Abstract
Shigella flexneri is a facultative intracellular pathogen that relies on a type III secretion system and its associated effector proteins to cause bacillary dysentery in humans. The genes that encode this virulence system are located on a 230-kbp plasmid and are transcribed in response to thermal, osmotic, and pH signals that are characteristic of the human lower gut. The virulence genes are organized within a regulatory cascade, and the nucleoid-associated protein H-NS represses each of the key promoters. Transcription derepression depends first on the VirF AraC-like transcription factor, a protein that antagonizes H-NS-mediated repression at the intermediate regulatory gene virB. The VirB protein in turn remodels the H-NS–DNA nucleoprotein complexes at the promoters of the genes encoding the type III secretion system and effector proteins, causing these genes to become derepressed. In this study, we show that the VirB protein also positively regulates the expression of its own gene (virB) via a cis-acting regulatory sequence. In addition, VirB positively regulates the gene coding for the VirF protein. This study reveals two hitherto uncharacterized feedback regulatory loops in the S. flexneri virulence cascade that provide a mechanism for the enhanced expression of the principal virulence regulatory genes.
INTRODUCTION
Shigella flexneri is a Gram-negative facultative intracellular pathogen of humans and primates and is the causative agent of bacillary dysentery (20, 22, 48, 50, 51, 60). The invasion of host cells by S. flexneri requires the expression of virulence genes located on a 230-kb plasmid (Fig. 1) (12, 16, 61, 62). The products of these genes include the Ipa invasins, which mediate the invasion of the gut epithelium and macrophage apoptosis; the Mxi and Spa proteins, which form a type III secretion system (TTSS) (2–4, 9, 10, 49, 64, 75); and the IcsA, IcsB, and VirA proteins, which are responsible for the intercellular spreading of bacteria in the lower gut (7, 12, 29, 56, 63, 74). The virulence genes are expressed only under conditions approximating those found at the site of host cell invasion, i.e., at 37°C, at a pH of 7.4, and with moderate osmolarity (18, 38, 39, 45, 46, 52, 53). Modifications to local DNA topology in response to environmental stress play an important part in modulating S. flexneri virulence gene transcription (18, 42, 47, 72). The chromosomally encoded H-NS protein controls much of this expression through the repression of many of the virulence promoters under conditions that are inappropriate for invasion (6, 17, 34, 35, 39). Virulence gene activation occurs via a regulatory cascade mediated by the plasmid-encoded VirF and VirB proteins (1, 19, 20) (Fig. 1).
Fig 1.
Genetic map of the 230-kb large virulence plasmid of S. flexneri showing the positions of prominent regulatory and structural genes. Arrowheads represent individual genes, and the black disc at oriR is the origin of plasmid replication. The 31-kb entry region is enlarged to show the positions and orientations of the virulence genes. Hatched arrowheads show the genes encoding proteins of the type III secretion system machinery and its effector/translocator proteins and their chaperones. Gray arrowheads represent the genes encoding plasmid-partitioning proteins (parA and parB), and black arrowheads indicate regulatory genes (virF and mxiE). The virB gene is shown here as a gray-and-black arrowhead, as it encodes a ParB-like protein with gene regulatory functions. The virF and virB genes are labeled in large font to highlight their involvement in the regulatory cascade that overcomes the repressive effects of H-NS on virulence gene expression. The figure is not drawn to scale.
The transcriptional cascade is initiated with the activation of the virF gene, which encodes an AraC-like transcription activator (55). The H-NS protein binds to two sites at the virF promoter, one overlapping the transcription start site and the other centered upstream at position −250, separated by a region of DNA that is intrinsically curved (see Fig. 3) (26, 57). Repression involves the formation of a nucleoprotein complex in which H-NS binds cooperatively to these sites at virF (58). This nucleoprotein complex is temperature sensitive, forming at temperatures below 32°C but being disrupted by the changes in DNA topology that accompany a temperature upshift to 37°C (58). This disruption in local DNA architecture simultaneously reveals a binding site for the factor for inversion stimulation (FIS) nucleoid-associated protein within the promoter region of virF. The principal role of FIS at the virF promoter is to exercise direct positive transcriptional control at the permissive temperature of 37°C, once the H-NS-mediated repression of virF has been relieved (27). The integration host factor (IHF) architectural protein also facilitates the expression of virF: the transcription of virF is stimulated by IHF in both the logarithmic and the early stationary phases of growth (54). The regulation of virF gene expression also includes posttranscriptional controls that act to modulate the rate of mRNA translation (23, 24).
Fig 3.
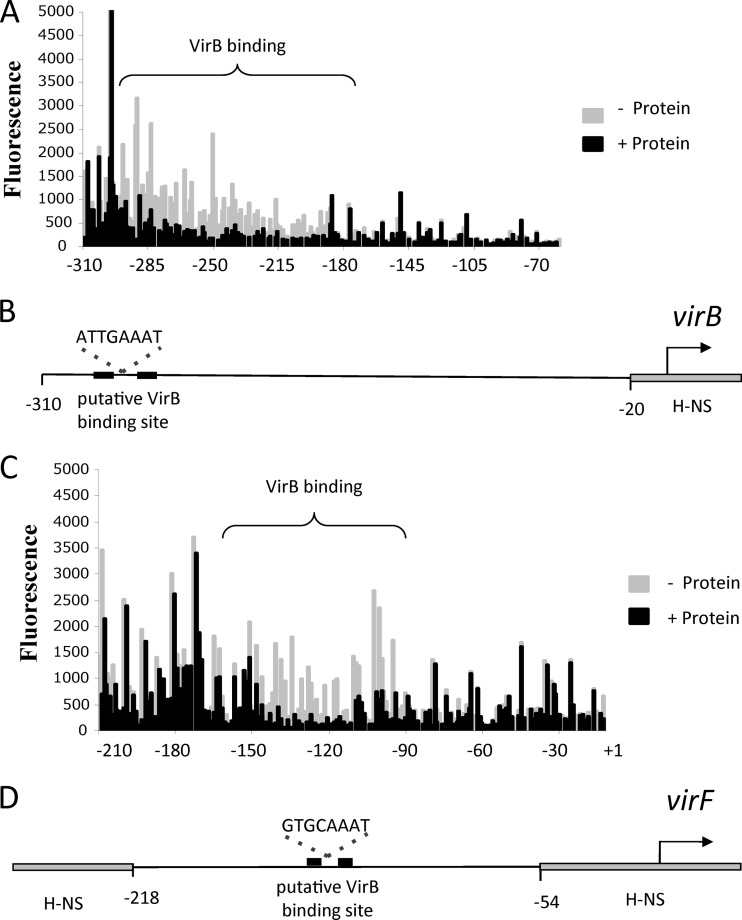
Autoregulation of virB and virF is dependent on cis-acting VirB binding sites. Summaries of the structures of various derivatives of the virB promoter region (A to D) and of the virF promoter region (E to G) are presented at the left; levels of expression of the virB-gfp (A to D) and the virF-gfp (E to G) transcriptional reporter fusions are given at the right. The data shown are relative to the level of fluorescence of a negative-control strain containing only the pZEP08 vector (Table 2). (A to D) The native virB promoter encompassing positions −310 to +30 (A), the virB promoter from positions −80 to +30 (B), the virB promoter with mutations made to the putative VirB site (C), and the virB promoter with the VirB binding site from the icsB promoter inserted at position −80 (D). (E to G) The native virF promoter encompassing positions −380 to +60 (E), the virF promoter from positions −280 to +30 (F), and the virF promoter with mutations made to the putative VirB site (G).
