from ParaBAD; Ampr | 8 |
| pCS26 | pSC101 ori, luxCDABE; Kanr | 3 |
| pCS26-flhD | flhD promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-fliE | fliE promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-fliL | fliL promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-flhB | flhB promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-flgB | flgB promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-flgA | flgA promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-fliF | fliF promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-fliA | fliA promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-fliC | fliC promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pCS26-motA | motA promoter from EDL933 fused to luxCDABE in pCS26; Kanr | This study |
| pFLAG-CTC | Cytoplasmic expression of C-terminal FLAG fusion protein under control of Ptac; Ampr | Sigma |
| pFLAG-Z0021 | pFLAG-CTC carrying Z0021 with native stop codon; Ampr | This study |
| pBAD33 | Arabinose-inducible expression vector; Cmr | 11 |
| pFLAG-Z0021-araC-PBAD | pFLAG-CTC carrying Z0021 with native stop codon at HindIII/KpnI sites and araC-PBAD cloned in at BglII/SalI sites; Ampr | This study |
| pFLAG-Z0021-flhDC | pFLAG-CTC carrying Z0021 with native stop codon and araC-PBAD-flhDC cloned in at BglII/SalI sites; Ampr | This study |
| Primers | ||
| Red-OI-1-F | GGAATGGTGAAATTTATCGCAGATAGCATTTCTTCTAAATTATAAGATGTGTAGGCTGGAGCTGCTTCG | |
| Red-OI-1-R | CTCATAAATTAAATTAATAAAAATTCATCAACTTCAAGGACTGATAATATGAATGTCCTCCTTA | |
| Red-Z0020-F | GCGCTTCCAGGGAAGTCATCTTATAATTTAGAAGAAGTGTAGGCTGGAGCTGCTTCG | |
| Red-Z0020-R | CGGCAGCTCCCCCAAAGTTAAGGTGGGGGAGATAGACATATGAATATCCTCCTTA | |
| Red-Z0021-F | CGATGAAAACAAAACATATATATGCCAGTAAAAGGAGTTTACTGTGTAGGCTGGAGCTGCTTCG | |
| Red-Z0021-R | GTATGGAATGGTGAAATTTATCGCAGATAGCATTTCTTCTAAACATATGAATATCCTCCTTA | |
| Red-Z0022-F | CATAGTAAAATCGCCGCGAGATAACAGGAAAAAGTCGTGTAGGCTGGAGCTGCTTCG | |
| Red-Z0022-R | TGTGATTAATAAAAGCCACTTCATAGTAAACTCCTTCATATGAATATCCTCCTTA | |
| Red-Z0022-25-F | TAAGGCTAACCTGACTGCACAGATCAACAAACTGGCTTAAGTGTAGGCTGGAGCTGCTTCG | |
| Red-Z0022-25-R | TAAAAGAATATAGACTCAATGTGATTAATAAAAGCCACTTCACATATGAATATCCTCCTTA | |
| conf-pKD4-F | CCTTCTTGACGAGTTCTTCT | |
| Conf-Z0020-R | AGTTAAGGTGGGGGAGATAG | |
| Conf-Z0021-F | CCTGTGGATTGACCAATGTC | |
| Conf-Z0021-R | ACGACTTCCGACTTTAAACC | |
| Conf-Z0022-25-R | GATTAATAAAAGCCACTTCA | |
| promflhD-F | GCGCGGATCCCATTATTCCCACCCAGAATAACC | |
| promflhD-R | GCGCCTCGAGGCATCCTGAATTAACTTATCAAG | |
| promfliE-F | GCGCGGATCCTATCGCTGACATTTTCATCTCCTG | |
| promfliE-R | GCGCCTCGAGTGGGCGTGAATATTACCGTTACC | |
| promfliL-F | GCGCCTCGAGGCCAGCATATTTCGCTGTTCACC | |
| promfliL-R | GCGCGGATCCGTAATCAGTCATGTGTTGCGGGTC | |
| promflhB-F | GCGCGGATCCCTCGTCAGACACGTCGCCAATCC | |
| promflhB-R | GCGCCTCGAGAGCCAGTCAGGATCAGGTGGACG | |
| promflgB-F | GCGCCTCGAGGGTGGAGGCTGTTGTTTTTGCCGCTC | |
| promflgB-R | GCGCGGATCCCTTATCGAGCATATCTCCTCCGCAG | |
| promflgA-F | GCGCGGATCCTATTGCCAGCATTTTCGCCCCCAG | |
| promflgA-R | GCGCCTCGAGATTGGCGATGTTTGCTGCCAGCAC | |
| promfliF-F | GCGCCTCGAGTGCGGCAGTGATTCCTGCGCACG | |
| promfliF-R | GCGCGGATCCAGTCGCATTCATCGCGCACCTCGTG | |
| promfliA-F | GCGCGGATCCGAGTGAATTCACGATAAACAGCCCTG | |
| promfliA-R | GCGCCTCGAGGCAACTCCTGCGACAACCACTCCAG | |
| promfliC-F | GCGCGGATCCGACTTGTGCCATGATTCGTTATCC | |
| promfliC-R | GGGCCTCGAGGCGATTTCCTTTTATCATTCGACA | |
| prommotA-F | GCGCGGATCCTAAGATAAGCACGACATCATCCTTC | |
| prommotA-R | GCGCCTCGAGCTTCGATGTTCTGTAATGCATGG | |
| qRT-flhD-F | ACCTCCGAGTTGCTGAAACAC | |
| qRT-flhD-R | TTGCTGGAGATCGTCAACGC | |
| qRT-flgM-F | CCGTTCAACCGCGCGAAACC | |
| qRT-flgM-R | TGCTGCCGGGTTGCATCAGT | |
| qRT-motA-F | GCGAACAGTCTGGCGCTGGT | |
| qRT-motA-R | TGTGCGATAAGCGCCCCCAG | |
| qRT-fliC-F | TGACGGTGCCTCTCTGACATTC | |
| qRT-fliC-R | AAGACTTCGCAGCATCACTGG | |
| qRT-icdA-F | ACGTGATTGCTGATGCATTCCTGC | |
| qRT-icdA-R | ACCGTTCAGGTTCATACAGGCGAT | |
| pSU315-flhC-F | CCACAACTGCTGGATGAACAGAGAGTACAGGCTGTTTATCCGTATGATGTTCCTGAT | |
| pSU315-flhC-R | GTTACCGCTGCTGGAATGTTGCGCCACACCGTATCAGCATATGAATATCCTCCTTAG | |
| conf-flhC-F | CAGAGCCAGCAGATCCATATAC | |
| conf-flhC-R | GTTTGTGTAATGGCGTCGATGC | |
| Z0021-F | GCGCAAGCTTATGAAGTGGCTTTTATTAATC | |
| Z0021-R | GCGCGAATTCTTATAAGATGACTTCCCTGGAAG | |
| flhDC-F | GCGCAAGCTTTTAAACAGCCTGTACTCTCTGTTC | |
| flhDC-R | GCGCGGTACCAATAAGGAGGAAAAAAAAGTGGGAATAATGCATACCTCCGAG | |
| araC-flhDC-F | GCGCAGATCTTTATGACAACTTGACGGCTACATC | |
| araC-flhDC-R | GCGCGTCGACCTGATTTAATCTGTATCAGGCTG |
Bold sequences are restriction sites. The underlined sequence is the optimal ribosome-binding site.
