Abstract
Bacterial differentiation is often associated with the asymmetric localization of regulatory proteins, such as histidine kinases. PdhS is an essential and polarly localized histidine kinase in the pathogenic alphaproteobacterium Brucella abortus. After cell division, PdhS is asymmetrically segregated between the two sibling cells, highlighting a differentiation event. However, the function(s) of PdhS in the B. abortus cell cycle remains unknown. We used an original approach, the pentapeptide scanning mutagenesis method, to generate a thermosensitive allele of pdhS. We report that a B. abortus strain carrying this pdhS allele displays growth arrest and an altered DivK-yellow fluorescent protein (YFP) polar localization at the restrictive temperature. Moreover, the production of a nonphosphorylatable PdhS protein or truncated PdhS proteins leads to dominant-negative effects by generating morphological defects consistent with the inhibition of cell division. In addition, we have used a domain mapping approach combined with yeast two-hybrid and fluorescence microscopy methods to better characterize the unusual PdhS sensory domain. We have identified a fragment of the PdhS sensory domain required for protein-protein interaction (amino acids [aa] 210 to 434), a fragment sufficient for polar localization (aa 1 to 434), and a fragment (aa 527 to 661) whose production in B. abortus correlates with the generation of cell shape alterations. The data support a model in which PdhS acts as an essential regulator of cell cycle progression in B. abortus and contribute to a better understanding of the differentiation program inherited by the two sibling cells.
INTRODUCTION
Bacterial adaptation relies on signal transduction systems that allow sensing of the environmental and intracellular conditions in order to engage an appropriate cellular response. The two-component systems are widespread response systems classically composed of a sensory histidine kinase (HK) and its cognate response regulator (RR). The transfer of a phosphoryl group from the HK activates the RR that acts as an effector protein (21). HKs are multidomain proteins that contain a variable N-terminal sensory domain (SD) and a structurally conserved C-terminal part (catalytic domain [CD]), which contains an ATPase domain and a short conserved domain that contains the phosphorylatable histidine (23). Differences between the input domains of various HKs reflect the diversity of stimuli sensed and the cellular functions of the HK within the cell. It is thus important to characterize the SD of HKs in order to gain insights into their specific cellular role. Several functional subdomains present in the SD have been identified (31), including PAS domains, which are particularly widespread in sensing proteins (23) and have been involved in small molecule binding and protein-protein interactions (27, 31).
Two-component systems are also used as regulatory networks involved in the control of the bacterial cell cycle and differentiation, as exemplified by studies on the free-living alphaproteobacterium model Caulobacter crescentus (5, 29). In this bacterium, two transmembrane HKs, PleC and DivJ, modulate the phosphorylation state of the response regulator DivK involved in the control of the developmental program inherited by the two sibling cells (25, 38). Three homologs of PleC and DivJ HKs are found in Brucella abortus (16, 18), a facultative intracellular pathogen responsible for a worldwide zoonosis called brucellosis (28). PdhS (PleC DivJ homologue sensor), a cytoplasmic homologue to both PleC and DivJ, was found to be essential and polarly localized and was proposed to be involved in the control of B. abortus cell cycle (18). Consistently, PdhS overproduction in B. abortus was found to cause morphological defects (18). By comparison with PleC and DivJ, PdhS displays unusual features: (i) the absence of predicted transmembrane segment, (ii) a large N-terminal domain (795 amino acids [aa]) without predicted function, except for the presence of a putative PAS domain, and (iii) the ability to specifically interact with the class II fumarase FumC, a metabolic enzyme of the citric acid cycle (26). PdhS localization is dynamic, and after cell division, it is found only at the old pole of the mother cell (18). The newly generated cell will acquire PdhS at its old pole prior to division, highlighting a differentiation event (18).
In order to gain insights into the biological role of PdhS during the B. abortus cell cycle, we generated a conditional allele of the pdhS essential gene. The PdhS loss of function results in a growth arrest and the delocalization of polar DivK-yellow fluorescent protein (YFP) fusion. In addition, the generation of aberrant morphologies by the production of a regulatory fragment or the presence of a nonphosphorylatable version of PdhS is compatible with a dominant-negative effect on cell division, since the incubation with mitomycin C generated similar phenotypes. To better characterize PdhS, we identified functional subdomains within its sensory domain. These data indicate that PdhS is involved in B. abortus cell cycle progression by regulating cell growth and division and highlight the complexity of this HK by associating functions with subdomains within the SD.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All Brucella strains used in this study were derived from B. abortus 544 Nalr (a spontaneous nalidixic acid-resistant mutant) and were routinely cultivated in 2YT rich medium (1% yeast extract, 1.6% peptone, 0.5% NaCl). The Escherichia coli strain used for the molecular cloning experiments was DH10B (Invitrogen Life Technologies), except for the ccdB-containing plasmids that were propagated in the DB3.1 strain (Invitrogen Life Technologies). All E. coli strains were grown in LB broth. Antibiotics were used at the following concentrations when appropriate: B. abortus and E. coli (for liquid and solid cultures): nalidixic acid, 25 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml; rifampin, 50 μg/ml; and spectinomycin, 100 μg/ml. The C. crescentus strain used in this study was CB15N grown in PYE medium (10), and the antibiotic concentrations were 1 μg/ml chloramphenicol and/or 15 μg/ml nalidixic acid for liquid cultures and 2 μg/ml chloramphenicol and/or 20 μg/ml nalidixic acid for solid cultures. The plasmids were mobilized from the E. coli S17-1 strain into B. abortus by conjugation. Mitomycin C was used at a final concentration of 2 μg/ml for 7 h in 2YT medium at 37°C with agitation.
Essentiality assays.
The essentiality of divK and pdhS was tested using a previously reported procedure applied to demonstrate synthetic lethality (26). The absence of deletants for the strain harboring the pdhSH805A allele (pdhS with an H→A change at position 805) was significantly different from the frequency (7/106) of deletion observed for the strain having a wild-type allele of pdhS (P = 0.0017 for a binomial variable, with n = 93 and a deletion probability of 7/106). The low frequency of pdhS deletion in the presence of pBBR1MCS-pdhS is probably due to the toxicity of pdhS overexpression. The absence of deletants for the strain harboring the divKD53A allele was significantly different from the frequency of deletion (14/29) observed for the strain having a wild-type allele of divK (P = 2.6 10−11 for a binomial variable, with n = 37 and a deletion probability of 14/29).
Molecular techniques.
The yeast two-hybrid (Y2H) assay was performed as previously described (36). The pVV213 plasmids (18) carrying the activating domain of Gal4p fused to PdhS fragments were introduced in yeast (Saccharomyces cerevisiae) strain MaV203 and pVV212 plasmids carrying the binding domain of Gal4p fused to PdhS or FumC were introduced in yeast strain MaV103. For the mating, yeast strains were plated on yeast extract-peptone-dextrose (YPD) medium overnight and then replicated on selective medium (i.e., depleted in uracil or histidine with 100 mM 3-aminotriazole) when appropriate, as previously described (7). For the localization screen, Gateway cloning with pools of each coding sequence (CDS) was used, as previously described for pools of multiple CDS (9).
