Abstract
We investigated the effect of HLA class I alleles on clinical parameters for HIV-1 disease progression in the Japanese population, where two strongly protective alleles, HLA-B*57 and HLA-B*27, are virtually nonexistent. HLA-B alleles showed a dominant role, primarily through HLA-B*67:01 and the HLA-B*52:01-C*12:02 haplotype. Neither a rare-allele nor a heterozygote advantage was found, suggesting that the effect of HLA alleles in the Japanese population is either different from those observed in Africans and Caucasians or undetectable due to limited power.
TEXT
The presence of particular HLA class I alleles is associated with the rate of progression to AIDS and/or with clinical markers of disease progression, such as viral load (VL) and CD4 T cell count (2, 3, 7, 11, 13). HLA-B*57 and HLA-B*27 are well-known to associate with successful control of HIV-1 or slow progression to disease in Caucasians and Africans (1, 4, 5, 9–11) but not in Asians, since the frequencies of these alleles are very low (<1%) in this population. There is growing evidence that HIV-1-specific CTL responses restricted by HLA-B allotypes play a key role in determining disease outcomes relative to HLA-A and -C (7, 9), but it remains possible that the association between HLA-B and HIV-1 control is biased by the extremely strong effects of only a few HLA-B alleles, particularly the protective HLA-B*57 and -B*27 alleles and the detrimental HLA-B*58:02 and -B*35:02/B*35:03 alleles (6–8). The exceptional protection afforded by HLA-B*57 and -B*27 may mask weaker effects of other HLA alleles, in part due to the potential for inappropriate correction for the effects of these two alleles. Since both of these alleles are virtually nonexistent (i.e., present in 0% and 0.5% of the population, respectively) in Japan (12), studies of Japanese cohorts provide an unbiased means of determining the effects of other HLA alleles on HIV control. Therefore, we analyzed the association of HLA alleles with markers of disease progression, VL and CD4 count, in Japanese individuals chronically infected with HIV-1.
We recruited 504 chronically HIV-1-infected, antiretroviral therapy (ART)-naïve Japanese individuals (464 men and 40 women) who visited our hospital during 2000-2010 and who did not meet any criteria for clinical AIDS. The median (interquartile ranges) VL and CD4 count are 35,000 (9,175/100,000) and 288 (183/402), respectively. HLA alleles of these individuals were determined at the 4-digit level. Associations between HLA alleles and VL or CD4 count obtained at the first visit were determined among these individuals. We considered 48 HLA class I alleles occurring at a frequency of greater than 1% (11 HLA-A, 22 HLA-B, and 15 HLA-C), which excluded both B*57 (0 identified) and B*27:05 (1 identified) (see Table S1 in the supplemental material). The differential contribution of HLA-A, -B, and -C alleles on VL in the cohort as a whole was determined using the Kruskal-Wallis statistic H, which is a nonparametric measure of variation between data groups (i.e., among alleles at a given locus). The range of effects (protective to susceptible) across alleles at each given class I locus on VL was largest for HLA-B (H = 81, P = 0.0005) but was also significant for both HLA-A (H = 37, P = 0.006) and HLA-C (H = 52, P = 0.0001) (Table 1). HLA-B also showed the greatest range of effects on CD4 counts, and for this outcome, it was the only locus that showed significance (H = 71, P = 0.004 for HLA-B; H = 18, P = 0.43 for HLA-A; H = 29, P = 0.084 for HLA-C). These results indicate that the HLA-B locus has the greatest effect on HIV-1 control in Japanese and confirmed previous findings in Caucasians and Africans.
Table 1.
Kruskal-Wallis test for associations of viral load and absolute CD4 count with alleles at three HLA class I loci in Japanese individuals chronically infected with HIV-1a
| Allele (n) | VL |
CD4 cell count |
||
|---|---|---|---|---|
| P | H | P | H | |
| HLA-A (504) | 0.006 | 37 | 0.43 | 18 |
| HLA-B (504) | 0.0005 | 81 | 0.004 | 71 |
| HLA-C (503) | 0.0001 | 52 | 0.084 | 29 |
Values in bold are statistically significant.