Construction of mutants and HA-tagged allelic variants.
The deletion of single or multiple genes within OI-1 was carried out as described by Datsenko and Wanner (8). Linear DNA was PCR amplified from pKD4 by using the primers listed in Table 1. E. coli O157:H7 strain EDL933 harboring pKD46 was transformed with the concentrated PCR product, plated onto LB supplemented with kanamycin, and incubated overnight at 37°C. All of the strains generated were confirmed by PCR. A similar strategy using pSU315 (37) was employed to create strains in which flhC had been tagged with tandem hemagglutinin (HA) epitopes at the carboxy terminus.
Motility assays.
Swimming motility was assessed by using 0.25% LB agar plates. Overnight cultures were standardized to an optical density at 600 nm (OD600) of 1.0, and 2 μl of culture was stabbed into the agar plates by using a sterile pipette tip. Ampicillin was added to the plates for maintenance of plasmids, and isopropyl-β-d-thiogalactopyranoside (IPTG) and arabinose were added to the plates for induction when necessary. The plates were incubated for 6 h at 37°C, after which time the diameter of the swimming zone around the inoculation site was measured.
TEM.
Wild-type EDL933 and ΔOI-1 and ΔZ0021 mutant strains were cultured on motility agar for 6 h at 37°C. Bacteria were adsorbed to carbon-stabilized Formvar supports on 200-mesh copper transmission electron microscopy (TEM) grids by floating the grids with the Formvar side down on a drop of culture for 30 s and then rapidly washing them with water. Bacteria on TEM grids were stained by submerging the grids for 10 s in 0.1% (wt/vol) uranyl acetate and then examined with a Philips CM10 transmission electron microscope at an operating voltage at 80 kV. Digital images of bacteria were captured with an Olympus Soft Imaging Systems Morada 11-megapixel charge-coupled device camera (Biophysics Interdepartment Group, University of Guelph, Guelph, Ontario, Canada).
Western blot analysis.
To examine the levels of H7 flagellin in the wild-type EDL933 and ΔOI-1 and ΔZ0021 mutant strains, cultures were grown in LB for 3, 6, or 24 h at 37°C. Whole-cell lysates were resolved on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore Corp.). The blots were incubated with a 1:2,000 dilution of rabbit polyclonal antiserum to H7 flagellin, followed by a 1:2,000 dilution of peroxidase-conjugated mouse anti-rabbit immunoglobulin G, and developed using a chemiluminescence detection system (Pierce Chemical Company, Rockford, IL). For Western blot analysis of FlhC-HA in the wild-type EDL933 and ΔZ0021 mutant strains, cultures were grown in LB at 37°C to an OD600 of ∼0.5. Whole-cell lysates were collected and probed using anti-HA (1:2,000) and anti-DnaK (1:10,000) antibodies. DnaK served as a loading control.
Transcriptional reporter assays.
Luciferase reporter constructs for class I, II, and III promoters were generated by PCR amplification of promoter regions from E. coli O157:H7 strain EDL933 genomic DNA by using primers listed in Table 1. The PCR products were cloned into pCS26 (3) and transformed into wild-type EDL933 and ΔZ0021 mutant strains. The sequences of all constructs were confirmed by sequencing. Overnight cultures were subcultured into LB to a starting OD600 of 0.005 and grown for 6 h with shaking. Luminescence of cultures was measured directly (EnVision; Perkin-Elmer), and output was relative light units normalized to the OD600. All experiments were performed in triplicate.
Quantitative real-time PCR (qRT-PCR) analysis.
Wild-type and ΔZ0021 mutant strains were grown in LB at 37°C to an OD600 of ∼0.6, and total RNA was isolated using a High Pure RNA isolation kit (Roche, Laval, Quebec, Canada). RNA was treated with DNase I (Ambion TURBO DNA-free kit, Applied Biosystems, Foster City, CA). First-strand cDNA was synthesized from 500 ng total RNA by using the Transcriptor First Strand cDNA synthesis kit (Roche). RT-PCR amplification was performed with 5 μl cDNA in a reaction mixture containing 1× LightCycler 480 CYBR Green I Master (Roche) and the forward and reverse primers at 500 nM (each) in a 20-μl volume. Relative quantification was performed using the LightCycler 480 Relative Quantification software (Roche) to compare the relative expression of selected gene targets to that of the icdA control gene. The primers designed to amplify regions of flhD, flgM, motA, fliC, and icdA are listed in Table 1. The amplification efficiencies of all primer sets were validated by standard curves. Assays were performed in triplicate with the LightCycler 480 II (Roche) instrument.