Protein purification and autophosphorylation assay.
Purification of PdhS derivatives was performed as previously described (7). Briefly, a His6 tag was fused to the carboxy-terminal side of PdhS and PdhSH805A on a pET15B vector (Novagen). After 3 h of induction with 24 μg/ml isopropyl-β-d-thiogalactopyranoside (IPTG) of the E. coli BL21(DE3) strain carrying pET15B-pdhS or pET15B-pdhSH805A, cells were lysed by sonication as previously described (35), and total cell extracts were placed on a Ni-nitrilotriacetic acid (NTA) His Bind Superflow (Novagen). After successive washings with 25, 50, and 75 mM imidazole and elution with 100 mM imidazole, the elution buffer containing the purified proteins was replaced by an autophosphorylation buffer (50 mM Tris-HCl, 25 mM NaCl, 25 mM KCl, 5 mM MgCl2) using a NAP5 column (GE Healthcare). The autophosphorylation assay was performed as previously described (6, 39), with purified proteins being incubated for 5 or 30 min with 1 μl of radioactive 3,000 Ci/mmol [γ-32P]ATP (PerkinElmer) in a 50-μl reaction volume.
Conserved motif prediction.
The first analysis of conserved domains in PdhS SD was performed using a simple BLAST (BLASTP) on the Conserved Domains Database (CDD). In the second analysis, three rounds of PSI-BLAST (i.e., two iterations after the first BLASTP) were made with the 661 first residues to generate a position-specific score matrix that was used for similarity search with the sequences extracted from the Protein Data Bank (PDB).
Domain mapping and PCR.
Fragments of the pdhS coding sequence (CDS) were amplified using the primers reported in Table 1. The amplified fragments were compatible with Gateway recombinational cloning (Invitrogen). Each CDS fragment was then transferred (i) to the RK2-based low-copy pRH008 plasmid for fusion with yellow fluorescent protein (YFP) (17) (9) or (ii) to pVV213 for the yeast two-hybrid assay (18). Each primer name corresponds to the CDS fragment to be amplified, with the “Up” primers hybridizing at the 5′ end of the CDS fragment and the “Down” primers hybridizing at the 3′ end of the CDS fragment.
Table 1.
Primer sequences used for domain mapping
| Primer name | Sequence (5′→3′) |
|---|---|
| F1_Up | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGTCAGGATCATACCCCTTCA |
| F1_Down | GGGGACCACTTTGTACAAGAAAGCTGGGTAGCTGTTATCGGGCATTTGTGTT |
| F2_Up | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGGCAACACAAATGCCCGATAACA |
| F2_Down | GGGGACCACTTTGTACAAGAAAGCTGGGTAGGTTTTCGGCCTGGGCTGTTC |
| F3_Up | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGGGCGACGCCCCAGCCCAGTT |
| F3_Down | GGGGACCACTTTGTACAAGAAAGCTGGGTATGGCGGTTTCGGCAGAACGG |
| F4_Up | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGGCCGGCGGTATTCCGCAAAAA |
| F4_Down | GGGGACCACTTTGTACAAGAAAGCTGGGTACGCGGTTTCGTCCGGCTGGA |
| F5_Up | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGCCCGTAAAAACACGCTCCGAA |
| F5_Down | GGGGACCACTTTGTACAAGAAAGCTGGGTACGCCTGCTTTTCCTCATCATTG |
| PAS_Up | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGCTCCCCGCCCCGGCCGAACT |
| PAS_Down | GGGGACCACTTTGTACAAGAAAGCTGGGTACTGGTTGGAGGCGCGCTCGG |
| CD_Up | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGGAGGCCGAGCGCGCCTCCAA |
| CD_Down | GGGGACCACTTTGTACAAGAAAGCTGGGTAGTCAGCCAGAACGCGGGTTG |
Microscopy.
Samples were examined with a Nikon 80i fluorescence microscope using a 100× objective, as previously described (18, 26). DAPI (4′,6′-diamidino-2-phenylindole) was used at 1 μg/ml to stain the DNA, and the mixture was incubated for 1 h at room temperature in phosphate-buffered saline (PBS) medium. The membrane-staining dye FM4-64 (Molecular Probes) was used at a final concentration of 1 μg/ml for 5 min in 2YT medium to observe global bacterial morphology. For the observations, bacteria were grown until they reached the stationary phase (optical density at 600 nm [OD600] of 1.2), and a culture sample (1.5 μl) was spread on the agarose pad, covered by a sealed coverslip. Time-lapse microscopy was performed by placing strains on a microscope slide that was layered with a pad of 1% agarose containing 2YT medium (700 μl per slide). The culture was resumed under the microscope at 23°C, and images were taken every hour. For scanning electron microscopy (SEM), bacteria were fixed with paraformaldehyde 4% for 15 min at 37°C and then placed on a coverslip covered with poly-l-lysine (Sigma), coated at 50 μg/ml, for 1 h at room temperature in water. Once the poly-l-lysine was removed, the samples were centrifuged at 1,500 rpm for 5 min and then dehydrated (successively for 5 min with 30, 50, 70, 85, and 100% ethanol. After the critical point was reached, gold particles were sprayed onto the samples with a metallizer (Balzers-Union). Scanning electron microscopic images were obtained using a JEOL device at 5,000× at 15 kV. When indicated, the average ± standard deviation was calculated from three independent experiments, with n representing the total number of counted bacteria.
Pentapeptide scanning mutagenesis.
