Abstract
Much of our knowledge of the initiation of DNA replication comes from studies in the Gram-negative model organism Escherichia coli. However, the location and structure of the origin of replication within the E. coli genome and the identification and study of the proteins which constitute the E. coli initiation complex suggest that it might not be as universal as once thought. The archetypal low-G+C-content Gram-positive Firmicutes initiate DNA replication via a unique primosomal machinery, quite distinct from that seen in E. coli, and an examination of oriC in the Firmicutes species Bacillus subtilis indicates that it might provide a better model for the ancestral bacterial origin of replication. Therefore, the study of replication initiation in organisms other than E. coli, such as B. subtilis, will greatly advance our knowledge and understanding of these processes as a whole. In this minireview, we highlight the structure-function relationships of the Firmicutes primosomal proteins, discuss the significance of their oriC architecture, and present a model for replication initiation at oriC.
INTRODUCTION
The Firmicutes are Gram-positive bacteria encompassing three major classes, Bacilli, Clostridia, and Mollicutes. They have a relatively low G+C content in their genomes and are morphologically and physiologically diverse. Rod-shaped bacilli, spherical cocci, and aerobic, anaerobic spore-forming, and non-spore-forming bacteria are found in this group. They likely represent the most ancestral phylum of prokaryotes, with high-G+C-content Gram-positive and Gram-negative bacteria having diverged from the Firmicutes at a later stage in evolution (13, 70). Bacillus subtilis is the best studied of the Firmicutes and is widely considered the Gram-positive model bacterium, but other bacilli, streptococci, staphylococci, and clostridia have been extensively studied because of their medical and industrial importance.
In addition to the universally conserved replication initiation protein DnaA (32, 65) and the replication restart protein PriA (17, 39), the low-G+C-content Firmicutes generally have two unique essential genes, dnaD and dnaB, coding for the replication initiation proteins DnaD and DnaB, respectively (Table 1) (note that DnaB in B. subtilis is unrelated to DnaB in Escherichia coli). In some cases—for example, in several Mollicutes—there are no distinct dnaD and dnaB genes, but there is, instead, a single gene annotated as dnaD-like which may combine both functions. No homologous proteins are found outside the Firmicutes, suggesting a replication initiation machinery distinctly different from those of other bacteria (31). In addition, there are a number of regulatory proteins which are not found in the Gram-negative model organism Escherichia coli, including YabA, Soj, SirA, and Spo0A, while other regulatory proteins are found in E. coli but not B. subtilis (these regulatory proteins have been subject to a recent review [29]). DnaA, DnaD, and DnaB, together with the helicase loader DnaI (called DnaC in E. coli), the replicative helicase DnaC (called DnaB in E. coli), and the primase DnaG, constitute the primosomal machinery in B. subtilis and related low-G+C-content Gram-positive bacteria (Table 1). Their primosomal activity was first described by the dependency of a single-stranded initiation site, ssiA, on these proteins to prevent accumulation of single-stranded DNA during rolling circle plasmid replication in B. subtilis and Staphylococcus aureus (7). During initiation of chromosomal replication in B. subtilis, they assemble at the origin, oriC, in a strictly hierarchical manner, starting with the association of the master replication initiation protein DnaA with oriC and the sequential recruitment of DnaD, DnaB, the DnaI-DnaC complex, and, finally, DnaG to complete the active primosome (9, 25, 36, 62, 72). The same ordered association of these proteins loads the replicative helicase during replication restart at sites other than oriC in a DnaA-independent but PriA-dependent manner (8, 9, 36, 41, 64). Also in Staphylococcus aureus, dnaD, dnaB, and dnaI were found to be important for replication initiation and restart (33, 34), though detailed information on the hierarchy in these processes is not available.
Table 1.
Comparison of proteins involved in replication initiation in the two model organisms Bacillus subtilis and Escherichia coli
| Function | Name(s) of protein(s) of the indicated type in each organism |
|
|---|---|---|
| B. subtilis | E. coli | |
| Replication initiation protein | DnaA | DnaA |
| Replicative helicase | DnaCa | DnaBa |
| Helicase loader | DnaIa | DnaCa |
| Primase | DnaG | DnaG |
| Accessory/remodeling protein | DnaB,a,b DnaDb | |
| Accessory protein | DiaA | |
| Remodeling factor | IHF, Fis | |
| Histone-like protein | HBsu | HU1, HU2 |
| Regulatory protein | Hda, SeqA | |
| Regulatory protein | YabA, Soj, SirA, Spo0A | |
| Primosomal protein | PriA | PriA |
| Primosomal accessory protein | PriB, PriC, DnaT | |
For historical reasons, the B. subtilis replicative helicase is called DnaC and the helicase loader is DnaI. B. subtilis DnaB and DnaC are unrelated to the E. coli DnaB and DnaC.
In several of the Mollicutes, there are not separate DnaB and DnaD proteins; there is a single DnaD-like protein which may combine functions from both.
MODULAR ARCHITECTURE OF THE DnaD AND DnaB PROTEINS
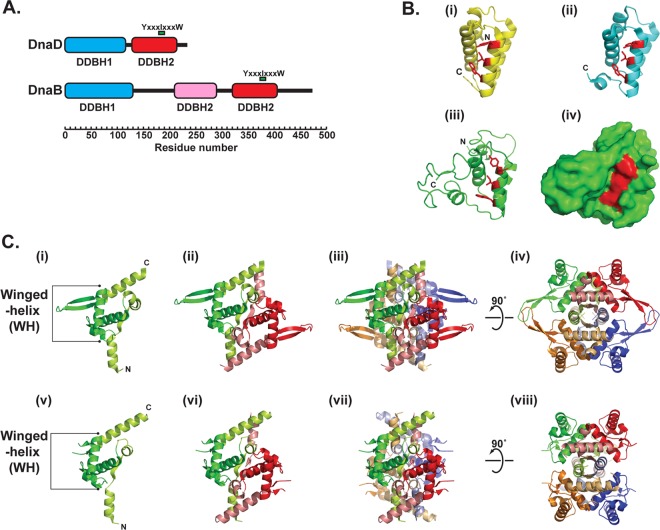
Bioinformatics analyses through hidden-Markov-model-based techniques revealed that DnaD and DnaB are structurally related (37). They have a modular architecture comprising two domains termed DDBH1 and DDBH2 (for DnaD DnaB homology 1 and 2, respectively). B. subtilis DnaD comprises two-domain DDBH1-DDBH2 architecture, whereas DnaB comprises three-domain DDBH1-DDBH2-DDBH2 architecture, with the middle DDBH2 domain being degenerate (20, 37, 46) (Fig. 1A). Atomic force and electron microscopy of B. subtilis DnaB revealed a tetrameric ring structure with one face of the ring comprising the DDBH1 domains forming a closed ring and the opposite face comprising the DDBH2 domains forming an open ring (46, 81).