Construction of pFLAG-Z0021 and pFLAG-Z0021-flhDC.
Z0021 and flhDC were amplified from E. coli O157:H7 strain EDL933 chromosomal DNA with primers Z0021-F and Z0021-R and primers flhDC-F and flhDC-R, respectively. Z0021 was cloned into the HindIII/KpnI sites of pFLAG-CTC to create pFLAG-Z0021, while flhDC was cloned into pBAD33 (11) at the KpnI/HindIII sites. A fragment containing araC-PBAD-flhDC was amplified from pBAD33-flhDC with primers araC-flhDC-F and araC-flhDC-R and cloned into the BglII/SalI sites of pFLAG-Z0021. The resulting plasmid, pFLAG-Z0021-flhDC, had an IPTG-inducible copy of Z0021 and a tightly regulated copy of flhDC under arabinose control.
Sequencing of Z0021 from an STEC strain collection.
The Z0021 gene was amplified from purified chromosomal DNA by PCR using forward primer 5′-AAG CGG ACG CTA TTA CAA TTA G-3′ and reverse primer 5′-GTC CCG ATG GTT CGC CAT TAA C-3′ to generate a 721-bp product. DNA was purified through Sephadex and sequenced using a BigDye Terminator sequencing kit (Applied Biosystems, Life Technologies, Carlsbad, CA) and an ABI 3730XL automated sequencer (Applied Biosystems). Sequencing of amplified fragments was performed in both directions and in duplicate. A consensus sequence was generated from four sequence reads per strain using Discovery Studio Gene software (Accelrys Software Inc., San Diego, CA).
RESULTS
Deletion of OI-1 increases motility and flagellin production in E. coli O157:H7.
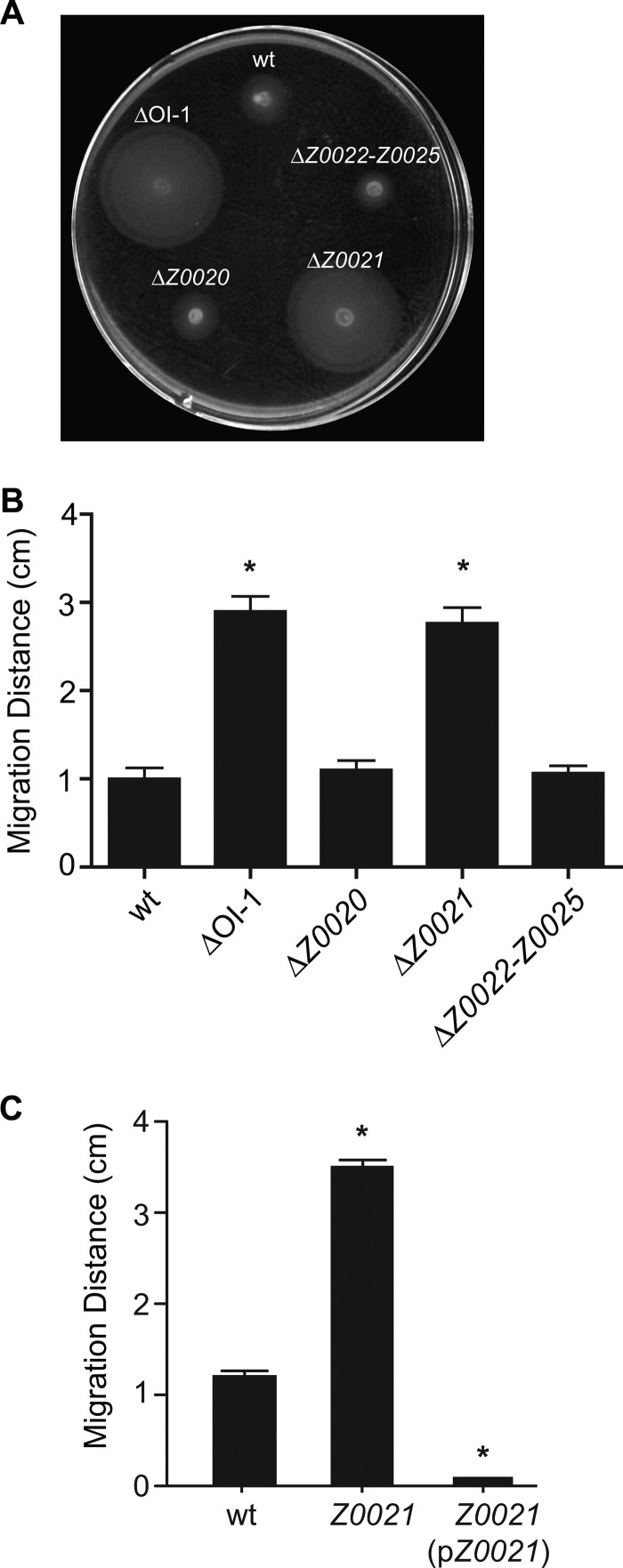
Given the existence of flagellar regulatory proteins encoded within fimbrial gene clusters in other bacteria (19, 25, 31), we sought to determine whether the putative fimbrial genomic island OI-1 encodes any repressors of motility. We created a ΔOI-1 mutant strain in which the genes Z0020 to Z0025 were replaced with a kanamycin resistance cassette and examined the swimming motility of the resulting mutant. In a standard soft-agar motility assay, the ΔOI-1 mutant strain was highly motile, in contrast to the wild-type EDL933 strain (Fig. 2A and B). To elucidate which gene(s) was responsible for this phenotype, a series of single- and multiple-deletion mutants was constructed and assayed for motility. The ΔZ0021 mutant strain showed enhanced swimming motility similar to that produced by the complete deletion of OI-1, while the motility of the ΔZ0020 and ΔZ0022-Z0025 mutants was comparable to that of wild-type EDL933 (Fig. 2A and B). All of the strains exhibited growth kinetics similar to those of the wild-type parental strain (data not shown). The enhanced-motility phenotype of the ΔZ0021 mutant strain was eliminated upon complementation with Z0021 on a plasmid (Fig. 2C). These data indicate that deletion of Z0021 is responsible for the increased-swimming phenotype of the ΔOI-1 mutant strain.