With the repression of virF overcome, the AraC-like protein VirF activates the transcription of the virB regulatory gene. VirF interacts directly with the virB promoter in a region spanning positions −17 to −105 (71). The VirF binding sites are located immediately upstream of the region bound by H-NS, which extends from positions −20 to +20 and includes the virB promoter. The inactivation of hns gene expression by mutation causes an increased expression level of the virB gene at 30°C, demonstrating that H-NS plays a role in controlling the thermal regulation of virB expression (6, 35). However, there is an absolute requirement for the presence of the VirF protein for the full activation of virB, indicating that the AraC-like VirF protein is not simply an antirepressor that displaces H-NS but has a positive role to play in virB promoter activation (53, 70–72). The current model predicts that VirF interacts physically with RNA polymerase to induce transcription and that this interaction is dependent on an increase in the local negative supercoiling of the DNA (55, 71, 72). The shift to an optimum DNA topology occurs in response to an increase in temperature from 30°C to 37°C and under conditions of high osmolarity (47, 52, 72). Thus, at 30°C, the presence of VirF alone is insufficient to activate virB, but at 37°C or under high-salt conditions, DNA topological changes at the promoter make interactions between VirF and RNA polymerase productive, leading to transcription activation. The binding of IHF to the region upstream of the VirF binding site further enhances the expression of virB (54). Like the virF gene, the virB gene is also the subject of posttranscriptional control, in this case via the iron-regulated small regulatory RNA RyhB (44).
The VirB protein acts as an antirepressor to alleviate the H-NS-mediated transcriptional silencing of the promoters that govern the expression of the operons on the 230-kbp plasmid that encode the type III secretion system and its effector proteins (Fig. 1) (6, 26, 27, 57, 58, 68, 73). VirB binds to a DNA sequence motif that is related to those parS sequences that are targeted by ParB-like proteins involved in plasmid partitioning (68, 73). Indeed, the VirB protein itself closely resembles a ParB-like protein in its amino acid sequence (1, 5, 41). If the virB gene played a plasmid-partitioning role in an earlier phase of the evolution of the plasmid, this is no longer the case (12, 65). The loss of this (hypothetical) plasmid-partitioning function may have made the VirB DNA binding protein available for the gene regulatory role that it performs in the modern plasmid. This role places it at a point that is intermediate between the primary regulator, VirF, and the promoters of the structural genes. The significance of having a two-stage VirF-VirB-dependent regulatory system for the S. flexneri virulence cascade is unclear but may reflect the evolution of a regulatory checkpoint within the cascade at virB (35, 73).
VirB regulates transcription via a relatively unsophisticated mechanism in which it uses the parS-like sequence as an initial binding and nucleation site and then polymerizes along the DNA to antagonize H-NS binding at adjacent A+T-rich DNA sequences; the associated remodeling of the nucleoprotein complex allows the silenced promoter to be derepressed (73). The VirB binding site on the 230-kbp plasmid with the best match to a complete parS-like sequence is located upstream of the icsB gene. Other functional VirB binding sites on the plasmid that have been characterized have the core elements of the parS sequence but lack the completeness of the icsB motif (14, 73).
The virB gene is separated from the icsB parS-like motif by the icsB-ipg-ipa-acp operon (Fig. 1). Since ParB-like proteins can autoregulate their own genes via adjacently located cis-acting parS sequences (8, 11, 21, 25, 28, 32, 37), it was hypothesized previously that virB may have been disconnected from the VirB binding site at icsB through the insertion of the icsB-ipg-ipa-acp operon during the evolution of the 230-kbp plasmid (35). Might this mean that VirB once had a role in controlling the expression of its own gene, and if so, has the modern plasmid evolved an alternative mechanism that restores the VirB-mediated control of virB transcription? This question seemed relevant given the many examples of bacterial regulatory proteins that govern the expression of the genes that encode them (33, 36, 59, 66, 69). Here we report the discoveries that VirB does indeed positively autoregulate virB transcription and that VirB also feeds back positively onto the transcription of the virF master regulatory gene.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
All of the bacterial strains used in the study were derivatives of Shigella flexneri serotype 2a strain 2457T or Escherichia coli K-12. They are listed in Table 1, together with genotypes and sources. The plasmids used in this study are listed in Table 2, together with genotypes, sources, and any details of plasmid construction. The sequences of all of the oligonucleotide primers used in the study are listed in Table 3.
Table 1.
Strains
Table 2.
Plasmids
| Plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| pZep08 | GFP reporter plasmid; Apr Cmr | 31 |
| pBAD33 | Vector with l-arabinose-inducible PBAD promoter; Apr | 6 |
| pBADvirB | virB gene expressed from l-arabinose-inducible PBAD promoter; Apr | 6 |
| pZep-virB-310 | Positions −310–+20 of PvirB cloned into pZep08 | This study |
| pZep-virB-310 SDM | pZep-virB-310 with putative VirB binding site mutated | This study |
| pZep-virB-150 | Positions −150–+20 of PvirB cloned into pZep08 | This study |
| pZep-virB-80 | Positions −80–+20 of PvirB cloned into pZep08 | This study |
| pZep-virB-80-icsB | Positions −240–−80 of PicsB cloned into pZep-virB-80 | This study |
| pZep-virF-380 | Positions −380–+64 of PvirF cloned into pZep08 | This study |
| pZep-virF-280 | Positions −280–+64 of PvirF cloned into pZep08 | This study |
| pZep-virF-280 SDM | pZep-virF-280 with putative virF binding site mutated | This study |
Table 3.