Fig 1.
Modular architecture of DnaD and DnaB. (A) Domain structure of DnaD and DnaB. The extents of the DDBH1 and DDBH2 homology domains are shown, and the position of the conserved sequence YxxxIxxxW is marked in the DDBH2 domains. The pink DDBH2 domain of DnaB is a degenerate domain with a degenerate YxxxIxxxW motif (Y239LYGIDPLQ247). (B) Structure of the DnaD DDBH2 domain. Ribbon structures of the DnaD C-terminal domains from Enterococcus faecalis (PDB code 2I5U) (i), Streptococcus mutans (PDB code 2ZC2) (ii), and B. subtilis (iii). (iv) The residues of the conserved motif YxxxIxxxW are shown in red. Surface representation of the B. subtilis DnaD C-terminal domain, showing the conserved motif YxxxIxxxW in red. (C) Structure of the DnaD DDBH1 domain. Ribbon structures showing the Bacillus subtilis DnaD N-terminal domain as a monomer (i), dimer (ii), and tetramer (iii and iv) and the Geobacillus kaustophilus DnaD N-terminal domain as a monomer (v), dimer (vi), and tetramer (vii and viii). The winged-helix (WH) fold is identified on the monomer structure and is shown in the darker color in all representations.
Two crystal structures of DDBH1 from the B. subtilis (PDB accession code 2V79) and Geobacillus kaustophilus (PDB accession code 2vn2) DnaD proteins revealed a typical winged-helix (WH) fold with two structural extensions, a helix-strand-helix at the N terminus, and a single helix at the C terminus (24, 59) (Fig. 1C). The structures revealed the formation of dimers and tetramers via the strands of the helix-strand-helix (Fig. 1C). These N-terminal extensions form a small β-sheet in the dimer which then tightly interacts with another β-sheet from a second dimer, forming a striking threonine tetrad in the tetramer (59). Although a model has been proposed for a DDBH1 dimer interacting with double-stranded DNA via the WH fold (24), only a very weak interaction of DDBH1 with DNA has been detected by single-molecule atomic force spectroscopy experiments (82). This weak DNA binding is likely to be a residual DNA-binding activity of the WH fold and physiologically insignificant. Instead, the WH fold seems to be involved in higher-order oligomerization, rather than DNA binding, as the DDBH1 domain of B. subtilis DnaD forms large scaffolds. Indeed, the WH of B. subtilis DnaD is pronounced and relatively long compared to WH motifs in other proteins, and a truncation of the wing resulted in defective higher-order tetramer-tetramer interactions (59). Hence the WH of DDBH1 is somewhat related to the structurally homologous WH (21, 59, 68) that mediates oligomerization of the ESCRT II domains of the ESCRT (endosomal sorting complex required for transport) machinery involved in membrane deformation and scission in cytokinesis (1). It remains to be established whether DnaD plays an additional role in cell division.
Strong single-stranded and double-stranded DNA-binding activities reside in the DDBH2 domains of B. subtilis DnaD (11) and DnaB (20), suggesting that the DDBH2 domain, rather than the DDBH1 domain, is responsible for DNA binding in replication initiation proteins in Firmicutes. DDBH2 is annotated in Pfam as the DnaB_2 family (accession number PF07261). It is found in many bacterial and phage proteins of unknown function. In several cases, two DDBH2 domains are in tandem or linked to other domains, such as Rep_3 (PF01051), TrmB (PF01978), IstB_IS21 (PF01695), and others. In fact, 16 different architectures that include one or more DDBH2 domains are listed in Pfam. Two crystal structures of DDBH2 from DnaD homologous proteins from Streptococcus mutans UA 159 (PDB accession code 2ZC2) and from Enterococcus faecalis (PDB accession code 2I5U) have been deposited by the Midwest Center for Structural Genomics (Fig. 1B). Based upon the former, a structural model for the B. subtilis DDBH2 domain of DnaD was built and verified by nuclear magnetic resonance (NMR) (37) (Fig. 1B). Bioinformatics analyses, combined with protein NMR, mutagenesis, and genetic complementation studies, revealed that a highly conserved sequence motif within DDBH2, YxxxIxxxW, together with an unstable α-helix and part of a flexible C-terminal region, contributes to DNA binding (37) (Fig. 1B). Binding of DDBH2 to DNA induces the formation of high-order oligomers (11). Interestingly, the cyanobacterial Ftn6 protein, which functions in the recruitment of FtsZ and regulates assembly of the Z ring during cell division, has a 77-amino-acid N-terminal domain structurally homologous to DDBH2 (35).
INTERACTIONS WITH DNA DURING REPLICATION INITIATION/REINITIATION
The B. subtilis DnaD and DnaB proteins possess DNA-remodeling activities. DnaB laterally compacts DNA (81), while DnaD bends DNA, eliminates writhe, and converts it to negative twist, i.e., duplex unwinding (71, 80, 81, 82). Binding of the DnaD DDBH2 to DNA untwists the double helix, whereas the scaffolds formed via the DDBH1 interactions appear to reinforce the helix-untwisting activity by forming anchorage points for the DDBH2-DNA complexes (80, 82). As DnaD interacts directly with DnaA (26), it is attractive to speculate that the duplex untwisting activity of DnaD may enhance or stabilize the melting of the chromosomal origin, oriC, through an oriC-DnaA-DnaD complex during initiation of DNA replication.