The previously reported pentapeptide scanning mutagenesis method (14) was applied to the generation of thermosensitive (TS) alleles for pdhS. We inserted the pdhS sequence by reverse BP (Invitrogen) from pVV212-pdhS (18) and pDONR223 to obtain the entry vector pDONR223-pdhS. This plasmid was used as a target for pdhS pentapetide scanning mutagenesis. The pDONR223-pdhS plasmid was introduced in E. coli strain DH5α, which contains pFH395, a plasmid carrying the replicative transposon Tn4430 and a kanamycin resistance gene. Tn4430-mediated cointegrates between pFH395 and pDONR223-pdhS were selected by mating pooled DH5α transformants with the rifampin-resistant E. coli strain HB101R. Briefly, 3,000 donor colonies were mixed with the E. coli HB101R recipients for 2 h at 37°C on an LB agar plate without antibiotics to allow the mating. The bacteria were then resuspended in 1 ml of LB and spread on LB agar plates supplemented with rifampin and spectinomycin to select transconjugants. Plasmid DNA was extracted from a pool of ∼30,000 transconjugants and transformed back into DH10B in order to isolate plasmids resulting from the resolution of the cointegrates by site-specific recombinations. Plasmid DNA from a pool of ∼ 300,000 clones was digested with KpnI to excise the bulk of the Tn4430, ligated with T4 DNA ligase under dilution conditions promoting self-ligation, and used for transformation in DH10B. The KpnI excision leaves a scar of 15 nucleotides (nt) in the target sequence, corresponding to the insertion of 5 codons in the gene of interest (12). A total of 100,000 clones, containing about 75% of the expected self-ligated plasmids, were obtained and pooled for plasmid extraction. This pool of plasmids was enriched in pDONR223 derivatives carrying a mutated allele of pdhS (pdhS*). The resulting pool of pdhS* alleles was transferred by recombinational (LR) cloning to a Gateway-compatible plasmid derived from pJQ200-uc1 (30) (in which the Gateway cassette was amplified by PCR and cloned at the NotI restriction site present on the pJQ200-uc1) to generate the pJQ200-uc1-pdhS* library. About 3,000 pJQ200-uc1-pdhS* candidates were used to perform allelic replacement of the wild-type pdhS gene in B. abortus strain 544, as previously described for fumC allelic replacement (26). A total of 1,650 sucrose-insensitive clones were individually spotted on solid 2YT medium and incubated at 36°C and 42°C. Among the 1,650 clones tested, 137 candidates with different levels of thermosensitivity were identified, 20 of them being completely unable to generate colonies on 2YT agar plates at 42°C while retaining this capacity at 36°C.
RESULTS
PdhS is required for bacterial growth.
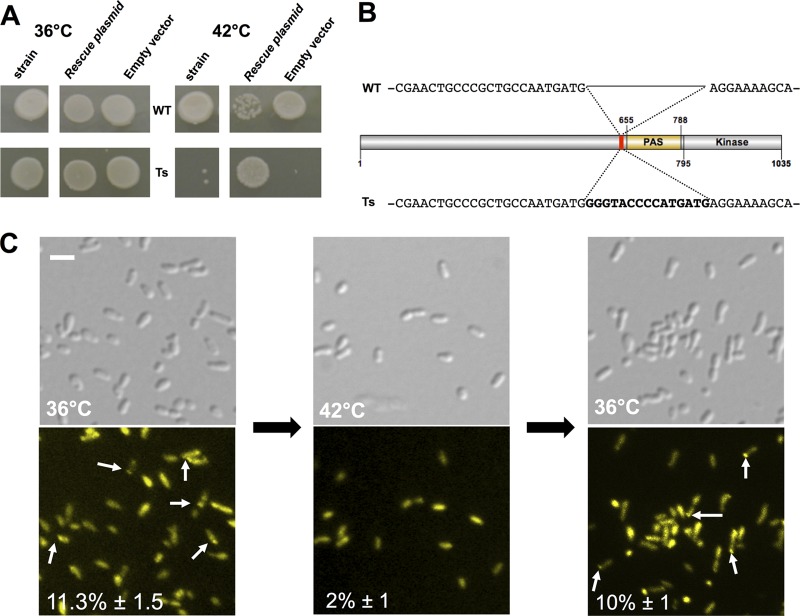
To observe phenotypes due to a loss of function of PdhS, we generated conditional alleles of the pdhS essential gene. We constructed a randomly mutated library of pdhS variants using the pentapeptide scanning mutagenesis method (14, 19, 20). This method relies on the insertion of variable 5-aa peptides at random positions of a protein of interest resulting from the integration or excision of the bacterial transposon Tn4430 into the corresponding target gene (see Materials and Methods). Previous studies have shown that the method often generates mutants with increased functional and structural flexibility, correlated with different levels of thermosensitivity in the mutated proteins (19). We thus applied this method to isolate B. abortus strains that would be thermosensitive (TS) because they are mutated in the pdhS coding sequence. After allelic replacement of wild-type pdhS with the mutant library, 1,650 clones were screened for their failure to form colonies on rich medium at 42°C (restrictive temperature) while still being able to generate colonies at 36°C (permissive temperature). A total of 137 candidates showing different levels of thermosensitivity were identified, of which 20 were completely unable to grow at the nonpermissive temperature. One specific clone was retained for further characterization (Fig. 1). The complementation assay showed that the selected TS mutant was able to generate colonies at the restrictive temperature only when a low-copy-number plasmid (pMR10cat) carrying the wild-type copy of the pdhS gene was present (Fig. 1A). The presence of the pMR10cat empty vector did not complement the thermosensitive phenotype. This shows that the thermosensitive phenotype does not result from polar effects and that it is specifically due to mutation in the pdhS gene. We sequenced the pdhS coding sequence in the temperature-sensitive strain and found the expected insertion of 15 bp located upstream of the predicted PAS domain (Fig. 1B).
Fig 1.
The thermosensitive strain highlights a PdhS loss of function. (A) Wild-type (WT) strain of B. abortus and the thermosensitive (Ts) strain grown at the permissive temperature (36°C) or restrictive temperature (42°C), with a plasmid carrying the wild-type pdhS sequence (rescue plasmid) or the empty vector. (B) Sequence of the 15-bp scar at the insertion site of Tn4430. (C) DivK-YFP localization in the B. abortus TS strain. Differential interference contrast (DIC) images are shown in the upper panels, and YFP fluorescence is shown in the bottom panels. White arrows show polar DivK-YFP. The percentage in each panel indicates the proportion of bacteria in which a polar DivK-YFP was detected. Scale bar, 2 μm.
The optical density of an exponentially growing culture of the TS strain remained stable after being switched from the permissive temperature (36°C) to the restrictive temperature (42°C). This suggests that pdhS is required for normal growth. Viability of the wild type and the TS strain was not affected at the restrictive temperature for 90 min or 6 h. Indeed, at 42°C in rich medium, 99% (n = 512) and 99.3% (n = 440) of the wild-type bacteria were alive, while the proportions of live bacteria of the TS strain were 98.8% (n = 388) and 99.5% (n = 414) after 90 min and 6 h of incubation, respectively. Interestingly, at the restrictive temperature for 90 min, 3 h, or 6 h in liquid culture, the morphology of the TS strain is very similar to the that of the wild-type strain (Fig. 1C) (data not shown); i.e., there were no morphological alterations. These data indicate that PdhS is required for growth, and its loss of function leads to a cell cycle arrest without generating aberrant morphologies in B. abortus, unlike in the pdhS overexpression strain (18).
PdhS controls DivK phosphorylation in vivo.
We previously proposed that PdhS could be involved in the B. abortus cell cycle regulation through the control of the response regulator DivK (18). In the predivisional cell of C. crescentus, the polar localization of DivK relies upon its phosphorylation state (25). To study the effect of PdhS on DivK control in B. abortus, we used the TS strain to observe the dynamic localization of DivK-YFP under different conditions. We introduced the low-copy-number plasmid pMR10cat-divK-yfp into the TS strain. At permissive temperature (36°C), the resulting strain showed 11.3% ± 1.5% (average ± standard deviation; n = 107) of polar DivK-YFP fusion (Fig. 1C). However, after 90 min at the restrictive temperature (42°C), the DivK-YFP polar localization is strongly decreased (2% ± 1%; n = 107). When the same culture returned to the permissive temperature (36°C), the DivK-YFP fusion regained its polar localization in 10% ± 1% of the bacteria (n = 129). As a control, we introduced the pMR10cat-divK-yfp plasmid into the wild-type strain and showed that DivK-YFP polar localization was retained at 42°C (data not shown).