Primers
| Oligonucleotide | Sequence (5′–3′)a |
|---|---|
| icsB fw.TAG | TGTACCTCGTGAGCATATGTAGT |
| icsB rev.TAG | GGGGCATTGATTGCAGTTTT |
| virB.rev.+190.XbaI | TTTTCTAGATTTTGGTTGACGAAGGTTAAATC |
| virB.fw.-80.NotI | TTTGCGGCCGCTTGCATCAATCCAGCTATTAAA |
| virB.fw.-310.NotI | TTTGCGGCCGCGCCGATTCTCTTTCTCTGATTC |
| virB.RT.rev | TTTTGGTTGACGAAGGTTAAATC |
| virB.RT.fw.+2 | ACTGCATTTAACTTTGTCAATA |
| virB.fw.SDM | GATTCTCTTTCTCTGgctgggcGCTGGATATGATTTAG |
| virB.rev.SDM | CTAATCATATCCAGCgcccagcAGAGAAAGAGAATC |
| virF.rev.+160.XbaI | TTTTCTAGAAATGACGGTTAGCTCAGGCA |
| virF.fw.-280.NotI | TTTGCGGCCGCTGGAGCCTCCAGTCTGAAG |
| virF.fw.-380.NotI | TTTGCGGCCGCCTTCACGATCGCAATATGGA |
| virF.RT.fw | GGTTCGCTTGCATAACTATA |
| virF.RT.rev | ATAAAAGCAATTTGCCCTTC |
| virF.fw.SDM | TATGGTTATAGTCCCgggCAGTGCgggcACTTAGCTTGTTGCA |
| virF.rev.SDM | TGAACAAGCTACGTgcccGCACTGcccGGGACTATAACCATA |
| pZec-6FAM.F | FAM-ACAAATCCGCCCTCTAGCAGCCCG |
| pZec-confirm.R | ACGGGAAAAGCATTGAACAC |
Lowercase letters in sequences represent bases introduced during site-directed mutagenesis. Underlining indicates the locations of recognition sequences for restriction enzymes.
DNA manipulations.
Plasmid DNA was routinely purified from 3-ml cultures of E. coli or S. flexneri strains by using the HiYield plasmid minikit according to the manufacturer's instructions (RBC Bioscience). Bacterial chromosomal DNA was isolated by using the Puregene DNA purification kit (Gentra). Linear DNA fragments, either PCR products or restriction-endonuclease-cleaved DNA, were purified for cloning or for the preparation of labeled probes by using the HiYield gel/PCR DNA fragment extraction kit (RBC Bioscience). Restriction endonuclease cleavage of circular plasmids or linear DNA involved incubation with 1 to 2 U of the restriction enzyme in a 20-μl volume containing the appropriate reaction buffer. Reaction mixtures were incubated at the recommended temperature for 1 to 2 h. Following digestion, the reaction mixtures were usually heated to >60°C for 20 min to denature the enzyme. Bacteriophage T4 DNA ligase (Roche) was used when DNA fragments were cloned into the plasmid vectors. Site-directed mutagenesis (SDM) was performed by using the QuikChange II SDM kit according to the manufacturer's recommendations (Stratagene).
Growth conditions.
All experiments were performed, unless otherwise stated, at 30°C, a temperature that is nonpermissive for virB gene expression. The ectopic expression of the virB gene was achieved by using a pBAD33-derived plasmid in which the virB open reading frame was transcribed from the PBAD promoter. This promoter was induced by using 0.2% l-arabinose and repressed by using 0.2% d-glucose. Cultures were grown overnight with antibiotic selection and aeration in LB broth and then diluted 1:100 into 25 ml of fresh prewarmed broth containing a selective antibiotic and either 0.2% d-glucose or 0.2% l-arabinose in a 250-ml flask and grown for a further 18 h, by which time they had reached the stationary phase. Samples were then removed for analysis.
Immunodetection of the VirB protein.
Total protein extracts were separated through a 12% SDS-PAGE gel. The separated proteins were transferred by electroblotting onto a nitrocellulose membrane for 1 h at 80 V using the Bio-Rad MiniProtean II system. Nitrocellulose membranes were stained with Ponceau red (0.2% Ponceau dye, 3% trichloroacetic acid) to check the efficiency of transfer before being blocked overnight with 5% dried skimmed milk in phosphate-buffered saline (PBS). The detection of VirB was performed with PBS containing 1% dried skimmed milk with primary polyclonal anti-VirB antiserum (1:500) and a secondary goat anti-rabbit horseradish peroxidase-conjugated antiserum (1:10,000). Membranes were developed by using the chemiluminescent Pierce West Pico Super Signal kit. The vector-only control contained the pBAD33 plasmid without the virB insert. Equal protein loading onto gels used for immunoblotting was ensured by monitoring the presence of the DnaK protein using rabbit monoclonal anti-DnaK antiserum at a dilution of 1:200,000 (Enzo Life Sciences).
Real-time quantitative PCR.
Total RNA was isolated from cultures by using the SV Total RNA isolation system (Promega), and purity and quality were assessed by electrophoresis in 1% agarose (1× Tris-acetate-EDTA [TAE]). Residual DNA contamination was removed by treatment with DNase I (Ambion DNA-free kit). Copy DNA templates were synthesized by random priming with 1 μg of RNA in a 20-μl reaction mixture using the GoScript reverse transcription system (Promega). Primers for quantitative real-time PCR (qRT-PCR) are listed in Table 3. PCRs were carried out in duplicate with the primer set on an ABI 7500 sequence detection system (Applied Biosystems), using FastStart SYBR Green (Rox; Roche). Standard curves were generated for each primer set by using five serial 10-fold dilutions of chromosomal DNA and were included in every run.
Quantification of green fluorescent protein (GFP) reporter constructs.
The bacterial culture to be assayed was harvested and immediately fixed in 4% formaldehyde in PBS and then stored at 4°C in the dark. Prior to analysis, samples were diluted to a concentration of approximately 106 bacteria per ml and then analyzed with an Epics-XL flow cytometer (Beckman Coulter). Approximately 10,000 bacteria per sample were assayed, and the results were expressed as mean channel fluorescence after analysis using EXPO-XL software. Each sample was measured in duplicate, and the mean values were determined from the results of at least three independent experiments.
DNase I footprinting using FAM-labeled primers.
The use of fluorescently labeled primers eliminates the need for radioactively labeled nucleotides, as well as slab gel electrophoresis, and takes advantage of commonly available automated fluorescent capillary electrophoresis instruments. With fluorescently labeled primers and dideoxynucleotide DNA sequencing, the terminal base of each digested fragment can be accurately identified with a capillary-based instrument (13). PCR was performed with a 6-carboxyfluorescein (FAM)-labeled primer to amplify the target promoter region of interest. This PCR product was then incubated with purified VirB, or bovine serum albumin (BSA) as a control, and then partially digested with DNase I. Briefly, bait DNA was prepared by using primers pZec.6FAM.F and pZec.comfirm.R (Table 3). The reactions were then conducted with a 15-μl reaction mixture volume consisting of 1× DNase I buffer (Roche) (40 mM Tris-HCl, 10 mM NaCl, 6 mM MgCl2 [pH 7.9]), 0.001 mM dithiothreitol, 100 ng/μl BSA, 50 nM bait DNA, and 50 μM VirB-His. This mixture was allowed to equilibrate at 37°C for 15 min, and 1 μl (0.0015 units) of DNase I was then added, mixed gently, and then incubated at 37°C for 10 min. The digestion reaction was stopped by the addition of 2 μl EDTA (100 mM) to the mixture, followed by vigorous vortexing and heat denaturation at 95°C for 10 min. Digestion products were desalted by using MicroSpin G-50 columns (GE Healthcare) and then analyzed by use of an ABI 3130 genetic analyzer along with GeneScan 500-Liz size standards (Applied Biosystems).