It is not immediately obvious how the lateral DNA compaction activity of DnaB contributes to its putative function in a dual-helicase-loader system with DnaI (25, 72). Its association with oriC is dependent on a disordered C-terminal region (20) which may be subject to proteolysis or susceptible to degradation in a growth phase-dependent manner. Truncated DnaB is depleted from the B. subtilis oriC, while intact DnaB binds to oriC asymmetrically at the DNA-unwinding element (DUE) half, consistent with a role in helicase loading (20). Whether this is regulatory proteolysis and, if so, how it occurs is not clear. However, it might affect the double-stranded DNA-binding site located near the C terminus (between residues 365 and 428) that encompasses a strictly conserved YxxxIxxxW motif (residues 374 to 382) shown to be important in DNA binding in DnaB (our unpublished data) and DnaD (20, 37). DnaB interacts with DnaD (9, 56, 57) and with the replicative helicase DnaC (72). The latter interaction is also preserved when DnaC is in complex with the helicase loader DnaI (72), and DnaB may therefore act to bridge the oriC-DnaA-DnaD complex with the DnaC-DnaI complex during replication initiation. Loading of DnaC appears to be mediated by a ring assembly mechanism rather than through a preformed ring opening-closing mechanism, and DnaB is essential in this process, acting in a dual-loader system together with DnaI (25, 65, 72). Their functional cooperation is reflected by the genetic juxtaposition of the two genes within the same operon (6, 20).
FUNCTIONS RELATED TO DnaA
At least in B. subtilis, DnaD and DnaB are recruited to DnaA-binding sites outside the chromosomal origin of replication, whereas the DnaC helicase is not (63). The hierarchy at these secondary sites is conserved, with DnaA binding first and DnaD and DnaB binding thereafter (63). Though it is possible that the recruitment of the primosomal proteins is a consequence of their functional interaction at oriC, it is equally conceivable that this may serve a regulatory role. DnaA can act as a transcription factor at the secondary sites (5, 19), and perhaps the interaction with the primosomal proteins is important for this activity. In E. coli, the datA locus regulates replication by titrating away replication initiation proteins, and one can envisage that the secondary sites in B. subtilis may serve a similar regulatory role under conditions of replication stress. It remains to be established whether DnaD and DnaB also interact with DnaA at secondary sites in other organisms.
MEMBRANE ATTACHMENT
It has long been known that DNA replication in bacteria is a membrane-associated process (38), and the association of oriC with the membrane appears to be critical for replication initiation (4). In E. coli, DnaA is thought to bind directly to membranes (18, 54), although membrane association of oriC is mediated primarily via SeqA and SeqB (60). In contrast, membrane association in B. subtilis is dependent on DnaB (23, 66, 74), and there is no evidence for a direct association between DnaA and the membrane. Western blotting studies revealed that DnaB colocalizes with the membrane after fractionation (20, 56), and a mutation in dnaB that suppresses temperature sensitivity of dnaD and dnaB mutant cells was found to constitutively recruit DnaD to the membrane (56). No in vivo growth-dependent degradation of the C-terminal part of membrane-bound DnaB was observed, in contrast to cytosolic DnaB, for which degradation caused depletion from oriC (20). Its C terminus therefore seems protected in membrane-associated DnaB, and it is likely that this region contains the site of membrane attachment. Localization studies of green fluorescent protein fusions of DnaB and DnaD by fluorescence microscopy revealed a distinct membrane-proximal location for both proteins (40). Although DnaD is predominantly cytosolic (20), it has been proposed that membrane-bound DnaB recruits DnaD to the membrane, which, in turn, recruits the DnaA-oriC complex to the membrane, and this provides another level of control for replication initiation (56).
DNA REPAIR
The abundance of DnaD molecules in vivo, 3,000 to 5,000 molecules per cell for B. subtilis (9), suggests that DnaD may also play additional roles beyond its essential role in DNA replication. In many Firmicutes with dnaD-like genes, including bacilli, lactobacilli, staphylococci, acholeplasma, enterococci, streptococci, and clostridia, dnaD is juxtaposed in the same operon to an nth gene coding for an endonuclease III of the Nth/MutY family of DNA repair endonucleases involved in base excision repair (BER) (14). Deletion of nth in B. subtilis results in an H2O2-sensitive phenotype, whereas the dnaD operon is transiently activated by H2O2 in both B. subtilis and S. aureus and the dnaD mRNA levels remain relatively high compared to the marked reduction of the dnaB and dnaI mRNA levels upon H2O2 treatment (12, 14), suggesting a possible regulatory link between DnaD and DNA repair. DnaD is indirectly involved in the Nth-mediated repair of abasic DNA sites, as it alters the DNA topology, which stimulates the activity of Nth (14). Furthermore, mutations in S. aureus dnaD result in sensitivity to the DNA-cross-linking agent mitomycin C and to UV (33). The latter was correlated with a defect in the elongation step of DNA replication, likely linked to defects in replication restart. Based on these findings, it seems reasonable to assume that across species, DnaD may play a role in DNA repair as well as replication.