Fig 2.
Deletion of OI-1 increases swimming motility. (A) Wild-type (wt) EDL933 and ΔOI-1, ΔZ0020, ΔZ0021, and ΔZ0022-Z0025 mutant strains were assessed for swimming motility in a standard soft-agar motility assay. (B) Migration distance in motility agar plates was quantified after 6 h at 37°C. (C) Complementation of the Z0021 deletion restores the nonmotile phenotype. All data are means and standard deviation from three independent experiments. *, P < 0.001 (compared to the wild type).
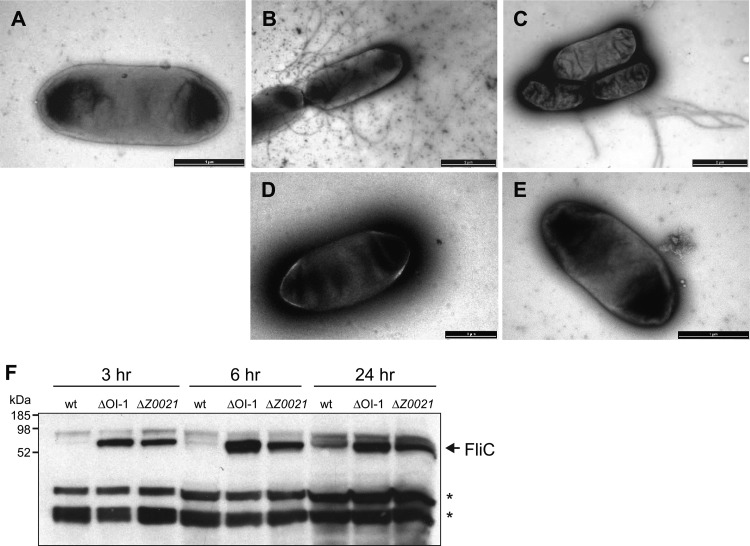
We then investigated whether the enhanced motility of the ΔOI-1 and ΔZ0021 mutants was due to an increase in flagellar motility function or to an increase in flagellar biosynthesis. Bacteria were harvested from motility plates and examined by TEM to visualize surface flagella. While the majority of the wild-type EDL933 bacteria were either nonflagellated or possessed a limited number of surface flagella, the ΔOI-1 and ΔZ0021 mutants displayed a high number of flagella on their surface (Fig. 3A to C), suggesting that loss of Z0021 derepresses the biosynthesis of flagella under these conditions. The complemented ΔOI-1 and ΔZ0021 mutants possessed a limited number of or no surface flagella, similar to wild-type EDL933 bacteria (Fig. 3D and E). To further confirm that the enhanced motility of the mutant strains was due to an increase in flagellar biosynthesis, the amount of flagellin produced by the Z0021 and OI-1 deletion mutants was compared to that of the wild type at 3, 6, and 24 h of growth. As shown in Fig. 3F, the ΔOI-1 and ΔZ0021 mutants showed higher levels of flagellin production than the wild type at all of the time points examined. Taken together, these data are consistent with OI-1-encoded Z0021 as a repressor of motility in E. coli O157:H7 strain EDL933.
Fig 3.
Deletion of Z0021 increases flagellum production in E. coli O157:H7 strain EDL933. (A to E) Transmission electron micrographs of wild-type E. coli O157:H7 strain EDL933 (A; scale bar, 1 μm), a ΔOI-1 mutant strain (B; scale bar, 2 μm), a ΔZ0021 mutant strain (C; scale bar, 2 μm), a ΔOI-1(pZ0021) mutant strain (D; scale bar, 1 μm), and a ΔZ0021(pZ0021) mutant strain (E; scale bar, 1 μm). Cells were harvested from motility agar after 6 h at 37°C, stained with 0.1% uranyl acetate, and viewed by TEM. (F) Western blot detection of flagellin (FliC) in the wild-type (wt) and ΔOI-1 and ΔZ0021 mutant strains. Asterisks indicate nonspecific cross-reacting bands used as a loading control.
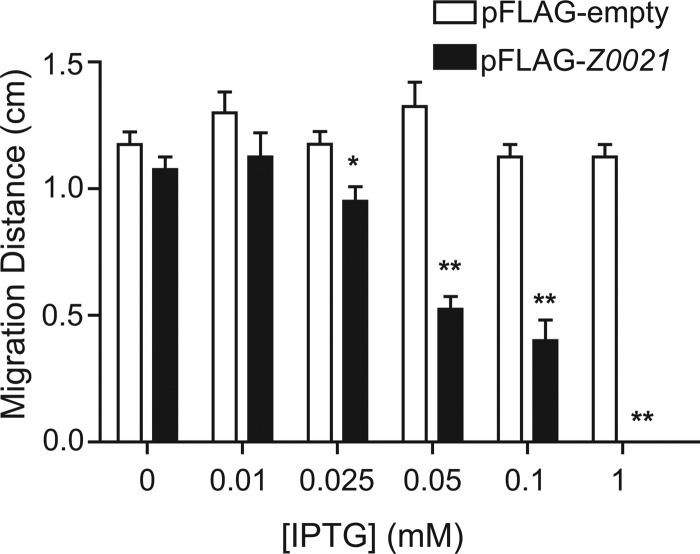
Overexpression of Z0021 inhibits motility in E. coli O157:H7.