RESULTS
Ectopic expression of the VirB protein at 30°C can alleviate the repressive effects of H-NS at the virB and virF promoters.
The thermoregulation of the virB and virF promoters involves transcriptional repression by H-NS at nonpermissive temperatures, i.e., 30°C (1). VirB-dependent promoters of virulence structural genes can be derepressed under these conditions by the ectopic overexpression of the VirB protein in trans (6). The effect of VirB on its own promoter and on that of virF was tested by using a similar approach. The PBAD promoter, which is induced by l-arabinose and repressed by d-glucose, was used to manipulate the levels of the VirB protein in the cell, and VirB protein levels were monitored by Western blotting (Fig. 2A and B) The levels of each transcript specified by the virB and virF promoters were then analyzed by qRT-PCR, using oligonucleotide primers specific for each promoter. In the case of the virB transcript, a forward primer (virB.RT.fw.+2) (Table 3) was designed to anneal immediately after the native virB promoter but preceding the start codon of the virB open reading frame. This is a region that was not included when expression plasmid pBADvirB was constructed (6). When used in combination with the reverse primer virB.RT.rev (Table 3), this facilitated discrimination between the native virB transcript and that expressed by the PBAD promoter in plasmid pBADvirB.
Fig 2.
Artificial expression of the VirB protein at 30°C can alleviate the repressive effects of H-NS at the virB and virF promoters. (A) Western blot detection of the VirB protein in wild-type strain BS184 at 30°C and 37°C and in BS184 harboring l-arabinose-inducible plasmid pBADvirB at 30°C in the presence of d-glucose (the PBAD promoter is repressed) or l-arabinose (the PBAD promoter is induced). An arrow indicates the 36.5-kDa VirB protein. (B) Western blot showing that in the presence of only the pBAD33 vector (lacking the virB open reading frame), treatment with neither d-glucose nor l-arabinose causes the VirB protein to appear. Successful protein transfer in the immunoblotting apparatus was confirmed by the detection of the 69-kDa DnaK protein. (C) qRT-PCR analysis of transcripts from the native virB and virF promoters in the presence of VirB ectopically expressed from pBADvirB (induced by l-arabinose) or in its absence (repressed by d-glucose). The expression levels of virB and virF were also monitored in the presence of the pBAD33 vector alone and in an hns null mutant. These experiments were performed at a growth temperature of 30°C, which is repressive for the expressions of the virB and virF genes in wild-type strain BS184.
Under conditions of low-level VirB protein expression (i.e., in the presence of d-glucose), the promoters of both virB and virF were only weakly active, as was expected under these repressive conditions. However, with the increase in the level of VirB protein production in the culture to which l-arabinose was added, both the virB and virF promoters were derepressed (Fig. 2C). The expression level of virB increased 5-fold upon the induction of VirB protein expression, while the virF expression level increased 3-fold. In an hns null mutant, both promoters produced results similar to those seen following VirB-mediated derepression. This was consistent with the established role of VirB as an H-NS antagonist (35, 73).
Autoregulation of virB is dependent on a cis-acting VirB binding site.
Further investigation of VirB autoregulation was conducted with a virB-gfp reporter fusion. A region of 330 bp of the virB promoter was inserted upstream of the green fluorescent protein gene (gfp) to monitor virB promoter activity. This construct incorporated the promoter, the H-NS binding sites spanning positions −20 to +20, and the upstream region extending to position −310. The experiments were conducted at a nonpermissive temperature of 30°C, a temperature at which virB was normally repressed, using cultures grown aerobically to the stationary phase. The PBAD vector system was used to manipulate the levels of the VirB protein in the cell with the addition of either 0.2% d-glucose to repress transcription (−VirB) or 0.2% l-arabinose to induce transcription (+VirB) (Fig. 3). In the presence of d-glucose, the virB-gfp fusion remained repressed, but in the presence of l-arabinose, the increased expression level of the VirB protein resulted in a 5-fold increase in the virB-gfp transcription level (Fig. 3A). In an hns null mutant, the level of expression of the virB-gfp fusion increased 5- to 6-fold despite the repressive temperature, showing that the virB-gfp fusion showed a response profile similar to that of the native virB gene (Fig. 2C and 3A).
The truncation of the virB promoter region to include only the segment between positions −80 and +190 abolished the sensitivity of the virB construct to the presence of the VirB protein (Fig. 3B). This finding suggested that virB autoregulation was dependent on the region extending from positions −310 to −80. The truncated virB construct retained most of the region known to be bound by VirF (71) and so could be activated by this essential transcription factor upon the removal of H-NS from the promoter. This was demonstrated by monitoring the expression of the construct in an hns mutant, where a 5-fold increase in promoter activity was detected (Fig. 3B). These data suggested the possibility of a binding site for the VirB protein in the region spanning positions −310 to −80 of the virB promoter. A previously reported analysis of the icsB, spa15, and virA promoters of Shigella sonnei established a consensus binding site for VirB as being 5′-(A/G)(A/T)G(G)AAAT-3′ (68). These sites are not always found as inverted repeats as was the case for icsB (73). Bioinformatic analysis of the virB promoter revealed a close match to this sequence, centered at position −280. The putative VirB binding site, 5′-ATTGAAAT-3′, matched the consensus at seven of eight positions. This site was mutagenized in accordance with previously reported data relating to the binding specificity of VirB (73), to generate the mutated derivative 5′-GCCTGGGC-3′. When monitored in the presence or absence of the VirB protein during growth at a repressive temperature (30°C), the VirB-dependent upregulation of the virB promoter was diminished in the mutant with the altered site, suggesting that this was indeed a binding site for VirB (Fig. 3C).
Interestingly, the insertion of the well-characterized VirB binding site from the icsB promoter at position −80 of the mutated virB promoter restored this VirB-dependent derepression of the virB promoter (Fig. 3D). This demonstrated that the VirB binding site at the virB promoter could be removed and functionally replaced by a VirB binding site native to another VirB-regulated promoter. It also showed that the location of the site can be altered without a loss of function, in keeping with our previous findings (35).