OriC ARCHITECTURE
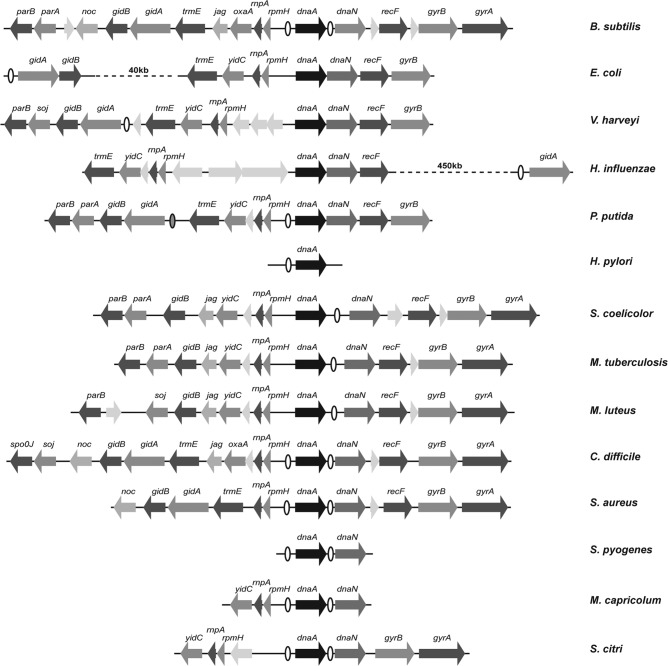
Chromosomal replication in bacteria starts from a single origin, oriC. The position of oriC on the bacterial chromosome is conserved across bacterial species and classes, with most found in close proximity to the dnaA gene encoding the DNA replication initiation protein DnaA (49, 77). The genes surrounding oriC and dnaA are also commonly conserved, consisting of the gene cluster rnpA-rpmH-dnaA-dnaN-recF-gyrB-gyrA, with oriC located in an intergenic region adjacent to dnaA (Fig. 2). DnaA binds to 9-bp conserved sequences, with multiple such sites forming DnaA-box clusters which are typically found at the origin as DnaA binds to these regions to initiate replication (Fig. 3). Origins also contain an AT-rich DUE close to the DnaA-box clusters where the initial unwinding occurs, allowing the loading of two bidirectional helicases, one on each strand, around which two complete replisomes subsequently assemble (65). B. subtilis has the consensus origin gene structure and has DnaA-box clusters in the intergenic regions both upstream and downstream of the dnaA gene, i.e., in the rpmH-dnaA and dnaA-dnaN regions, with the DUE in the dnaA-dnaN region (45, 47) (Fig. 2 and 3). Both upstream and downstream DnaA-box clusters are essential for origin function in B. subtilis (43). The DnaA-box cluster in the promoter region upstream of the dnaA gene also enables gene expression to be autoregulated: binding of DnaA to this DnaA-box cluster has been shown to decrease transcription and expression of the dnaA gene (50). B. subtilis has six other DnaA-box clusters (51, 63), one of which is close to the dnaA gene, 3 kb upstream in the thdF-jag intergenic region (77), but they are not essential for origin function. The consensus origin organization with the three DnaA-box clusters in B. subtilis is often described as the primordial DNA replication origin. This makes it an excellent model for understanding origin structure and regulation.
Fig 2.
Conservation of gene organization surrounding the dnaA/oriC region in different bacteria. Open ellipses indicate the DnaA-box clusters which form the origins of replication. The additional DnaA-box cluster which can function as an ars in P. putida is indicated by a filled ellipse.
Fig 3.

Map of the B. subtilis oriC region. Positions of the consensus DnaA-boxes (5′-TTATCCACA-3′) are shown as blue triangles, and DnaA-boxes with one mismatch are shown as green triangles. DnaA-boxes on the upper and lower strands are indicated as above and below the line, respectively. The position of the dnaA promoter and direction of transcription are identified by the bent arrow. The minimal region required for oriC function covers the three 16-bp AT-rich sequences upstream of dnaA (small yellow diamonds) to the 27-bp AT-rich DNA-unwinding element downstream of dnaA (large yellow diamond).
Replication initiation in E. coli is the most studied system, but it may not provide a good model for bacteria in general. The origin in E. coli and in closely related Gram-negative species, including Vibrio harveyi and Haemophilus influenzae, has undergone a major rearrangement so that the functional origin is no longer proximal to the dnaA gene, although the gene organization around dnaA has been retained (3, 48) (Fig. 2). A small DnaA-box cluster directly upstream of the dnaA gene is also retained: this has been shown to enable autoregulation of dnaA expression (2, 73). Gene analysis suggests that the E. coli origin is related to the nonessential DnaA-box cluster in the thdF-jag region of the B. subtilis chromosome. The Gram-negative bacteria Pseudomonas putida and Pseudomonas aeruginosa have their origins immediately upstream of the dnaA gene, in the rpmH-dnaA intergenic region, and have a much-shortened dnaA-dnaN intergenic region containing no DnaA-box clusters (61, 76) (Fig. 2). Interestingly, the DnaA-box cluster linked to thdF in P. putida and P. aeruginosa can function as an autonomously replicating sequence (ars) when subcloned onto a plasmid. The Helicobacter pylori origin region has undergone considerable rearrangement relative to the consensus, but oriC is still located directly upstream of the dnaA gene (79) (Fig. 2). The Gram-positive Actinobacteria (high G+C content) conform to the consensus origin structure found in B. subtilis. Studies have shown that the replication origins in Streptomyces coelicolor (10), Mycobacterium tuberculosis (53), and Micrococcus luteus (16) contain DnaA-box clusters in both the rpmH-dnaA and dnaA-dnaN intergenic regions, and the site of initiation is in the dnaA-dnaN region (Fig. 2). However, only the dnaA-dnaN region was required to achieve a functional origin when subcloned onto a plasmid, although the rpmH-dnaA DnaA-box clusters did have an effect on origin replication. As before, dnaA transcription was autoregulated by binding of DnaA to DnaA-box clusters in the dnaA promoter (28).