PdhS is encoded by an essential gene, while the two other HKs potentially involved in DivK phosphorylation are encoded by nonessential genes (i.e., pleC and divJ) (16). Thus, if the essential feature of PdhS relies on DivK phosphorylation state, the divK gene should be essential, like divK in C. crescentus, and the phosphorylation of PdhS and DivK should also be essential. We constructed a nonphosphorylatable version of PdhS in which the evolutionarily conserved histidine (H805) was replaced by an alanine residue (H805A). Using an autophosphorylation assay in the presence of [γ-32P]ATP, we showed that the purified wild-type recombinant PdhS-His6 protein was able to autophosphorylate (Fig. 2A). On the contrary, the purified PdhSH805A-His6 protein did not incorporate radioactivity (Fig. 2A). This shows that PdhSH805A-His6 lost its autophosphorylation capacity while being soluble, suggesting that it was folded. Using the rescue plasmid carrying the wild-type copy of pdhS (pBBR1MCS-pdhS), we were able to obtain pdhS deletion (ΔpdhS) strains, which was not the case with the empty vector pBBR1MCS or the rescue plasmid pBBR1MCS-pdhSH805A (Table 2), showing that PdhS and its phosphorylation are essential. divK deletion (ΔdivK) strains were obtained with a rescue plasmid carrying the wild-type copy of divK (pMR10cat-divK), and no deletion events were obtained with the empty vector, showing that divK is essential in B. abortus (Table 2). The divKD53A copy (16) carried on a pMR10cat cannot rescue a divK deletion strain, demonstrating the essentiality of DivK phosphorylation in B. abortus. Altogether, this shows that divK gene is essential and that phosphorylation of both PdhS and DivK proteins is essential under the tested conditions. Those data are consistent with a model in which PdhS could regulate the B. abortus cell cycle through its ability to control the phosphorylation level of DivK in vivo.
Fig 2.
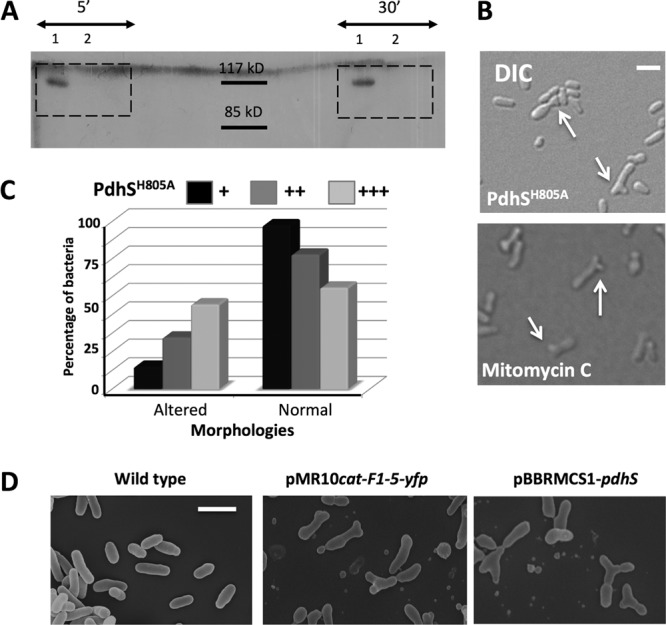
Several alterations of PdhS generate morphologies similar to cell division defects. (A) Autophosphorylation assay on PdhS and PdhSH805A. Recombinant PdhS-His6 (no. 1) and PdhSH805A-His6 (no. 2) proteins were used in the autophosphorylation assay, with incubations of 5 and 30 min with radioactively labeled [γ-32P]ATP as indicated. The expected size of PdhS-His6 is 111 kDa. (B) Altered morphologies generated by the overproduction of the nonphosphorylatable version of PdhS (PdhSH805A [upper panel]) and mitomycin C (lower panel), which blocks bacterial division. DIC, differential interference contrast. Scale bar, 1 μm. (C) Proportions of altered and normal morphologies when PdhSH805 is carried on an integrative plasmid (pSK; +) or on replicative plasmids, with either low (pMR; ++) or medium (pBBR; +++) copy numbers. (The number of bacteria counted for each condition was >400.) (D) Scanning electron microscopy of wild-type B. abortus (left panel), of cells producing PdhS F1-5 fused to YFP (middle panel; plasmid pMR10cat-F1-5-yfp), and cells overproducing PdhS (right panel; plasmid pBBR1MCS-pdhS). Scale bar, 2 μm.
Table 2.
Essentiality tests for the pdhS and divK genes
| Strain | No. of clones obtained for strain type with plasmida: |
|||||
|---|---|---|---|---|---|---|
| pBBR1MCS |
pMR10cat |
|||||
| −pdhS | +pdhS | +pdhSH805A | −divK | +divK | +divKD53A | |
| Wild type | 14 | 99 | 93 | 38 | 29 | 37 |
| ΔpdhS mutant | 0 | 7 | 0 | |||
| ΔdivK mutant | 0 | 14 | 0 | |||
The number of clones obtained for the three strains (either wild type, ΔpdhS, or ΔdivK), transformed with plasmids carrying (+) or not (−) pdhS or divK, is indicated. The pdhSH805A and divKD53A alleles correspond to unphosphorylatable versions of PdhS and DivK, respectively.
Production of a nonphosphorylatable PdhS protein generates dominant-negative effects.
PdhS loss of function results into a bacterial growth arrest. PdhS phosphorylation being essential, we attempted to observe phenotypes resulting from dominant-negative effects on PdhS activity. We characterized the effect of the expression of a pdhS allele inactivated by a point mutation. We inserted the pdhSH805A allele, from which a nonphosphorylatable version of PdhS is produced, on a pMR10cat vector in fusion with yfp. The PdhSH805A-YFP fusion was polarly localized in B. abortus and C. crescentus (data not shown), suggesting that PdhSH805A-YFP is properly folded in these bacteria. These data suggest that phosphorylation of PdhS is not essential for its polar localization.