A cis-acting VirB binding site facilitates VirB-mediated derepression of virF.
A virF-gfp transcriptional fusion was constructed to examine the effect of increasing the concentration of the VirB protein on a repressed virF promoter in bacteria growing at 30°C (Table 2). The region of the virF promoter placed upstream of gfp spanned positions −380 to +64 and included all of the virF DNA that is known to be bound by the H-NS and FIS proteins (58). Once again, the experiments were conducted at a repressive growth temperature (30°C), and the intracellular concentration of the VirB protein was manipulated by using the l-arabinose-inducible PBAD promoter. The virF regulatory region of the virF-gfp fusion maintained H-NS-mediated repression at 30°C in the absence of VirB; however, upon the induction of VirB protein expression, the virF-gfp fusion was upregulated to levels similar to those obtained with an hns mutant (Fig. 3E). The shortening of the promoter region to remove DNA upstream of the H-NS binding site (position −278) did not inhibit the VirB-mediated derepression of the virF promoter (Fig. 3F). This finding indicated that any potential VirB binding site either overlapped the H-NS binding region or was located at a site within the 160 bp of the DNA connecting the two H-NS binding sites. An inspection of the DNA sequence of this region revealed a close match to the consensus for VirB binding sites at position −134. The DNA sequence of the putative VirB binding site, 5′-GTGCAAAT-3′, matched the consensus sequence at seven of eight positions. This placed it almost exactly at the midpoint between the two H-NS binding sites. The DNA sequence of the VirB binding site was mutated to 5′-GCTCGGGC-3′, and this resulted in a reduced level of derepression in the presence of the VirB protein. Full derepression was detected only in an hns mutant (Fig. 3G).
VirB interaction with the regulatory regions of the virB and virF promoters.
Bioinformatic analysis and assays of mutagenized promoters identified possible VirB binding sites in the regulatory regions of the virB and the virF promoters. Preliminary evidence obtained from electrophoretic mobility shift assays (EMSAs) suggested that the VirB protein bound directly to both the virB and virF promoters (data not shown). To obtain more precise information about the VirB binding sites, DNase I protection assays were conducted in vitro with purified VirB protein at each of the promoters.
The DNase I footprinting experiments were performed by using an automated capillary DNA sequencer. With this method, the DNA was amplified with a fluorescent dye, rather than a radioactive label, that could easily be detected with an automated fluorescent DNA analysis instrument that resolves oligonucleotides by capillary electrophoresis. The virF and virB promoter regions used in the EMSA studies were PCR amplified by using FAM-labeled primers and incubated with or without purified VirB protein. These mixtures were then subjected to DNase I digestion. The reactions were stopped after 10 min, and the fluorescently labeled digestion products were analyzed. In the case of the virB regulatory region, a zone of protection was detected between positions −290 and −180 (Fig. 4A and B). This location corresponded to the VirB binding-site consensus sequence revealed by bioinformatic and site-directed mutagenesis studies (Fig. 3C). For the virF region, an area of protection associated with VirB binding was observed between positions −160 and −90 (Fig. 4C and D), again corresponding to the location of the putative VirB binding-site consensus sequence (Fig. 3G).
Fig 4.
VirB requires cis-acting sequences for efficient binding at virB and at virF. (A) Plot showing the protection pattern of the virB promoter region after digestion with DNase I following incubation in the presence (black) or absence (gray) of the VirB protein. The labeled VirB site shows an area with a significant difference in the peak pattern. (B) Schematic representation of the virB promoter region shown at the same scale below the DNase I footprinting data to show the relationship of the VirB binding site to the virB transcription start site. (C) Plot showing the protection pattern of the virF promoter region after digestion with DNase I following incubation in the presence (black) or absence (gray) of the VirB protein. The labeled VirB site shows an area with a significant difference in the peak pattern. (D) Schematic representation of the virF promoter region shown at the same scale below the DNase I footprinting data to show the relationship of the VirB binding site to the virF transcription start site.
VirB is required for the thermal induction of the virB and virF promoters.
An increase of the temperature to 37°C, indicative of entry into the human host, is the key environmental signal resulting in an invasive phenotype of S. flexneri (70). The above-described data suggested that VirB could exert an effect at 30°C when overexpressed ectopically by using an inducible promoter. It was possible that this VirB-dependent derepression of H-NS-mediated transcriptional silencing at the virB and virF promoters aided the upregulation of these genes, and, hence, the virulence genes, following a thermal upshift from 30°C to 37°C.
To test this hypothesis, cells of wild-type strain BS184, harboring the virB-gfp fusion on plasmid pZep08, and virB null mutant strain CJD1018, harboring the same virB-gfp fusion, were grown overnight at 30°C. The cells were then diluted 1:100 and grown at 30°C until the early exponential phase (optical density at 600 nm [OD600] of 0.1), when the cultures were divided, and one-half was shifted to a temperature of 37°C. Samples were taken at regular time points, and the level of GFP fluorescence was measured. It was found that in the virB null mutant, the induction of the virB-gfp fusion was delayed and reduced compared to the induction of the virB-gfp fusion in a wild-type background with a functional copy of the virB gene (Fig. 5). Therefore, VirB contributes to the full induction of the virB promoter in response to a thermal upshift, presumably by aiding in the removal of H-NS from the virB promoter region.
Fig 5.
Thermal induction of the virB and virF promoters. (A) Growth of wild-type (WT) strain BS184 and its virB mutant at 30°C and 37°C over a 24-h period. (B) Expression of a virB-gfp fusion as a function of growth phase in a wild-type or virB mutant background at 30°C or 37°C. Samples were taken at regular intervals, and GFP fluorescence was measured. (C) Expression of the virF-gfp fusion as a function of growth phase in a wild-type or virB mutant background at 30°C or 37°C. Samples were taken at regular intervals, and GFP fluorescence was measured. Error bars show the standard deviations from the means.
Similar experiments were conducted with the virF-gfp promoter fusions in wild-type and virB mutant backgrounds. The results showed that while the initial induction of the virF promoter was reduced in strains lacking the VirB protein, the ultimate level of virF transcription after 6 h of growth was similar to that observed for the wild type. This finding suggested that while VirB may play a role in boosting the initial activation of transcription of virF in response to a thermal upshift, this VirB dependency ceased to be relevant upon continued growth at the inducing temperature, at least under laboratory growth conditions (Fig. 5).