In addition to B. subtilis, the origins of chromosomal replication from several other Firmicutes have been characterized. The consensus origin organization was seen in the closely related Bacilli species Streptococcus pyogenes (67) and the more-distantly related Mollicutes species Mycoplasma capricolum (42) and Spiroplasma citri (55, 75). The identification of the minimal ars by subcloning onto a plasmid showed that both the rpmH-dnaA and dnaA-dnaN DnaA-box clusters are required for a functional origin in these bacteria, as is the case with B. subtilis, and it is postulated that this may be a common requirement of all the Firmicutes replication origins. However, the role of this “split origin” is unclear. Electron microscopy studies show that when B. subtilis DnaA (BsDnaA) was bound to the DnaA-box clusters upstream and downstream of the dnaA gene in B. subtilis oriC, the two regions interacted and caused the DNA to “loop” (30). This looping occurred both with a 2.5-kb linear origin fragment containing the two DnaA-box clusters separated by the dnaA gene and when the origin was on a supercoiled circular plasmid. The parallel experiment with a fragment of the E. coli origin showed a similar result, with loops forming between E. coli DnaA (EcDnaA) bound to a DnaA-box cluster at the origin and to a DnaA-box cluster consisting of two DnaA boxes 500 bp downstream of oriC. Surprisingly, EcDnaA was able to bind to the DnaA-box clusters in the B. subtilis origin to promote looping and at a much lower protein/origin ratio (20-fold less) than the cognate BsDnaA. In the reciprocal experiment, BsDnaA was able to bind to both DnaA-box clusters in the E. coli origin fragment but could not sustain a looped structure.
It is unlikely that looping at a split origin is absolutely essential for DnaA-mediated unwinding of the AT-rich DUE region. An open complex was observed when BsDnaA bound to a plasmid containing B. subtilis oriC with deleted upstream DnaA-box clusters (30), and experiments in other bacterial systems have demonstrated that the DnaA nucleoprotein filament alone is normally sufficient for the formation of an open complex (15, 52). Of course, this does not directly imply a functional open complex formed by DnaA alone and does not preclude a functional role for the split origin during DUE unwinding in vivo in B. subtilis and other Firmicutes. It has been suggested that the looped structures may be part of a regulatory system for initiation control (78), as observed in the replication of the plasmid R6K (58). However, the absolute requirement for both sets of DnaA-box clusters to form a functional plasmid replication origin would allude to more than a purely regulatory function. It may be that the two DnaA–DnaA-box cluster complexes are involved in loading the two replisomes, with one loading the first replisome on the leading strand and the other loading the second replisome on the lagging strand. Whatever the purpose of the split origin, it is unlikely that random diffusion would be sufficient to efficiently bring the distant DnaA filaments together to form a functional origin in vivo.
The correlation between the conservation of DnaD, a protein which both interacts with DnaA and bends DNA to form loops, and the split-origin structure in the Firmicutes may be more than coincidence, especially in light of recent studies on the HobA protein of Helicobacter pylori. HobA is a DnaA-binding protein which is a functional counterpart of diaA in E. coli, and it has been proposed to form a molecular scaffold onto which regular oligomers of DnaA can assemble (69). DnaD may function in a similar manner to bridge the DnaA molecules bound to the DnaA-box clusters in the Firmicutes split origin, bringing them together to form the functional loop structure (Fig. 4). In addition, the C-terminal domain of DnaD binds directly to DNA, enabling it to form a scaffold bridging different areas of the DNA to create loops and stabilize loops once they have formed. This would form a functional open complex at oriC, increase the frequency and stability of loop formation, and, thus, promote efficient chromosome replication. This model depends on not only the high intracellular concentration of DnaD, estimated at 3,000 to 5,000 molecules per cell (9), but also the efficient targeting of DnaD to the origin, initially via its interaction with DnaA and then via the growing DnaD scaffold.
Fig 4.

Speculative model of possible roles for DnaD in the initiation of chromosome replication of B. subtilis and other Firmicutes. (i) DnaA binds to the DnaA-boxes in the origin region. DnaA-bound DnaD may assist in the recruitment of additional DnaA and stabilize the binding to nonperfect DnaA boxes. (ii) DnaD bound to the DnaA nucleoprotein filament may recruit additional DnaD which invades and binds to the adjacent DNA region. (iii) As the DnaD scaffold extends into the DNA between the DnaA-boxes, it may cause the DNA to bend. (iv) This would have the effect of bringing the two DnaA nucleoprotein filaments together such that they can interact. DnaD may stabilize this interaction, both by forming bridges between the two DnaA complexes and by stabilizing the looped DNA structure through an extended supporting scaffold. (v) DnaB is recruited to the DnaA complex, and the AT-rich region forming the DUE begins to unwind. DnaD may assist in the recruitment of DnaB and stabilize this interaction. The DnaD-induced conversion of DNA writhe into negative twist may also assist in the melting of the DUE region. (vi) The DnaC-DnaI complex is recruited to the unwound DNA by DnaA and DnaB, and replication is initiated. A possible explanation for the requirement for the two DnaA nucleoprotein filaments is that one filament is responsible for loading DnaC-DnaI onto one unwound DNA strand in cis while the other is responsible for loading DnaC-DnaI onto the other unwound DNA strand in trans.
THREE PROTEINS ARE BETTER THAN ONE?
It is interesting to consider why the Firmicutes have the complexity of the additional proteins DnaB and DnaD and a dual origin when DnaA alone and a single origin are sufficient for E. coli. The most likely explanation is that a three-protein system provides additional levels of regulation and control which may be advantageous. The timing of replication initiation in E. coli appears to be directly controlled by the levels of intracellular DnaA (29). Recent studies with a small-cell mutant of E. coli with a delayed replication cycle showed that increasing DnaA levels alleviated the delay (22). In contrast, replication initiation was neither delayed in a small-cell mutant of B. subtilis nor advanced by additional DnaA. The prevailing opinion is that the DnaA concentration is not the limiting factor in B. subtilis (44, 48, 51), and increasing intracellular DnaA may actually inhibit replication (48). If the timing of replication initiation is no longer directly dependent on DnaA, then additional factors, such as DnaB and DnaD, may have a role in controlling this critical process. DnaA may also function as a master regulator: the expression of 34 genes (including dnaA) throughout the B. subtilis genome has been shown to be regulated by DnaA binding at DnaA-box clusters outside the oriC region (27). These genes include some associated with tolerance to replication stress, while others are associated with the control of the sporulation cycle. The fact that DnaB and DnaD are recruited to many of these sites suggests that they may also modulate DnaA activity at the non-oriC DnaA-box clusters (63). The additional levels of control exerted by DnaB and DnaD on DnaA activity, both at the origin and elsewhere on the genome, may well increase the ability of Firmicutes such as B. subtilis to thrive in a variety of environments and respond to a wider range of conditions where E. coli cannot.