Having identified Z0021 as a potential negative regulator of flagellum-based motility, we investigated whether elevated expression of Z0021 would repress the motility of E. coli O157:H7 strain EDL933. Z0021 was cloned under the control of the tac promoter into pFLAG-CTC, and the motility of wild-type EDL933 carrying pFLAG-Z0021 was assessed at various concentrations of IPTG. Increasing inducer concentrations from 0.05 to 1 mM led to a marked decrease in the swimming motility of E. coli O157:H7 strain EDL933, which was completely abolished at 1 mM IPTG (Fig. 4). Western blot analysis confirmed that these motility patterns correlated with elevated levels of Z0021 (data not shown).
Fig 4.
Overexpression of Z0021 represses swimming motility in E. coli O157:H7 strain EDL933. The motility of E. coli O157:H7 harboring an empty pFLAG vector or a pFLAG-Z0021 vector was assessed at various concentrations of IPTG. The migration distances on motility plates were quantified, and the data represent means and standard deviations. *, P < 0.01; **, P < 0.001 (compared to the empty-vector control).
Deletion of Z0021 increases the transcription of class II and III promoters.
The enhanced swimming motility of the ΔZ0021 mutant was associated with an increase in flagellum production. To examine where in the flagellar activation cascade this point of regulation occurs, we cloned promoter regions corresponding to all three of the promoter classes as transcriptional fusions to luxCDABE and measured the luciferase activities in wild-type EDL933 and ΔZ0021 mutant cells. While the flhDC promoter activity of the ΔZ0021 mutant strain was comparable to that of the wild-type strain, there was a significant increase in the activity of the transcriptional reporters for all of the class II and III promoters tested in the ΔZ0021 mutant strain compared to that in the wild-type strain (Fig. 5A). These data suggest that Z0021 acts downstream of the transcription of flhDC and prior to the activation of the middle genes. To elucidate whether the increase in class II/III promoter expression was due to increased FlhD and FlhC production, we first used qRT-PCR to measure flhD, flgM, motA, and fliC transcript levels in the wild-type and ΔZ0021 mutant strains. Deletion of Z0021 led to a substantial increase in all middle and late flagellar gene transcripts but did not alter the level of the flhD transcript (Fig. 5B). We then replaced the chromosomal copy of flhC in the flhDC operon with a C-terminally HA-tagged variant and determined the levels of FlhC-HA in the wild-type and ΔZ0021 mutant backgrounds by Western blot analysis. As expected from the reporter data, the levels of FlhC in the ΔZ0021 mutant were comparable to the levels in the wild type (Fig. 5C). These results suggest that Z0021 regulates flagellar synthesis after the translation of the FlhD and FlhC subunits.
Fig 5.
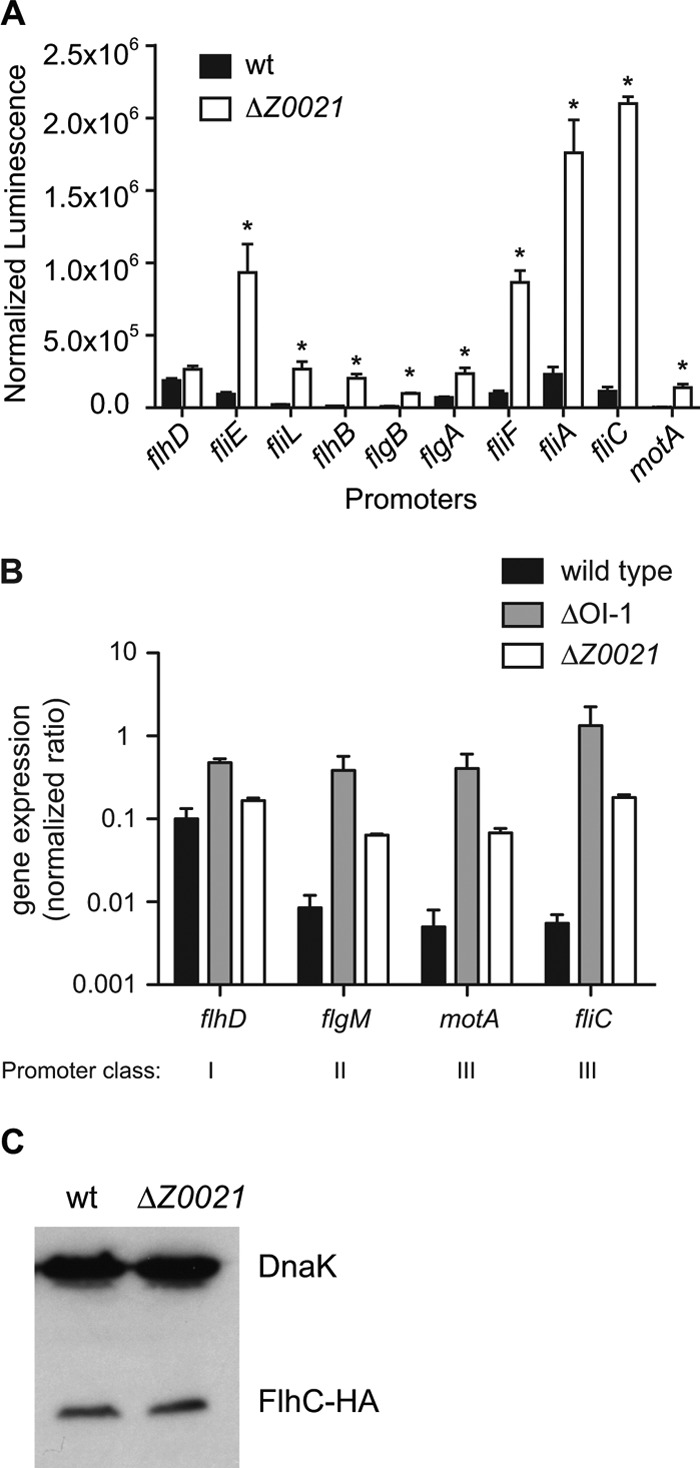
Deletion of Z0021 disrupts flagellar biosynthesis at the level of class II/III activation. (A) Transcriptional reporter activity for class I, II, and III promoters was determined in the wild-type (wt) EDL933 and ΔZ0021 mutant strains. Luminescence was measured after 6 h of growth at 37°C and normalized to the OD600. Data represent means and standard deviations. *, P < 0.001 (compared to the wild-type control). (B) qRT-PCR analysis of flhD, flgM, motA, and fliC in the wild-type and ΔZ0021 mutant strains. All assays were performed independently with three technical replicates, and the levels of all transcripts were normalized to that of icdA mRNA. The data represent the means and standard deviations of two independent experiments. (C) Wild-type EDL933 and ΔZ0021 mutant whole-cell lysates were collected at an OD600 of ∼0.5, and the levels of FlhC-HA were determined by Western blotting. DnaK served as a loading control.