DISCUSSION
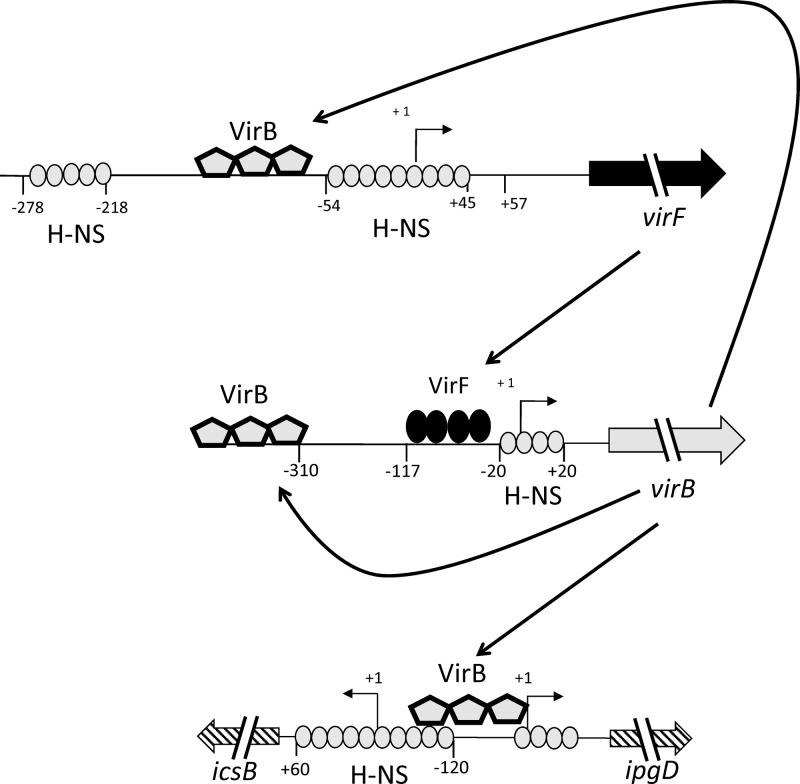
The regulatory cascade of Shigella flexneri is a paradigmatic example of the integration of environmental cues to elicit the expression, assembly, and activation of essential virulence factors (16, 19, 20, 30, 53). While once thought to be a linear, step-by-step process, in which environmentally wrought DNA topological adjustments activate virF, which in turn activates virB, which then derepresses the structural and effector protein genes, the results reported here reveal a much more sophisticated mechanism of transcriptional control. Our data demonstrate for the first time the ability of the VirB protein to upregulate both H-NS-repressed virB and virF promoters even during growth at the nonpermissive temperature of 30°C. These results show that virB, the regulatory gene located at an intermediate position in the wiring diagram of the S. flexneri virulence cascade, not only is autoregulated but also controls the expression of the primary regulatory gene virF, which has hitherto been ranked above it (Fig. 6).
Fig 6.
Revised schematic overview of the interplay of transcription factors in the virulence gene regulatory cascade. The VirB protein (pentagon) binds to its own promoter, antagonizing repression mediated by H-NS (oval). The VirB protein also feeds back onto virF, binding to its promoter and antagonizing H-NS-mediated repression.
The mechanism by which VirB regulates transcription is noteworthy for its relative mechanistic simplicity. VirB is an antirepressor whose chief task is to antagonize the transcriptional silencing activity of the H-NS protein. It does so by remodeling the H-NS-DNA nucleoprotein complex to facilitate the transcription of the silenced gene(s), and its ability to do so is linked directly to its intracellular concentration. The artificial adjustment of the concentration of the VirB protein upwards in bacteria growing at temperatures that are usually nonpermissive for virulence gene expression results in the derepression of those genes (6, 35, 73). Even at nonpermissive temperatures, low levels of the VirB protein can sometimes be detected in wild-type S. flexneri, suggesting that the protein concentration, while measurable, is below a threshold required for structural gene derepression (6, 53). The results described in this study reveal that the virB gene is itself a target for VirB-mediated transcriptional derepression. This provides a mechanism for boosting the rate of accumulation of the VirB protein for the positive control of the virulence cascade once the appropriate profile of environmental signals has been detected by the system. However, this simple VirB-virB positive feedback loop is complicated by its dependency on VirF, the AraC-like transcription factor that is essential for the full activity of the virB promoter (1, 71, 72). Our discovery that VirB also feeds back positively onto virF transcription addresses this complication by providing a mechanism to boost VirF and VirB levels simultaneously. In this way, both regulatory protein requirements of the virB promoter are met: VirB is available (presumably) to antagonize H-NS-mediated transcription silencing, and VirF contributes to the same antisilencing process and activates the virB promoter, in keeping with its role as a conventional transcription activator. The improvement in virF transcription due to the presence of VirB is significant but relatively small under the conditions used in our study (Fig. 5). This may reflect the fact that the in vitro growth regimen fails to capture all of the elements of the environment that characterize the site of epithelial cell invasion in the lower intestine of the host.
The two VirB binding sites that were characterized bioinformatically, genetically, and biochemically in this study expand further the list of such sites within the S. flexneri regulatory cascade. All of these sites share strong similarity to the box A motif of cis-acting parS sequences that are bound by ParB-like plasmid-partitioning proteins. The example of the icsB promoter remains the most parS-like of them all, and this may reflect an earlier role for this particular DNA sequence in plasmid segregation. For these DNA sequences to participate in antirepression at H-NS-silenced promoters, they have to bind VirB and to be appropriately positioned with respect to the H-NS–DNA nucleoprotein complex (35). The meaning of “appropriate positioning” is likely to vary from promoter to promoter, depending on which other regulatory factors are involved. The virF promoter experiences numerous controlling influences, including those of H-NS and its antagonists, the positively acting FIS and IHF proteins. The center of our newly discovered VirB binding site is located at position −134, flanked by the two sites that are bound and bridged by H-NS (Fig. 6) (17, 58). Extrapolating from the example of the icsB promoter, where the DNA-dependent polymerization of the VirB protein is known to coat about 100 bp of DNA (73), a similar behavior at virF would result in VirB binding extending right up to the H-NS-occupied sites, facilitating antirepression. Previously reported data obtained with a synthetic proU promoter harboring a VirB binding site also support this proposal (35).
As stated above, any model of VirB–H-NS antagonism at virB must also take into consideration the requirement for VirF for full activation. It may not be necessary for both VirF and VirB to be bound to the virB promoter simultaneously; perhaps VirB acts initially to prepare the promoter for VirF-mediated activation by first dislodging the H-NS repressor. More work is required to test this possibility.
The relevance of virB positive autoregulation and positive feedback onto virF becomes clear when the thermal induction kinetics of both promoters are observed (Fig. 6). Without autoregulation, the virB promoter has a delayed response to its thermal environmental cue and maintains a reduced level of transcription. The virF promoter is also delayed in activation; however, this is eventually overcome, most likely by conformational changes in the DNA, which are known to displace H-NS and permit activation by FIS (57, 58). The intermediate regulatory step of virB expression in the regulatory cascade has been a matter of some speculation. It is assumed that it provides a regulatory checkpoint prior to full commitment to the expression of the elaborate type III secretion system (16, 35). However, this additional regulation may impose a cost on the bacterium by postponing the assembly of the essential secretion system, while virB is derepressed, transcribed, and translated into a functioning antagonist of H-NS. Perhaps one purpose of positive feedback in the regulatory cascade is to ensure a transcriptional surge in virB expression, and, to a lesser extent, virF expression, to allow a “jump start” in TTSS gene expression in response to the appropriate environmental signals (Fig. 6).