Interestingly, a moderate overproduction of PdhSH805A-YFP led to generation of altered morphologies in B. abortus (Fig. 2B), while the wild-type copy of pdhS fused to yfp on the same vector generated wild-type morphologies (18). To see whether the level of PdhSH805A-YFP production could differently affect the B. abortus cell cycle, we integrated the pdhSH805A-yfp fusion as a single copy at the pdhS locus of B. abortus genome as well as on a low-copy-number plasmid and a medium copy-number plasmid, predicted to give moderate and high expression of pdhSH805A-yfp, respectively. The proportion of aberrant morphologies correlated with the predicted production level of the protein (Fig. 2C). It is interesting to note that the morphological alterations generated by the production of PdhSH805A are very similar to the cell shape produced by incubation of B. abortus with mitomycin C (Fig. 2B), a compound known to indirectly block cell division by cross-linking DNA (32). Those data are consistent with a dominant-negative effect of pdhSH805A-yfp on bacterial cell cycle progression, and particularly cell divison, through alteration of the endogenous PdhS function (see Discussion).
Defining putative subdomains in PdhS.
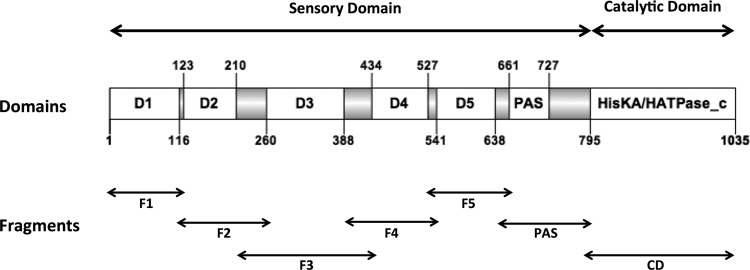
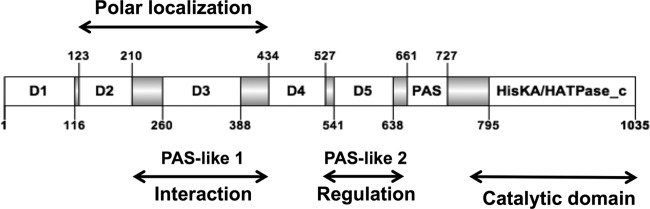
Because the dominant-negative effects on bacterial morphology were observed independently of the PdhS phosphorylation capacity, we attempted to identify the minimal PdhS portion that was still able to generate aberrant morphologies. The PdhS SD is unusually large compared to other HKs, being 795 aa long, whereas the SDs of PleC and DivJ from B. abortus are 543 aa and 154 aa long, respectively (16). While searching for clearly conserved motifs in the SD, a PAS domain (20) was found (residues 661 to 727, E value, 1.13e−9) (Fig. 3). Thus, to better characterize PdhS in B. abortus, we attempted to identify functional subdomains in its SD using a domain mapping approach. We performed multiple sequence alignments with B. abortus PdhS homologs in other alphaproteobacteria (Brucella ovis, Ochrobactrum anthropi, Agrobacterium tumefaciens, Sinorhizobium meliloti, and Mesorhizobium loti), and we identified a total of 7 conserved regions flanked by unconserved regions (UCRs) of different lengths (from 46 to 68 residues) with many insertions or deletions (Fig. 3). For the domain mapping approach, each conserved domain was cloned with its two flanking regions (altogether forming a “fragment”) in order to increase the probability of correct folding and stability. The combinations of domains are named from the first domain to the last. (For example, F1-3 contains the domains 1, 2, and 3, as well as all UCRs flanking them.)
Fig 3.
Structural organization of PdhS. Conserved domains are shown as open boxes, and the flanking unconserved regions (UCRs) are shown as gray boxes. The numbering of the residues flanking the different regions of the protein is indicated, as well as the position of the catalytic domain (HisKA/HATPase_c) and the PAS domain. D1 to D5 designate conserved subdomains without predicted functions. The protein fragments examined in the functional assays comprise the different conserved domains with their flanking UCRs.
Production of PdhS portions containing intact F5 generates aberrant morphologies.
The presence of the nonphosphorylatable version of PdhS results in dominant-negative effects on bacterial morphology, such as branched cells. We thus hypothesized that a regulation domain present in PdhS SD could alter endogenous PdhS activity. We attempted to identify more precisely the region responsible for this phenotype by expressing the cloned PdhS fragments fused to YFP on the low-copy-number vector pMR10cat in B. abortus. The production of fragments F1-5-YFP and F3-5-YFP generated aberrant bacterial shapes comparable to the altered morphologies resulting from the overexpression of pdhS gene, while fragments F1-4-YFP and F1-3-YFP had a wild-type phenotype (Fig. 2D) (data not shown), despite the polar localization of the expressed peptide (see Fig. 5). Moreover, the trimming of D5 in F1-5 (residues 1 to 613 instead of 1 to 661) did not generate numerous aberrant morphologies despite of its polar localization (18), further suggesting that intact or properly folded F5 could be necessary to modulate endogenous PdhS activity. Those data suggest that the presence of truncated PdhS proteins containing intact F5 correlates with the generation of altered morphologies and confirm the dominant-negative effect on PdhS that lead to cell division defects. We therefore propose that F5 contains a regulatory region of PdhS activity in B. abortus.
Fig 5.
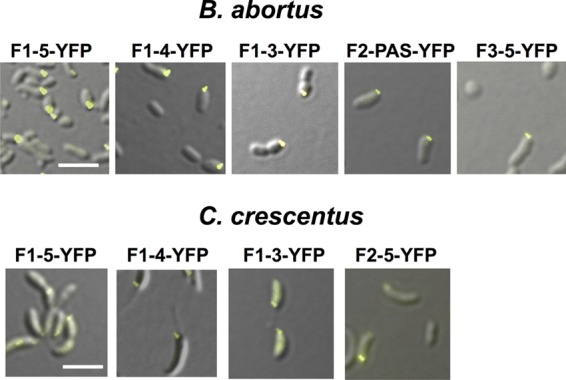
Localization assays. Shown are fragments of PdhS-YFP fusions that are able to polarly localize in B. abortus (upper panels) and in C. crescentus (lower panels). All fluorescence micrographs are merged differential interference contrast (DIC) and YFP images. Scale bar, 3 μm.
PdhS fragments interacting with PdhS and FumC.