FlhDC is a high-copy suppressor of the nonmotile phenotype associated with the overexpression of Z0021.
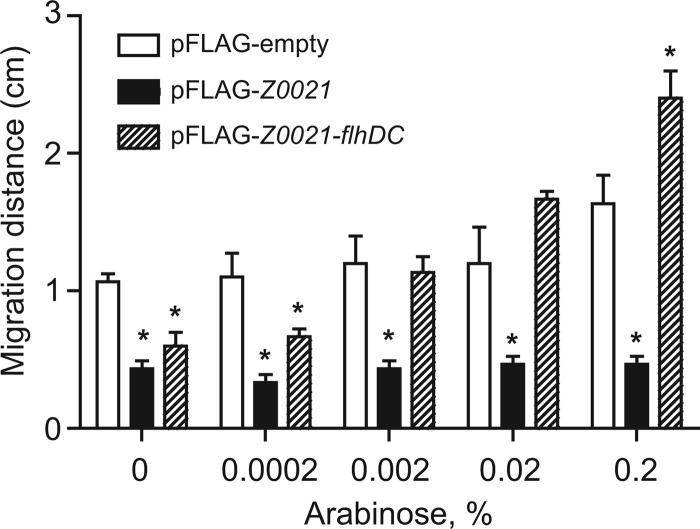
Since Z0021 exerts its regulatory effect on class II flagellar promoters without altering the levels of the master regulator, we considered that Z0021 was inhibiting the action of a functional FlhD4C2 complex. If this was the case, overexpression of flhDC should suppress the nonmotile phenotype associated with elevated levels of Z0021. To investigate this, we tested whether increasing expression of flhDC would restore motility to E. coli O157:H7 expressing Z0021. E. coli O157:H7 carrying pFLAG-Z0021-flhDC has a tightly regulated copy of flhDC under arabinose control and an IPTG-inducible copy of Z0021. The motility of this strain was assayed at various concentrations of arabinose and at a fixed concentration of 0.1 mM IPTG—a concentration which we showed impairs the motility of E. coli O157:H7 strain EDL933 harboring pFLAG-Z0021 (Fig. 4). In our soft-agar motility assay, an arabinose concentration of 0.002% restored the motility of E. coli O157:H7 strain EDL933 harboring pFLAG-Z0021-flhDC to that of the empty-vector control, while arabinose concentrations of ≥0.02% increased the motility of E. coli O157:H7 strain EDL933 harboring pFLAG-Z0021-flhDC beyond that of the empty-vector control (Fig. 6). Taken together, these data show that the motility defects caused by Z0021 can be suppressed by increasing flhDC expression, implying a genetic link between Z0021 and the FlhD4C2 regulatory complex.
Fig 6.
Increased expression of flhDC suppresses the nonmotile phenotype associated with Z0021. The motility of E. coli O157:H7 strain EDL933 harboring an empty pFLAG vector, pFLAG-Z0021-araC-PBAD, or pFLAG-Z0021-flhDC in 0.25% LB agar containing ampicillin and 0.1 mM IPTG was assessed. Arabinose was varied from 0 to 0.2%, and migration distance was measured after 6 h of growth at 37°C. Data represent means and standard deviations. *, P < 0.01 (compared to the pFLAG-empty vector).
Prevalence of Z0021 among STEC strains.