The detailed dissection of the S. flexneri virulence regulatory cascade illustrates the flexibility of bacterial gene regulatory circuits and their capacity to adapt through evolution to meet the challenges of an ever-changing environment. Bacteria have evolved a diverse set of regulatory pathways that govern various adaptive responses to survive under rapidly changing environmental conditions (40), and feedback loops are common elements of cellular regulatory circuits (15). Feedback regulation is critical for virulence in bacterial pathogens (76), and positive feedback is thought to shape response timing to allow the rapid expression of necessary genes when activated by certain stimuli (43, 67). These criteria certainly apply in the case of the S. flexneri virulence gene regulatory cascade, where the role of positive feedback at the level of transcription had hitherto been undiscovered.
ACKNOWLEDGMENTS
We thank Andrew Cameron for helpful advice on the DNase I footprinting experiments and the other members of the Dorman laboratory for useful discussions.
This work was supported by grant 07/IN1/B918 from Science Foundation Ireland.
Footnotes
Published ahead of print 20 July 2012
REFERENCES
- 1. Adler B, et al. 1989. A dual transcriptional activation system for the 230-kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627–635 [DOI] [PubMed] [Google Scholar]
- 2. Allaoui A, Ménard R, Sansonetti PJ, Parsot C. 1993. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect. Immun. 61:1707–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allaoui A, Sansonetti PJ, Parsot C. 1993. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasins. Mol. Microbiol. 7:59–68 [DOI] [PubMed] [Google Scholar]
- 4. Andrews GP, Maurelli AT. 1992. mxiA of Shigella flexneri 2a, which facilitates export of invasion plasmid antigens, encodes a homolog of the low-calcium-response protein, LcrD, of Yersinia pestis. Infect. Immun. 60:3287–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beloin C, McKenna S, Dorman CJ. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 277:15333–15344 [DOI] [PubMed] [Google Scholar]
- 6. Beloin C, Dorman CJ. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825–838 [DOI] [PubMed] [Google Scholar]
- 7. Bernardini ML, Mounier J, d'Hauteville H, Coquis-Rondon M, Sansonetti PJ. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. U. S. A. 86:3867–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bignell C, Thomas CM. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1–34 [DOI] [PubMed] [Google Scholar]
- 9. Blocker A, et al. 1999. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147:683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blocker A, et al. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652–663 [DOI] [PubMed] [Google Scholar]
- 11. Bouet J-Y, Funnell BE. 1999. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 18:1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchrieser C, et al. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760–771 [DOI] [PubMed] [Google Scholar]
- 13. Cameron ADS, Dorman CJ. 2012. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet. 8:e1002615 doi:10.1371/journal.pgen.1002615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Castellanos MI, et al. 2009. VirB alleviates H-NS repression of the icsP promoter in Shigella flexneri from sites more than one kilobase upstream of the transcription start site. J. Bacteriol. 191:4047–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cosentino Lagomarsino M, Jona P, Bassetti B, Isambert H. 2007. Hierarchy and feedback in the evolution of the Escherichia coli transcription network. Proc. Natl. Acad. Sci. U. S. A. 104:5516–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dorman CJ. 2009. The virulence plasmids of Shigella flexneri, p 151–170 In Schwartz E. (ed), Microbial megaplasmids. Springer, Heidelberg, Germany [Google Scholar]
- 17. Dorman CJ, Kane KA. 2009. DNA bridging and anti-bridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally-acquired genes. FEMS Microbiol. Rev. 33:587–592 [DOI] [PubMed] [Google Scholar]
- 18. Dorman CJ, Ní Bhriain N, Higgins CF. 1990. DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature 344:789–792 [DOI] [PubMed] [Google Scholar]
- 19. Dorman CJ, Porter ME. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29:677–684 [DOI] [PubMed] [Google Scholar]
- 20. Dorman CJ, McKenna S, Beloin C. 2001. Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Int. J. Med. Microbiol. 291:89–96 [DOI] [PubMed] [Google Scholar]
- 21. Dunham TD, Xu W, Funnell BE, Schumacher MA. 2009. Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J. 28:1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DuPont HL, Levine MM, Hornick RB, Formal SB. 1989. Inoculum size in shigellosis and implications for expected mode of transmission. J. Infect. Dis. 159:1126–1128 [DOI] [PubMed] [Google Scholar]
- 23. Durand JMB, Dagberg B, Uhlin BE, Björk GR. 2000. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 35:924–935 [DOI] [PubMed] [Google Scholar]
- 24. Durand JMB, Björk GR. 2003. Putrescine or a combination of methionine and arginine restores virulence gene expression in a tRNA modification-deficient mutant of Shigella flexneri: a possible role in adaptation of virulence. Mol. Microbiol. 47:519–527 [DOI] [PubMed] [Google Scholar]
- 25. Eberl L, Givskov M, Schwab H. 1992. The divergent promoters mediating transcription of the par locus of plasmid RP4 are subject to autoregulation. Mol. Microbiol. 6:1969–1979 [DOI] [PubMed] [Google Scholar]
- 26. Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17:7033–7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Falconi M, Prosseda G, Giangrossi M, Beghetto E, Colonna B. 2001. Involvement of Fis in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol. Microbiol. 42:439–452 [DOI] [PubMed] [Google Scholar]
- 28. Friedman SA, Austin SJ. 1988. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid 19:103–112 [DOI] [PubMed] [Google Scholar]
- 29. Goldberg MB. 2001. Actin-based motility of intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 65:595–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hale TL. 1991. Genetic basis of virulence in Shigella species. Microbiol. Rev. 55:206–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hautefort I, Proenca MJ, Hinton JC. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hayes F, Barillá D. 2010. Extrachromosomal components of the nucleoid: recent developments in deciphering the molecular basis of plasmid segregation, p 49–70 In Dame RT, Dorman CJ. (ed), Bacterial chromatin. Springer Press, Dordrecht, Netherlands [Google Scholar]
- 33. Hermsen R, Ursem B, ten Wolde PR. 2010. Combinatorial gene regulation using auto-regulation. PLoS Comput. Biol. 6:e1000813 doi:10.1371/journal.pcbi.1000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hromockyj AE, Tucker SC, Maurelli AT. 1992. Temperature regulation of Shigella virulence: identification of the repressor gene virR, an analog of hns, and partial complementation by tyrosyl transfer RNA (tRNA1Tyr). Mol. Microbiol. 6:2113–2124 [DOI] [PubMed] [Google Scholar]