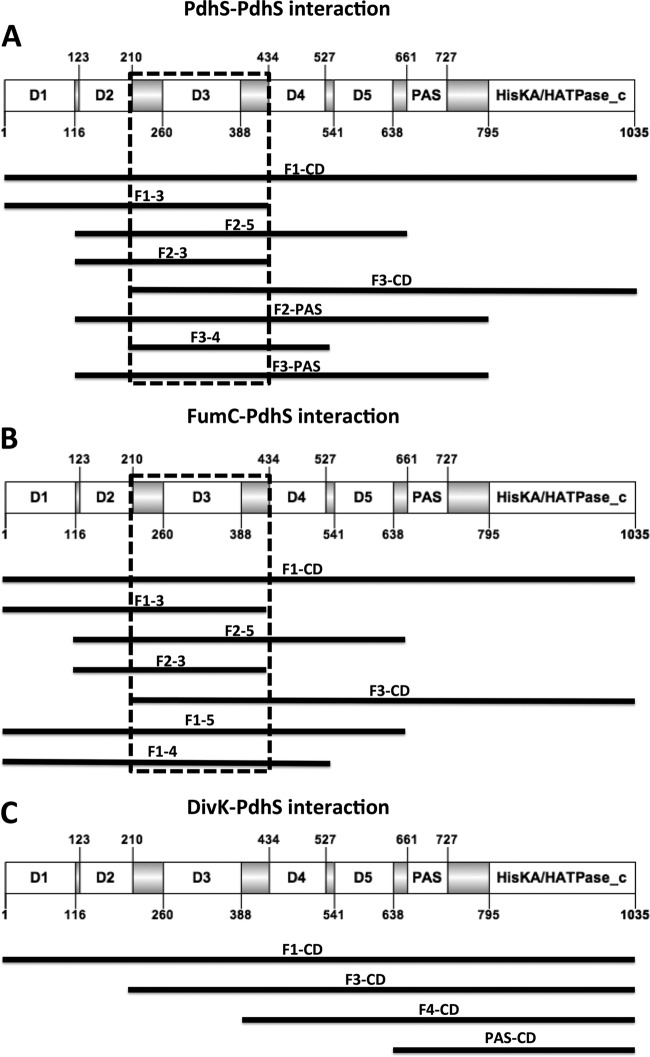
Using the yeast two-hybrid (Y2H) assay, a large portion of the PdhS SD (from residues 1 to 613) was previously shown to interact with the full-length PdhS protein (18) but also with the class II fumarase FumC (26). To better characterize the PdhS SD, we used the PdhS fragments to identify the minimal SD fragment required for these interactions. We fused all cloned PdhS fragments to the activating Gal4 activation domain, while the complete PdhS or FumC proteins were separately fused to the DNA binding domain of Gal4. The Y2H interactions were tested using HIS3 and URA3 reporters, on media lacking histidine or uracil, respectively (see Materials and methods).
The full-length PdhS (F1-CD) was used as a positive control. F3-CD, F2-5, F1-3, F2-PAS, F3-4, F3-PAS, and F2-3 gave a positive signal when PdhS itself was used as the prey in the Y2H assays (Fig. 4A). The common part of all of these fragments was F3, indicating that this portion of PdhS SD could be required for PdhS multimerization. However, when tested alone, F3 was negative in the Y2H assay (data not shown), a result that could be due to incorrect folding of the protein fragment or to the fact that F3 by itself is not sufficient for the interaction with PdhS.
Fig 4.
Interaction assay using the yeast two-hybrid (Y2H) approach. PdhS fragments that gave a positive interaction result are shown in black lines scaled to the PdhS primary structure. (A) PdhS fused to the DNA binding domain of the transcription factor Gal4 interacts with F1-CD (whole PdhS protein used as a positive control, from domain 1 to the catalytic domain), F1-3, F2-5, F2-3, F3-CD, F2-PAS, F3-4, and F3-PAS. The common fragment to all interacting fragments was F3 (residues 210 to 434). (B) FumC fused to the DNA binding domain of Gal4 interacts with the whole PdhS (F1-CD) and F1-3, F2-5, F2-3, F3-CD, F1-5, and F1-4. The common portion of the PdhS fragments interacting with FumC is F3 (residues 210 to 434). (C) PdhS fragments tested for their interaction with the response regulator DivK, reported to bind to the catalytic domain (18).
For the PdhS-FumC interaction assay, peptides corresponding to F1-CD, F1-5, F3-CD, F1-3, F2-3, F1-4, and F2-5 gave a positive interaction signal (Fig. 4B) which again identified the F3 as the common minimal fragment of PdhS that could be required to interact with FumC.
The RR DivK was shown to interact with the last 424 C-terminal residues of PdhS (18). We used this interaction as a positive control to test the production and the stability of truncated PdhS fragments in the Y2H assay (Fig. 4C). As expected, fragments containing the CD interacted with DivK, confirming that these fragments were properly produced and folded under the tested conditions. The fact that F1-CD and F3-CD retained the ability to interact with both PdhS and FumC while F4-CD and PAS-CD did not is consistent with the specific requirement of F3 for PdhS multimerization and FumC interaction.
PdhS minimal regions for polar localization.
We have shown that the phosphorylation capacity of PdhS was not required for its polar localization in B. abortus and C. crescentus. In addition, a portion of PdhS SD (residues 1 to 613) fused to the YFP produced from a low-copy-number replicative plasmid (pMR10cat) is polarly localized in B. abortus (18), indicating that this fragment contains the required information sufficient for this specific subcellular localization of the protein. To further restrict the protein region involved in polar localization, PdhS fragments were fused to YFP and expressed in B. abortus strain 544 from the pMR10cat plasmid. YFP fusions carrying the F1-5, F1-4, and F1-3 PdhS fragments were polarly localized in B. abortus (Fig. 5), showing 68%, 65%, and 64% polar localization, respectively (n > 200). This shows that subdomains D4 and D5 and PAS domains are not required for polar localization in B. abortus. Less frequent polar localization patterns were also observed with the production of fragments F2-PAS-YFP and F3-5-YFP. Taken together, the data suggest that the minimal protein determinant for PdhS polar localization in B. abortus lies within the F1-3 or the F3-5 fragments.
However, in B. abortus, endogenous polar PdhS may recruit the tested F1-3-YFP and F3-5-YFP since they contain the F3 segment, which was proposed to allow interaction with PdhS (Fig. 4A). To circumvent this possibility, we took advantage of the fact that B. abortus PdhS SD fused to YFP is polarly localized in C. crescentus (18), which does not have a homologue with an SD similar to PdhS. To test the minimal PdhS part required for polar localization in an in vivo background devoid of endogenous PdhS, we thus determined the localization of PdhS fragments in C. crescentus. Similarly to B. abortus, fragments F1-5-YFP, F1-4-YFP, and F1-3-YFP were polarly localized in C. crescentus. However, no polar fluorescent foci were observed when fragment F3-5-YFP was produced in C. crescentus. This indicates that the minimal portion of PdhS that is still able to be polarly localized in B. abortus and C. crescentus lies in the F1-3 fragment. The polar localization of F3-5-YFP in B. abortus but not in C. crescentus could be attributed to the presence of the F3 multimerization segment, allowing its recruitment by the endogenous PdhS. Additionally, fragment F2-5-YFP was polarly localized in C. crescentus. That fragment F2-PAS is polarly localized in B. abortus suggests the D1 domain is not strictly required for polar localization. We thus propose that the minimal portion of PdhS that polarly localized the protein in both B. abortus and C. crescentus resides within the F2-3 fragment, while the F1, F4, F5, and PAS fragments of the SD are not strictly required for polar localization. Altogether, the data generated by the domain mapping approach contribute to a better characterization of the PdhS SD.
DISCUSSION
We used the pentapeptide scanning mutagenesis method on an essential gene present in a class III pathogen to obtain a validated TS strain. We observed PdhS loss of function and a dominant-negative effect in B. abortus. The data presented here show that PdhS is involved in the control the bacterial cell cycle progression by regulating growth and division processes in B. abortus.