ACKNOWLEDGMENTS
Research in the P.S. lab is supported by a Wellcome Trust grant (091968/Z/10/Z). W.K.S. is supported by a Veni Fellowship of the Netherlands Organization for Scientific Research (NWO-ZonMW) and a Gisela Thier Fellowship of the LUMC.
Biographies

Geoffrey S. Briggs obtained his B.Sc. (Biochemistry) from the University of York in 1990 and his Ph.D. from the University of Kent at Canterbury in 1994. He held positions as a postdoctoral research associate at the University of Nottingham and the University of Leicester before joining the group of Professor Bob Lloyd at Nottingham in 2001, where he investigated the structure-function relationships of proteins involved in DNA recombination, repair, and replication in Escherichia coli. In 2011, he moved to the group of Professor Panos Soultanas, where he continued studies in the same general area, with the emphasis on the proteins and processes involved in the initiation of DNA replication in the bacterium Bacillus subtilis.

Wiep Klaas Smits obtained his M.Sc. and Ph.D. degrees from the Faculty of Mathematics and Natural Sciences of the University of Groningen, the Netherlands, in 2002 and 2007, respectively. After obtaining his Ph.D., he worked as a postdoctoral associate and fellow at the Department of Biology of the Massachusetts Institute of Technology in Cambridge (MA), where he studied DNA replication initiation in Bacillus subtilis. Since 2010, he has worked as a principal investigator at the Leiden University Medical Center in Leiden, the Netherlands, where one of his research lines involves the identification and characterization of the DNA replication machinery of Clostridium difficile in relation to that of other Firmicutes.

Panos Soultanas is a Professor of biochemistry and biological chemistry. He obtained his B.Sc. (biochemistry/microbiology) and Ph.D. degrees from the Department of Molecular Biology and Biotechnology at the University of Sheffield, United Kingdom, in 1987 and 1991, respectively. He worked as a postdoctoral research assistant at the University of Bristol (1991 to 1994), the University of Crete (1995 to 1996), and Oxford University (1996 to 2000), where he carried out research on site-specific recombination, DNA helicases, DNA primases, and DNA replication. In 2000, he was appointed to a Lectureship at the School of Chemistry, University of Nottingham, United Kingdom. He was promoted to a Readership in 2004 and to a Chair in 2008. In 2012, he was elected as a Fellow of the Society of Biology (FSB). His main area of research is structure-function relationships of DNA replication enzymes and DNA replication and repair in the Gram-positive model of the Firmicutes species Bacillus subtilis.
Footnotes
Published ahead of print 13 July 2012
REFERENCES
- 1. Adell MAY, Teis D. 2011. Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett. 585:3191–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atlung T, Clausen ES, Hansen FG. 1985. Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet. 200:442–450 [DOI] [PubMed] [Google Scholar]
- 3. Berenstein D, Olesen K, Speck C, Skovgaard O. 2002. Genetic organization of the Vibrio harveyi dnaA gene region and analysis of the function of the V. harveyi DnaA protein in Escherichia coli. J. Bacteriol. 184:2533–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boeneman K, Crooke E. 2005. Chromosomal replication and the cell membrane. Curr. Opin. Microbiol. 8:143–148 [DOI] [PubMed] [Google Scholar]
- 5. Breier AM, Grossman AD. 2009. Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J. Bacteriol. 191:486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruand C, Ehrlich SD. 1995. The Bacillus subtilis dnaI gene is part of the dnaB operon. Microbiology 141:1199–1200 [DOI] [PubMed] [Google Scholar]
- 7. Bruand C, Ehrlich SD, Janniére L. 1995. Primosome assembly site in Bacillus subtilis. EMBO J. 14:2642–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruand C, Farache M, McGovern S, Ehrlich SD, Polard P. 2001. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol. Microbiol. 42:245–255 [DOI] [PubMed] [Google Scholar]
- 9. Bruand C, et al. 2005. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol. Microbiol. 55:1138–1150 [DOI] [PubMed] [Google Scholar]
- 10. Calcutt MJ, Schmidt FJ. 1992. Conserved gene arrangement in the origin region of the Streptomyces coelicolor chromosome. J. Bacteriol. 174:3220–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carneiro MJVM, et al. 2006. The DNA-remodelling activity of DnaD is the sum of oligomerization and DNA-binding activities on separate domains. Mol. Microbiol. 60:1365–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang W, Small DA, Toghrol F, Bentley WE. 2006. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188:1648–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ciccarelli FD, et al. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287 [DOI] [PubMed] [Google Scholar]
- 14. Collier C, Machón C, Briggs GS, Smits WK, Soultanas P. 2012. Untwisting of the DNA helix stimulates the endonuclease activity of Bacillus subtilis Nth at AP sites. Nucleic Acids Res. 40:739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erzberger JP, Mott ML, Berger JM. 2006. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13:676–683 [DOI] [PubMed] [Google Scholar]
- 16. Fujita MQ, Yoshikawa H, Ogasawara N. 1990. Structure of the dnaA region of Micrococcus luteus: conservation and variations among eubacteria. Gene 93:73–78 [DOI] [PubMed] [Google Scholar]
- 17. Gabbai CB, Marians KJ. 2010. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair 9:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garner J, Durrer P, Kitchen J, Brunner J, Crooke E. 1998. Membrane-associated release of nucleotide from an initiator of chromosomal replication, Escherichia coli DnaA, occurs with insertion of a distinct region of the protein into the lipid bilayer. J. Biol. Chem. 273:5167–5173 [DOI] [PubMed] [Google Scholar]