- 35. Kane KA, Dorman CJ. 2011. Rational design of an artificial genetic switch: co-option of the H-NS-repressed proU operon by the VirB virulence master regulator. J. Bacteriol. 193:5950–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keseler IM, et al. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39:D583–D590 doi:10.1093/nar/gkq1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwong SM, Yeo CC, Poh CL. 2001. Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site, parS. Mol. Microbiol. 40:621–633 [DOI] [PubMed] [Google Scholar]
- 38. Maurelli AT, Blackmon B, Curtiss R., III 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43:195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maurelli AT, Sansonetti PJ. 1988. Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc. Natl. Acad. Sci. U. S. A. 85:2820–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McAdams HH, Srinivasan B, Arkin AP. 2004. The evolution of genetic regulatory systems in bacteria. Nat. Rev. Genet. 5:169–178 [DOI] [PubMed] [Google Scholar]
- 41. McKenna S, Beloin C, Dorman CJ. 2003. In vitro DNA binding properties of VirB, the Shigella flexneri virulence regulatory protein. FEBS Lett. 545:183–187 [DOI] [PubMed] [Google Scholar]
- 42. McNairn E, Ní Bhriain N, Dorman CJ. 1995. Over-expression of the Shigella flexneri genes coding for DNA topoisomerase IV compensates for loss of DNA topoisomerase I: effect on virulence gene expression. Mol. Microbiol. 15:507–517 [DOI] [PubMed] [Google Scholar]
- 43. Mitrophanov AY, Hadley TJ, Groisman EA. 2010. Positive autoregulation shapes response timing and intensity in two-component signal transduction systems. J. Mol. Biol. 401:671–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murphy ER, Payne SM. 2007. RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect. Immun. 75:3470–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakayama S, Watanabe H. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177:5062–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakayama S, Watanabe H. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella flexneri virF gene. J. Bacteriol. 180:3522–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ní Bhriain N, Dorman CJ. 1993. Identification and characterization of a topA mutation in Shigella flexneri. Mol. Microbiol. 7:351–358 [DOI] [PubMed] [Google Scholar]
- 48. Niyogi SK. 2005. Shigellosis. J. Microbiol. 43:133–143 [PubMed] [Google Scholar]
- 49. Parsot C, Ménard R, Gounon P, Sansonetti PJ. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16:291–300 [DOI] [PubMed] [Google Scholar]
- 50. Parsot C. 2005. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 252:11–18 [DOI] [PubMed] [Google Scholar]
- 51. Phalipon A, Sansonetti PJ. 2007. Shigella's ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol. Cell Biol. 85:119–129 [DOI] [PubMed] [Google Scholar]
- 52. Porter ME, Dorman CJ. 1994. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 176:4187–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Porter ME, Dorman CJ. 1997. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol. Gen. Genet. 256:93–103 [DOI] [PubMed] [Google Scholar]
- 54. Porter ME, Dorman CJ. 1997. Positive regulation of Shigella flexneri virulence genes by integration host factor. J. Bacteriol. 179:6537–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Porter ME, Dorman CJ. 2002. In vivo DNA-binding and oligomerization properties of the Shigella flexneri AraC-like transcriptional regulator VirF as identified by random and site-specific mutagenesis. J. Bacteriol. 184:531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Prévost MC, et al. 1992. Unipolar reorganization of F-actin layer at bacterial division and bundling of actin filaments by plastin correlate with movement of Shigella flexneri within HeLa cells. Infect. Immun. 60:4088–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Prosseda G, et al. 1998. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res. Microbiol. 149:15–25 [DOI] [PubMed] [Google Scholar]
- 58. Prosseda G, et al. 2004. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 51:523–537 [DOI] [PubMed] [Google Scholar]
- 59. Salgado H, et al. 2001. RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29:72–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sansonetti PJ. 2006. Shigellosis: an old disease in new clothes? PLoS Med. 3:e354 doi:10.1371/journal.pmed.0030354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sansonetti PJ, Kopecko DJ, Formal SB. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sansonetti PJ, et al. 1983. Alterations in the pathogenicity of Escherichia coli K-12 after transfer of plasmid and chromosomal genes from Shigella flexneri. Infect. Immun. 39:1392–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sansonetti PJ, Mounier J, Prévost MC, Mege RM. 1994. Cadherin expression is required for the spread of Shigella flexneri between epithelial cells. Cell 5:829–839 [DOI] [PubMed] [Google Scholar]
- 64. Sasakawa C, Komatsu K, Tobe T, Suzuki T, Yoshikawa M. 1993. Eight genes in region 5 that form an operon are essential for invasion of epithelial cells by Shigella flexneri 2a. J. Bacteriol. 175:2334–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sergueev K, Dabrazhynetskaya A, Austin S. 2005. Plasmid partition system of the P1par family from the pWR1000 virulence plasmid of Shigella flexneri. J. Bacteriol. 187:3369–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shen-Orr SS, Milo R, Mangan S, Alon U. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31:64–68 [DOI] [PubMed] [Google Scholar]
- 67. Shin D, Lee J, Huang H, Groisman EA. 2006. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314:1607–1609 [DOI] [PubMed] [Google Scholar]
- 68. Taniya T, et al. 2003. Determination of the InvE binding site required for expression of IpaB of the Shigella sonnei virulence plasmid: involvement of a ParB BoxA-like sequence. J. Bacteriol. 185:5158–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thieffry D, Huerta AM, Perez-Rueda E, Collado-Vides J. 1998. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays 20:433–440 [DOI] [PubMed] [Google Scholar]
- 70. Tobe T, et al. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5:887–893 [DOI] [PubMed] [Google Scholar]
- 71. Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. 1993. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J. Bacteriol. 175:6142–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tobe T, Yoshikawa M, Sasakawa C. 1995. Thermoregulation of virB transcription in Shigella flexneri by sensing of changes in local DNA superhelicity. J. Bacteriol. 177:1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Turner EC, Dorman CJ. 2007. H-NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. J. Bacteriol. 189:3403–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vasselon T, Mounier J, Hellio R, Sansonetti PJ. 1992. Movement along actin filaments of the perijunctional area and de novo polymerization of cellular actin are required for Shigella flexneri colonization of epithelial Caco-2 cell monolayers. Infect. Immun. 60:1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Venkatesan MM, Buysse JM, Oaks EV. 1992. Surface presentation of Shigella flexneri invasion plasmid antigens requires the products of the spa locus. J. Bacteriol. 174:1990–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Williams CL, Cotter PA. 2007. Autoregulation is essential for precise temporal and steady-state regulation by the Bordetella BvgAS phosphorelay. J. Bacteriol. 189:1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]