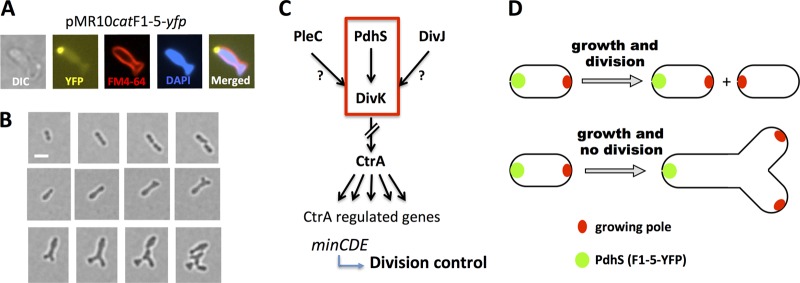
The aberrant morphologies generated by the presence of the nonphosphorylatable version of PdhS (PdhSH805A) can be due to the residual phosphatase activity since the mutation of the phosphorylatable conserved histidine has been shown to disrupt kinase activity but not the phosphatase activity of several HKs (22, 25). Moreover, the fragments containing F5 and the PdhSH805A form could titer PdhS partners in the cell. This dominant-negative effect can be correlated with the presence of the interacting fragment 3 that can permit the mutated protein to multimerize with endogenous wild-type protein. Overexpression of pdhS and production of fragment F1-5 or the H805A form all resulted in various morphological alterations, including the formation of Y-shaped bacteria (Fig. 2), which highlights a dominant-negative effect on cell division. Indeed, this shape is compatible with a cell division defect (37) without alteration of the growth capacity, since mitomycin C, a compound known to indirectly block cell division by cross-linking DNA, produced this effect as well (Fig. 2B). This observation is consistent with the previously reported formation of branches and swelling when cell division is impaired in Rhizobium meliloti (24). The TS strain at the restrictive temperature does not display these aberrations, probably because the growth problem (Fig. 1) prevents the generation of morphological alterations under these conditions. The mechanism allowing PdhS to control cell division is unknown, but one could predict that if PdhS controls DivK function, it could also indirectly control CtrA abundance and/or activity. In agreement with this hypothesis, CtrA abundance is decreased in a pdhS overexpression strain (18). Moreover, biochemical studies showed that CtrA∼P is able to bind to the minCDE promoter (1), and the MinD-YFP fusion protein was shown to oscillate in B. abortus (17), suggesting that it acts as an oscillator for FtsZ positioning, like in E. coli (8). Thus, by controlling DivK, CtrA, and Min, PdhS could indirectly control cell division in B. abortus (Fig. 6C). The recent report of unipolar growth in B. abortus and several other alphaproteobacteria (4) allows the generation of a simple model for the interpretation of the effect of mitomycin C and (over)expression of pdhS alleles. In this model (Fig. 6D), instead of having two growing poles in two sibling cells (thus one per cell, at the new pole, after cell division), the two growing foci remain in the nondividing mother cell, which starts to grow at two different places in the same cell, preferentially at the new pole (opposite to PdhS) (Fig. 6A, B, And D), therefore generating the Y-shaped bacteria.
Fig 6.
Generation of Y-shaped cells in B. abortus. (A) Differential interference contrast (DIC) and fluorescence microscopy of a representative Y-shaped cell generated by the production of F1-5-YFP. Cells were labeled with the FM4-64 and DAPI compounds (staining bacterial membranes and DNA, respectively) and observed using the fluorescent signals of YFP, FM4-64, and DAPI as indicated. (B) Time-lapse microscopy. Bacteria were placed on a slide layered with rich medium (2YT), and micrographs were taken every hour. B. abortus wild-type strain (upper panels) and B. abortus expressing the F1-5-YFP, Y-shaped cell generation (middle panels), and Y-shaped cell growth and division (lower panels). Scale bar, 1 μm. (C) Model of the regulatory pathway proposed for the interpretation of generation of altered morphologies. The red frame indicates the proposed essentiality pathway. (D) In a normal situation, without alteration of growth or division, each sibling cell inherits a new growing pole after division (shown above). If division is inhibited and growth still takes place, at a similar time or cell volume, two growing poles are also generated, but in the same cell and at the new pole, generating a Y shape (shown below).
We contributed to the characterization of the B. abortus essential HK PdhS using a domain mapping approach. We identified new parts in the SD required for (i) PdhS multimerization, (ii) PdhS interaction with the fumarase FumC, (iii) B. abortus cell cycle perturbation, and (iv) PdhS polar localization in B. abortus and C. crescentus (Fig. 7). This illustrates the complexity of the molecular functions of PdhS. A first observation of the domain mapping is the abundance of UCRs proposed to be linkers between domains and their length. This is striking for UCRs flanking D3, which are 50 and 46 aa long. It is of course not excluded that UCRs, and not only conserved domains, play a functional role. We could not rule out that a fragment folded in one assay is also folded in another one. This is why only PdhS fragments positive by at least one assay were taken into account for data interpretation, in an attempt to eliminate incorrectly folded or proteolyzed fragments that could give false-negative results.
Fig 7.
Fragments associated with PdhS functions. PdhS fragments proposed to be required for (i) polar localization, (ii) interaction with PdhS and FumC (“Interaction”), and (iii) generation of altered morphologies (“Regulation”) are indicated. Two additional PAS-like domains are predicted, one within D3 and one within D5. The catalytic domain interacts with DivK (18).
Fragment F3 (residues 210 to 434) is proposed to play a major role since it is required for PdhS multimerization, interaction with FumC, and localization. If the association with a polar structure mediates localization, then fragment 3 could be seen as an interaction platform in PdhS. The structure of D3 is unknown, but a weak similarity (E value of 10−3 after PSI-BLAST) was found with the PAS-B domain of the MmoS redox sensor of Methylococcus capsulatus (34). In MmoS, flanin adenine dinucleotide (FAD) is bound to the PAS-A domain but not to the PAS-B domain (34). Because PdhS is involved in B. abortus cell cycle regulation, the interaction between this HK and the citric acid cycle enzyme FumC is interesting since it could link cell cycle progression with the metabolic state of the bacterium. However, up to now this hypothesis has not been supported by experimental data. The presence of the fragments containing F5 correlates with the generation of altered morphologies in B. abortus. The D5 domain is weakly similar (E value of 4 × 10−4) to the LOV1 domain of Phot1 from Chlamydomonas reinhardtii, a flavin mononucleotide (FMN) binding domain (11). However, none of the residues involved in FMN binding are conserved, suggesting that D5 could have a similar fold without being able to bind FMN. The D5 domain is also weakly similar (E value of 10−3) to the 3MJQ structure in Protein Data Bank, an uncharacterized PAS domain with a fold resembling the one of the LOV1 domain. Structural predictions identified D3 and D5 as weak PAS-like domains, suggesting that there are at least three PAS-like domains in the SD of PdhS. Functional prediction is difficult because PAS-like domains fulfill highly diversified functions, therefore preventing the proposition of testable functional hypotheses about these domains.