- 19. Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. 2005. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. U. S. A. 102:12932–12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grainger WH, Machón C, Scott DJ, Soultanas P. 2010. DnaB proteolysis in vivo regulates oligomerization and its localization at oriC in Bacillus subtilis. Nucleic Acids Res. 38:2851–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hierro A, et al. 2004. Structure of the ESCRT-II endosomal trafficking complex. Nature 431:221–225 [DOI] [PubMed] [Google Scholar]
- 22. Hill NS, Kadoya R, Chattoraj DK, Levin PA. 2012. Cell size and the initiation of DNA replication in bacteria. PLoS Genet. 8:e1002549 doi:10.1371/journal.pgen.1002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoshino T, McKenzie T, Schmidt S, Tanaka T, Sueoka N. 1987. Nucleotide sequence of Bacillus subtilis dnaB: a gene essential for DNA replication initiation and membrane attachment. Proc. Natl. Acad. Sci. U. S. A. 84:653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang CY, Chang YW, Chen WT. 2008. Crystal structure of the N-terminal domain of Geobacillus kaustophilus HTA426 DnaD protein. Biochem. Biophys. Res. Comm. 375:220–224 [DOI] [PubMed] [Google Scholar]
- 25. Ioannou C, Schaeffer PM, Dixon N, Soultanas P. 2006. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 34:52475258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishigo-Oka D, Ogasawara N, Moriya S. 2001. DnaD protein of Bacillus subtilis interacts with DnaA, the initiator protein of replication. J. Bacteriol. 183:2148–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishikawa S, et al. 2007. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 14:155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jakimowicz D, et al. 2000. Structure and regulation of the dnaA promoter region in three Streptomyces species. Mol. Gen. Genet. 262:1093–1102 [DOI] [PubMed] [Google Scholar]
- 29. Katayama T, Ozaki S, Keyamura K, Fujimitsu K. 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8:163–170 [DOI] [PubMed] [Google Scholar]
- 30. Krause M, Ruckert B, Lurz R, Messer W. 1997. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 274:365–380 [DOI] [PubMed] [Google Scholar]
- 31. Lemon KP, Moriya S, Ogasawara N, Grossman AD. 2002. Chromosome replication and segregation, p 73–86 In Sonenshein AL, Hock JA, Losick R. (ed), Bacillus subtilis and its closest relatives. ASM Press, Washington, DC [Google Scholar]
- 32. Leonard AC, Grimwade JE. 2011. Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 65:19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Y, et al. 2004. Identification of temperature-sensitive dnaD mutants of Staphylococcus aureus that are defective in chromosomal DNA replication. Mol. Genet. Genomics 271:447–457 [DOI] [PubMed] [Google Scholar]
- 34. Li Y, et al. 2007. DnaB and DnaI temperature-sensitive mutants of Staphylococcus aureus: evidence for involvement of DnaB and DnaI in synchrony regulation of chromosome replication. Microbiology 153:3370–3379 [DOI] [PubMed] [Google Scholar]
- 35. Marbouty M, Saguez C, Chauvat F. 2009. The cyanobacterial cell division factor Ftn6 contains an N-terminal DnaD-like domain. BMC Struct. Biol. 9:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marsin S, McGovern S, Ehrlich SD, Bruand C, Polard P. 2001. Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem. 276:45818–45825 [DOI] [PubMed] [Google Scholar]
- 37. Marston FY, et al. 2010. When simple sequence comparison fails: the cryptic case of the shared domains of the bacterial replication initiation proteins DnaB and DnaD. Nucleic Acids Res. 38:6930–6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marvin DA. 1968. Control of DNA replication by membrane. Nature 219:485–486 [DOI] [PubMed] [Google Scholar]
- 39. Masai H, Tanaka T, Kohda D. 2010. Stalled replication forks: making ends meet for recognition and stabilization. Bioessays 32:687–697 [DOI] [PubMed] [Google Scholar]
- 40. Meile JC, Wu LJ, Ehrlich SD, Errington J, Noirot P. 2006. Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory. Proteomics 6:2135–2146 [DOI] [PubMed] [Google Scholar]
- 41. Merrikh H, Machón C, Grainger WH, Grossman AD, Soultanas P. 2011. Co-directional replication-transcription conflicts lead to replication restart. Nature 470:554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miyata M, Sano K, Okada R, Fukumura T. 1993. Mapping of replication initiation site in Mycoplasma capricolum genome by two-dimensional gel-electrophoretic analysis. Nucleic Acids Res. 21:4816–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moriya S, Atlung T, Hansen FG, Yoshikawa H, Ogasawara N. 1992. Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol. Microbiol. 6:309–315 [DOI] [PubMed] [Google Scholar]
- 44. Moriya S, Imai Y, Hassan AKM, Ogasawara N. 1999. Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid 41:17–29 [DOI] [PubMed] [Google Scholar]
- 45. Moriya S, Ogasawara N, Yoshikawa H. 1985. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. III. Nucleotide sequence of some 10,000 base pairs in the origin region. Nucleic Acids Res. 13:2251–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Núñez-Ramírez R, et al. 2007. Loading a ring: structure of the Bacillus subtilis DnaB protein, a co-loader of the replicative helicase. J. Mol. Biol. 367:764–769 [DOI] [PubMed] [Google Scholar]
- 47. Ogasawara N, Moriya S, von Meyenburg K, Hansen FG, Yoshikawa H. 1985. Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 4:3345–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogasawara N, Moriya S, Yoshikawa H. 1991. Initiation of chromosome replication: structure and function of oriC and DnaA protein in eubacteria. Res. Microbiol. 142:851–859 [DOI] [PubMed] [Google Scholar]
- 49. Ogasawara N, Yoshikawa H. 1992. Genes and their organization in the replication origin region of the bacterial chromosome. Mol. Microbiol. 6:629–634 [DOI] [PubMed] [Google Scholar]
- 50. Ogura Y, Imai Y, Ogasawara N, Moriya S. 2001. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 183:3833–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Okumura H, et al. 2012. Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 40:220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ozaki S, et al. 2008. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283:8351–8362 [DOI] [PubMed] [Google Scholar]