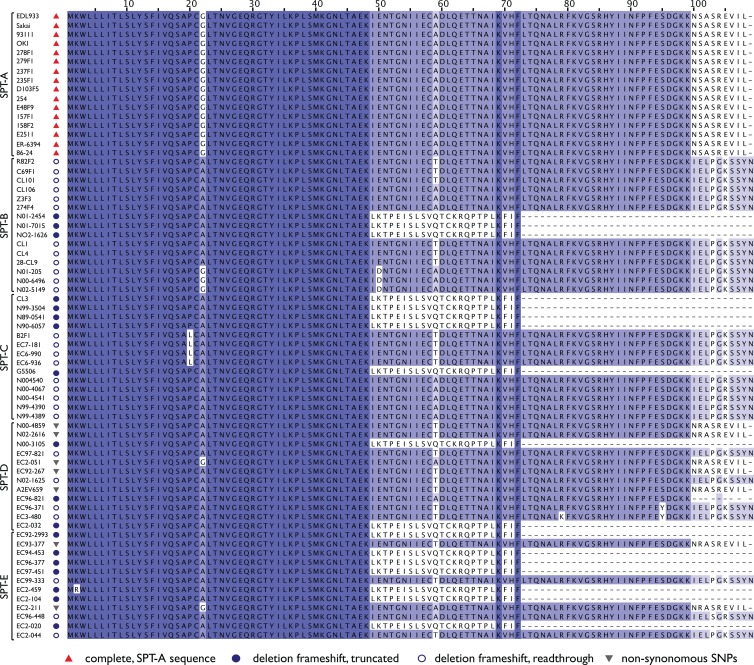
OI-1 was previously identified in seropathotype A strains of STEC. To examine the distribution of Z0021 among the other four seropathotypes of non-O157:H7 STEC, we screened the 69 strains from the original seropathotype collection of STEC strains (15) by PCR and sequenced the positive Z0021 gene products. Only the O157:H7 strains in the seropathotype A group had the same Z0021 sequence as the EDL933 reference strain with 100% conservation (Fig. 7). Although strains of the other seropathotypes harbored the Z0021 gene, the gene contained either a frameshift mutation at nucleotide position 148 resulting in a downstream premature stop codon at position 73 or a deletion mutation at position 299 resulting in a frameshift starting at amino acid position 100 (Fig. 7). Interestingly, these two gene variants appear to have evolved a number of times independently in different serotypes that assort to different seropathotype classes. Only seven other strains in the collection had a Z0021 sequence similar to that of the EDL933 reference strain; however, in all of the cases, these sequences had at least one nonsynonymous substitution. Thus, only the O157:H7 strains in our collection had a Z0021 sequence that was identical to that of the EHEC reference strain. This Z0021 sequence from all seropathotype A strains was distinguished from those of all other serotypes by unique frameshifts or nonsynonymous substitutions.
Fig 7.
Prevalence of Z0021 in STEC strains. Z0021 genes were amplified from 69 STEC strains, including non-O157 STEC strains of all seropathotypes. Nucleotide sequences were translated in silico, and the amino acid sequences were aligned. Strain names are given to the left of the sequences and grouped into seropathotypes (SPTs). The alignment is shown as a heat map according to the conservation at each position, with dark blue representing higher conservation and light blue representing lower conservation among all of the sequences. SNPs, single-nucleotide polymorphisms.
DISCUSSION
Many regulators of the flagellar transcriptional hierarchy have been identified, and some of these are encoded within fimbrial gene clusters, presumably to control the opposing processes of adherence and motility. In the E. coli O157:H7 genome, the majority of the putative fimbrial gene clusters are uncharacterized and no fimbrial-operon-encoded regulators of motility have been identified to date. In this paper, we have reported on a novel repressor of E. coli O157:H7 motility encoded in the putative type 1-like fimbrial gene cluster OI-1 and have shown that Z0021 regulates flagellar synthesis through its influence on class II promoters via FlhD4C2.
E. coli type 1 fimbriae have an established role in mediating adherence to host cells, and the closely related type 1-like P fimbriae have been shown to be important for the pathogenesis of uropathogenic E. coli (6, 23, 29). Since E. coli O157:H7 strains are unable to produce type 1 fimbriae because of a deletion in the fim regulatory region (28), novel type 1 or type 1-like fimbriae may play a role in the process of intestinal colonization. OI-1 is one of four fimbria-encoding OIs that are found in seropathotype A strains of O157 STEC and may be a key determinant of the unique pathogenesis of such strains (30). OI-1 may encode type 1-like fimbriae on the basis of its similarity to the E. coli type 1 fimbrial locus. In comparison to the LPF clusters (OI-141 and OI-154) which contain mutations in their export machinery (20), Z0022 and Z0023 of OI-1 encode potentially functional usher and chaperone proteins, respectively. OI-1 could therefore produce fimbriae without complementation from another fimbrial gene cluster. The putative adhesin gene in OI-1, Z0020, is not similar to characterized adhesin genes, suggesting that it may encode a novel adhesin or that other components of the fimbriae produced by OI-1 confer binding specificity (20). Future studies are required to structurally and functionally characterize the fimbriae produced by OI-1 and to determine the importance of this OI in O157 STEC pathogenesis.
In an analysis of the expression of 16 putative fimbrial gene clusters in E. coli O157:H7, OI-1 showed no or minimal expression under the in vitro conditions tested (20). While the expression of OI-1 in vivo has not been examined, our work has shown that the expression of OI-1, and in particular Z0021, is important for the repression of flagellar synthesis in E. coli O157:H7. The expression of flagella has been shown to vary during the course of an infection; flagella are produced extensively by attaching bacterial cells in the early stages of infection but are not present in bacterial microcolonies that form at later time points (22). We therefore speculate that the gene products of OI-1 may be of significance following the early stages of infection at a time when flagellar expression is diminished.
Z0021 was found to exert its regulatory effects on class II flagellar promoters downstream of flhDC transcription and translation. It is therefore conceivable that Z0021 could regulate flagellar synthesis by (i) targeting FlhD and/or FlhC for degradation or (ii) preventing a functional FlhD4C2 complex from binding to class II promoters. The ClpXP protease has been shown to degrade FlhD4C2 in both S. enterica serovar Typhimurium and E. coli O157:H7 (18, 34). Given that the transcript and protein levels of FlhD and FlhC were unchanged in the wild-type and ΔZ0021 mutant strains, this suggests that Z0021 regulates flagellar synthesis through a mechanism independent of ClpXP. The mechanism of action of Z0021 may be more similar to that of S. enterica serovar Typhimurium YdiV, which, in addition to repressing flagellar synthesis via a ClpXP-dependent pathway (33), also acts via a ClpXP-independent pathway by interacting with the FlhD4C2 complex and preventing its binding to class II promoters (38). However, we have been unable to identify motifs shared by Z0021 and YdiV and the two proteins lack sequence conservation at the amino acid level. The latter proposed mechanism for Z0021-mediated repression is supported by the ability of flhDC to suppress the nonmotile phenotype associated with the expression of Z0021, indicating that the effects of Z0021 are reversible and dependent on the levels of the FlhD and/or FlhC subunits.