The data presented here indicate that PdhS is involved in B. abortus cell cycle progression by regulating cell growth and division and highlight the complexity of this HK by associating functions with subdomains within the SD. The morphological asymmetry and the regulatory network involving the RR DivK and CtrA are conserved among several alphaproteobacteria (2, 3, 15, 33). Interestingly, the symbiotic bacterium Sinorhizobium meliloti contains a PdhS homolog named CbrA (12, 13). PdhS and CbrA share similarities in the SD, and suggested functions for PdhS activities and subdomains could be used to generate hypotheses about functions in CbrA. Because PdhS is asymmetrically localized at the time of division in B. abortus, being concentrated at the old pole of the mother cell (16), the two daughter cells could display differences regarding cell growth and cell division control.
ACKNOWLEDGMENTS
We thank the electron microscopy service of the University of Namur for help and advice.
C.V.D.H. and J.M. are FRIA-FNRS-FRS fellows. This work was funded by Concerted Research Actions 04/09-325 and 08/13-015, FRFC funds of the FRS-FNRS (grants 2.4521.04 and 2.4541.08), and the University of Namur.
Footnotes
Published ahead of print 27 July 2012
REFERENCES
- 1. Bellefontaine AF, et al. 2002. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol. Microbiol. 43:945–960 [DOI] [PubMed] [Google Scholar]
- 2. Bergmiller T, Ackermann M. 2011. Pole age affects cell size and the timing of cell division in Methylobacterium extorquens AM1. J. Bacteriol. 193:5216–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brilli M, et al. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst. Biol. 4:52 doi:10.1186/1752-0509-4-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown PJ, et al. 2012. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc. Natl. Acad. Sci. U. S. A. 109:1697–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown PJ, Hardy GG, Trimble MJ, Brun YV. 2009. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv. Microb. Physiol. 54:1–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. 2009. Dynamics of two phosphorelays controlling cell cycle progression in Caulobacter crescentus. J. Bacteriol. 191:7417–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Barsy M, et al. 2011. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell. Microbiol. 13:1044–1058 [DOI] [PubMed] [Google Scholar]
- 8. de Boer PA, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641–649 [DOI] [PubMed] [Google Scholar]
- 9. Dotreppe D, Mullier C, Letesson JJ, De Bolle X. 2011. The alkylation response protein AidB is localized at the new poles and constriction sites in Brucella abortus. BMC Microbiol. 11:257 doi:10.1186/1471-2180-11-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Evinger M, Agabian N. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fedorov R, et al. 2003. Crystal structures and molecular mechanism of a light-induced signaling switch: the Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys. J. 84:2474–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibson KE, Barnett MJ, Toman CJ, Long SR, Walker GC. 2007. The symbiosis regulator CbrA modulates a complex regulatory network affecting the flagellar apparatus and cell envelope proteins. J. Bacteriol. 189:3591–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gibson KE, Campbell GR, Lloret J, Walker GC. 2006. CbrA is a stationary-phase regulator of cell surface physiology and legume symbiosis in Sinorhizobium meliloti. J. Bacteriol. 188:4508–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hallet B, Sherratt DJ, Hayes F. 1997. Pentapeptide scanning mutagenesis: random insertion of a variable five amino acid cassette in a target protein. Nucleic Acids Res. 25:1866–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hallez L, Touyeras F, Hihn JY, Klima J. 2007. Energetic balance in an ultrasonic reactor using focused or flat high frequency transducers. Ultrason. Sonochem. 14:739–749 [DOI] [PubMed] [Google Scholar]
- 16. Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. 2004. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 12:361–365 [DOI] [PubMed] [Google Scholar]
- 17. Hallez R, Letesson JJ, Vandenhaute J, De Bolle X. 2007. Gateway-based destination vectors for functional analyses of bacterial ORFeomes: application to the Min system in Brucella abortus. Appl. Environ. Microbiol. 73:1375–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hallez R, et al. 2007. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J. 26:1444–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayes F, Hallet B. 2000. Pentapeptide scanning mutagenesis: encouraging old proteins to execute unusual tricks. Trends Microbiol. 8:571–577 [DOI] [PubMed] [Google Scholar]
- 20. Hayes F, Hallet B, Cao Y. 1997. Insertion mutagenesis as a tool in the modification of protein function. Extended substrate specificity conferred by pentapeptide insertions in the omega-loop of TEM-1 beta-lactamase. J. Biol. Chem. 272:28833–28836 [DOI] [PubMed] [Google Scholar]
- 21. Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165–170 [DOI] [PubMed] [Google Scholar]
- 22. Hsing W, Silhavy TJ. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 179:3729–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krell T, et al. 2010. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 64:539–559 [DOI] [PubMed] [Google Scholar]
- 24. Latch JN, Margolin W. 1997. Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J. Bacteriol. 179:2373–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. 2004. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118:579–590 [DOI] [PubMed] [Google Scholar]
- 26. Mignolet J, et al. 2010. PdhS, an old-pole-localized histidine kinase, recruits the fumarase FumC in Brucella abortus. J. Bacteriol. 192:3235–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Möglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moreno E, et al. 2006. The genus Brucella. Prokaryotes 5:315–456 [Google Scholar]
- 29. Purcell EB, Boutte CC, Crosson S. 2008. Two-component signaling systems and cell cycle control in Caulobacter crescentus. Adv. Exp. Med. Biol. 631:122–130 [DOI] [PubMed] [Google Scholar]
- 30. Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 31. Szurmant H, White RA, Hoch JA. 2007. Sensor complexes regulating two-component signal transduction. Curr. Opin. Struct. Biol. 17:706–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomasz M. 1995. Mitomycin C: small, fast and deadly (but very selective). Chem. Biol. 2:575–579 [DOI] [PubMed] [Google Scholar]
- 33. Tomlinson AD, Fuqua C. 2009. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr. Opin. Microbiol. 12:708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ukaegbu UE, Rosenzweig AC. 2009. Structure of the redox sensor domain of Methylococcus capsulatus (Bath) MmoS. Biochemistry 48:2207–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van der Henst C, et al. 2010. Overproduced Brucella abortus PdhS-mCherry forms soluble aggregates in Escherichia coli, partially associating with mobile foci of IbpA-YFP. BMC Microbiol. 10:248 doi:10.1186/1471-2180-10-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walhout AJ, Vidal M. 2001. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24:297–306 [DOI] [PubMed] [Google Scholar]
- 37. Wells VL, Margolin W. 2012. A new slant to the Z ring and bacterial cell branch formation. Mol. Microbiol. 84:199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wheeler RT, Shapiro L. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell 4:683–694 [DOI] [PubMed] [Google Scholar]
- 39. Wu J, Ohta N, Newton A. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 95:1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]