- 53. Qin MH, Madiraju MV, Rajagopalan M. 1999. Characterization of the functional replication origin of Mycobacterium tuberculosis. Gene 233:121–130 [DOI] [PubMed] [Google Scholar]
- 54. Regev T, Myers N, Zarivach R, Fishov I. 2012. Association of the chromosome replication initiator DnaA with the Escherichia coli inner membrane in vivo: quantity and mode of binding. PLoS One 7:e36441 doi:10.1371/journal.pone.0036441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Renaudin J, et al. 1995. Integrative and free Spiroplasma citri oriC plasmids: expression of the Spiroplasma phoeniceum spiralin in Spiroplasma citri. J. Bacteriol. 177:2870–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rokop M, Auchtung JM, Grossman AD. 2004. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 52:1757–1767 [DOI] [PubMed] [Google Scholar]
- 57. Rokop M, Grossman AD. 2009. Intragenic and extragenic suppressors of temperature sensitive mutations in the replication initiation genes dnaD and dnaB of Bacillus subtilis. PLoS One. 4:e6774 doi:10.1371/journal.pone.0006774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saxena M, Abhyankar M, Bastia D. 2010. Replication initiation at a distance: determination of the cis- and trans-acting elements of replication origin alpha of plasmid R6K. J. Biol. Chem. 285:5705–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schneider S, Zhang W, Soultanas P, Paoli M. 2008. Structure of the N-terminal oligomerization domain of DnaD reveals a unique tetramerization motif and provides insights into scaffold formation. J. Mol. Biol. 376:1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shakibai N, et al. 1998. High-affinity binding of hemimethylated oriC by Escherichia coli membranes is mediated by a multiprotein system that includes SeqA and a newly identified factor, SeqB. Proc. Natl. Acad. Sci. U. S. A. 95:11117–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith DW, Yee TW, Baird C, Krishnapillai V. 1991. Pseudomonad replication origins: a paradigm for bacterial origins? Mol. Microbiol. 5:2581–2587 [DOI] [PubMed] [Google Scholar]
- 62. Smits WK, Goranov A, Grossman AD. 2010. Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol. Microbiol. 75:452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smits WK, Merrikh H, Bonilla CY, Grossman AD. 2011. Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J. Bacteriol. 193:640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Soultanas P. 2011. The replication-transcription conflict. Transcription 2:140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Soultanas P. 2012. Loading mechanisms of ring helicases at replication origins. Mol. Microbiol. 84:6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sueoka N. 1998. Cell membrane and chromosome replication in Bacillus subtilis. Prog. Nucleic Acids Res. Mol. Biol. 59:35–53 [DOI] [PubMed] [Google Scholar]
- 67. Suvorov AN, Ferretti JJ. 2000. Replication origin of Streptococcus pyogenes, organization and cloning in heterologous systems. FEMS Microbiol. Lett. 189:293–297 [DOI] [PubMed] [Google Scholar]
- 68. Teo H, Perisic O, Gonzalez B, Williams RL. 2004. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell 7:559–569 [DOI] [PubMed] [Google Scholar]
- 69. Terradot L, Zawilak-Pawlik A. 2010. Structural insight into Helicobacter pylori replication initiation. Gut Microbes 1:330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tocheva EI, et al. 2011. Peptidoglycan remodelling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146:799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Turner IJ, Scott DJ, Allen S, Roberts CJ, Soultanas P. 2004. The Bacillus subtilis DnaD protein: a putative link between DNA remodelling and initiation of DNA replication. FEBS Lett. 577:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Velten M, et al. 2003. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell 11:1009–1020 [DOI] [PubMed] [Google Scholar]
- 73. Wang QP, Kaguni JM. 1987. Transcriptional repression of the dnaA gene of Escherichia coli by DnaA protein. Mol. Gen. Genet. 209:518–525 [DOI] [PubMed] [Google Scholar]
- 74. Winston S, Sueoka N. 1980. DNA-membrane association is necessary for initiation of chromosomal and plasmid replication in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 77:2834–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ye F, Renaudin J, Bove JM, Laigret F. 1994. Cloning and sequencing of the replication origin (oriC) of the Spiroplasma citri chromosome and construction of autonomously replicating artificial plasmids. Curr. Microbiol. 29:23–29 [DOI] [PubMed] [Google Scholar]
- 76. Yee TW, Smith DW. 1990. Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc. Natl. Acad. Sci. U. S. A. 87:1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yoshikawa H, Ogasawara N. 1991. Structure and function of DnaA and the DnaA-box in eubacteria: evolutionary relationships of bacterial replication origins. Mol. Microbiol. 5:2589–2597 [DOI] [PubMed] [Google Scholar]
- 78. Yoshikawa H, Wake RG. 1993. Initiation and termination of chromosome replication, p 507–528 In Sonenshein A, Hoch JA, Losick R. (ed), Bacillus subtilis and other Gram-positive bacteria: biochemistry, physiology and molecular genetics. ASM Press, Washington, DC [Google Scholar]
- 79. Zawilak A, et al. 2001. Identification of a putative chromosomal replication origin from Helicobacter pylori and its interaction with the initiator protein DnaA. Nucleic Acids Res. 29:2251–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang W, Allen S, Roberts CJ, Soultanas P. 2006. The Bacillus subtilis primosomal protein DnaD untwists supercoiled DNA. J. Bacteriol. 188:5487–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang W, et al. 2005. The Bacillus subtilis DnaD and DnaB proteins exhibit different DNA remodelling activities. J. Mol. Biol. 351:66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang W, et al. 2008. Single-molecule atomic force spectroscopy reveals that DnaD forms scaffolds and enhances duplex melting. J. Mol. Biol. 377:706–714 [DOI] [PMC free article] [PubMed] [Google Scholar]