Comparative sequence analysis of the Z0021 gene in STEC strains from our collection revealed a high degree of nucleotide sequence conservation among seropathotype A strains, including those which are nonmotile. FlhD4C2 not only acts as the master regulator of the flagellar cascade but has also been shown to bind to and activate nonflagellar genes (32). Z0021 may therefore be conserved among these nonmotile strains to exert a regulatory effect on nonflagellar genes via an interaction with FlhD4C2 or the FlhD and FlhC subunits. A highly conserved Z0021 gene is also present in some seropathotype B to E strains. Although OI-1 is found predominantly in seropathotype A strains, there is evidence of OI-1 in other seropathotypes, which may account for the wider distribution of Z0021 in non-O157 STEC strains (30).
In summary, we identified a novel repressor of motility in E. coli O157:H7 and showed that Z0021 negatively regulates flagellar synthesis at a stage prior to the transcription of the middle and late genes. The characterization of Z0021 is the first report of a fimbrial-operon-encoded gene product that represses flagellar synthesis in E. coli O157:H7, and consistent with this role, we propose that Z0021 be renamed fmrA (fimbrial-operon-encoded motility regulator A). Most importantly, our work has provided insight into the function of an OI that may be associated with the enhanced pathogenesis of seropathotype A strains of STEC.
ACKNOWLEDGMENTS
This work was funded by an operating grant to B.K.C. from the Canadian Institutes of Health Research (MOP-82704), an infrastructure grant from the Canada Foundation for Innovation, and the Canada Research Chairs Program from the Government of Canada. S.E.A. is the recipient of a Canada Graduate Scholarship from the CIHR. U.S. was supported by an NSERC Visiting Fellowship. B.K.C. is the Canada Research Chair in Infectious Disease Pathogenesis.
Footnotes
Published ahead of print 27 July 2012
REFERENCES
- 1. Akerley BJ, Cotter PA, Miller JF. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611–620 [DOI] [PubMed] [Google Scholar]
- 2. Bäumler AJ, Heffron F. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beeston AL, Surette MG. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Connell I, et al. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 93:9827–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coombes BK, Gilmour MW, Goodman CD. 2011. The evolution of virulence in non-O157 Shiga toxin-producing Escherichia coli. Front. Microbiol. 2:90 doi:10.3389/fmicb.2011.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doughty S, et al. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60–98 [DOI] [PubMed] [Google Scholar]
- 11. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayashi T, et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11–22 [DOI] [PubMed] [Google Scholar]
- 13. Karch H, Bielaszewska M, Bitzan M, Schmidt H. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229–243 [DOI] [PubMed] [Google Scholar]
- 14. Karmali MA. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karmali MA, et al. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karmali MA, et al. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775–782 [DOI] [PubMed] [Google Scholar]
- 17. Karmali MA, Petric M, Lim C, Fleming PC, Steele BT. 1983. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet ii:1299–1300 [DOI] [PubMed] [Google Scholar]
- 18. Kitagawa R, Takaya A, Yamamoto T. 2011. Dual regulatory pathways of flagellar gene expression by ClpXP protease in enterohaemorrhagic Escherichia coli. Microbiology 157:3094–3103 [DOI] [PubMed] [Google Scholar]
- 19. Li X, Rasko DA, Lockatell CV, Johnson DE, Mobley HL. 2001. Repression of bacterial motility by a novel fimbrial gene product. EMBO J. 20:4854–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Low AS, et al. 2006. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 8:1033–1047 [DOI] [PubMed] [Google Scholar]
- 21. Macnab RM. 1996. Flagella and motility, p 123–145 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 22. Mahajan A, et al. 2009. An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157:H7 with bovine intestinal epithelium. Cell. Microbiol. 11:121–137 [DOI] [PubMed] [Google Scholar]
- 23. Mulvey MA, et al. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494–1497 [DOI] [PubMed] [Google Scholar]
- 24. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pearson MM, Mobley HL. 2008. Repression of motility during fimbrial expression: identification of 14 mrpJ gene paralogues in Proteus mirabilis. Mol. Microbiol. 69:548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perna NT, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 27. Reiss DJ, Mobley HL. 2011. Determination of target sequence bound by PapX, repressor of bacterial motility, in flhD promoter using systematic evolution of ligands by exponential enrichment (SELEX) and high throughput sequencing. J. Biol. Chem. 286:44726–44738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roe AJ, Currie C, Smith DG, Gally DL. 2001. Analysis of type 1 fimbriae expression in verotoxigenic Escherichia coli: a comparison between serotypes O157 and O26. Microbiology 147:145–152 [DOI] [PubMed] [Google Scholar]
- 29. Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 3:65–72 [DOI] [PubMed] [Google Scholar]
- 30. Shen S, Mascarenhas M, Morgan R, Rahn K, Karmali MA. 2005. Identification of four fimbria-encoding genomic islands that are highly specific for verocytotoxin-producing Escherichia coli serotype O157 strains. J. Clin. Microbiol. 43:3840–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simms AN, Mobley HL. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect. Immun. 76:4833–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stafford GP, Ogi T, Hughes C. 2005. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology 151:1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takaya A, et al. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol. Microbiol. 83:1268–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tomoyasu T, et al. 2002. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Torres AG, et al. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Torres AG, Kanack KJ, Tutt CB, Popov V, Kaper JB. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238:333–344 [DOI] [PubMed] [Google Scholar]
- 37. Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 98:15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wada T, et al. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193:1600–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]