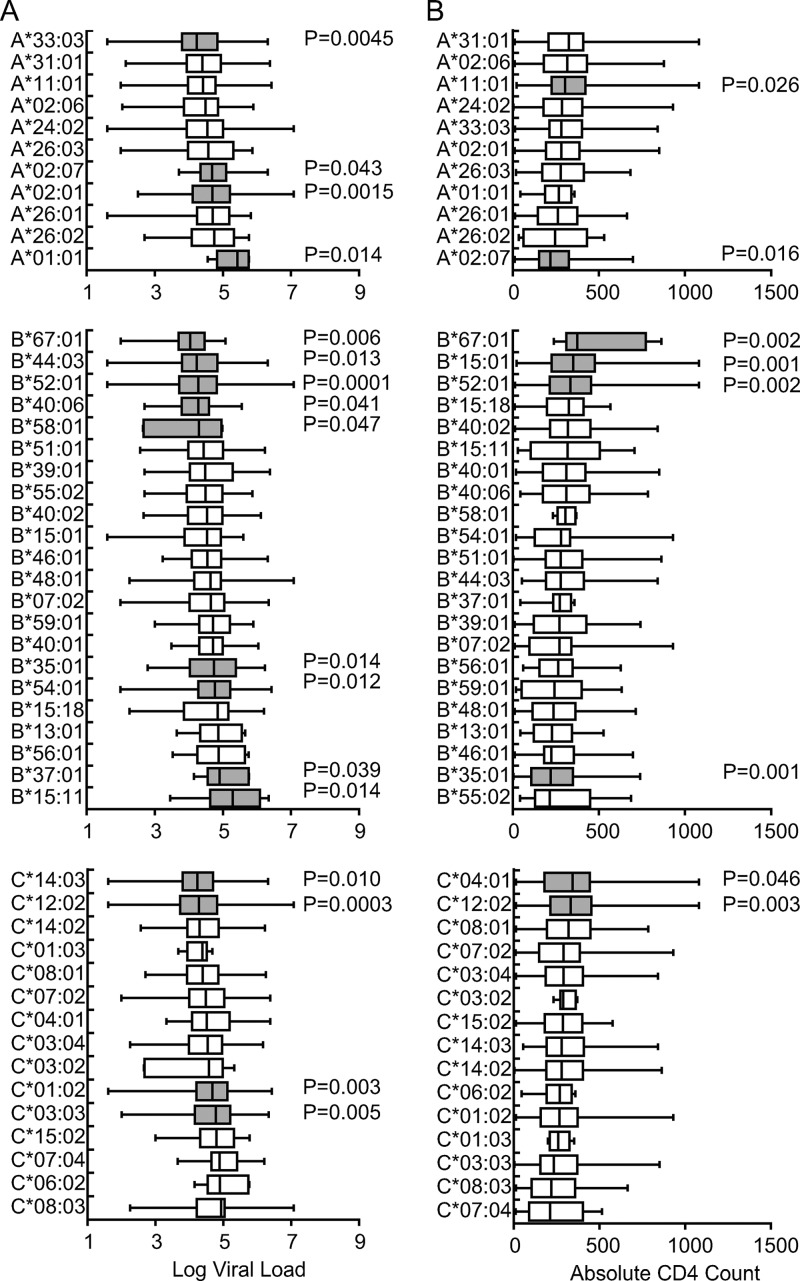
We found that 36% (4 of 11) of HLA-A alleles, 41% (9 of 22) of HLA-B alleles, and 27% (4 of 15) of HLA-C alleles were associated significantly with VL before correction for multiple tests (Fig. 1). Associations with CD4 count were observed for a more limited number of HLA class I alleles than those with VL: 18% (2 of 11) for HLA-A, 18% (4 of 22) for HLA-B, and 13% (2 of 15) for HLA-C (Fig. 1). Three alleles, HLA-B*67:01, -B*52:01, and -C*12:02, were significantly associated with both low VL and high CD4 count and two alleles, HLA-A*02:07 and -B*35:01, associated with both higher VL and lower CD4 count. After local false discovery rate estimation, the numbers of HLA alleles associated with VL and CD4 count were 14 and 5, respectively (see Table S2 in the supplemental material). HLA-B*52:01 and -C*12:02 form a known haplotype, which reaches a frequency of approximately 20% of the Japanese population (12), and given the protection associated with the individual alleles, it is possible that CTL or NK cell responses restricted by this haplotype play an important role in control of HIV-1 in Japanese. HLA-B*35:01 associated with high VL and low CD4 counts, which is consistent with data from Caucasians, in which this allele was more commonly observed among HIV noncontrollers (5). On the other hand, HLA-B*35:02 and -B*35:03 associate with rapid disease progression in Caucasians, whereas HLA-B*35:01 does not (6), highlighting distinctions across allelic associations depending on the clinical outcome being considered. HLA-A*02:07 (frequency = 0.073), an allele that is missing in Caucasians and Africans, also associated with susceptibility.
Fig 1.
Association of HLA allele expression with viral load and absolute CD4 count in Japanese individuals chronically infected with HIV-1. All HLA alleles occurring at a phenotypic frequency greater than 1% were examined for their associations with viral load (A) and absolute CD4 count (B) in a cohort of 504 chronically HIV-1-infected Japanese individuals recruited from 2000 to 2010. Associations with a P value of <0.05 are highlighted in gray.
An advantage of rare HLA alleles in HIV-1 disease progression has been reported previously (13). Therefore, we investigated the effect of HLA frequency on VL or CD4 count in our cohort by comparing these clinical measurements first to the sum of the frequencies of the two alleles at each HLA locus individually (HLA-A, -B, and -C separately) and second to the sum of the frequencies of all HLA class I alleles (HLA-A, -B, and -C combined) for each subject. No significant correlation was observed between the overall HLA allele frequency and VL or CD4 count (Fig. 2A and B), nor was there a correlation between HLA-A allele frequencies or HLA-C allele frequencies with these clinical measurements (Fig. 2C, D, G, and H). In contrast to previous observations, the frequencies of HLA-B alleles were positively and negatively associated with CD4 count and VL, respectively (Fig. 2E and F). We also analyzed the effect of HLA supertype frequency on VL and found no effect (see Fig. S1 in the supplemental material). These results indicate no advantage of rare alleles on VL and CD4 count in the Japanese cohort. Further, we detected no significant differences in either VL or CD4 count between heterozygotes and homozygotes at any individual HLA locus (Fig. 3A and B) or homozygosity at one, two, or all three class I loci (Fig. 3C and D). Thus, a heterozygote advantage of HLA class I was not observed in this cohort.
Fig 2.
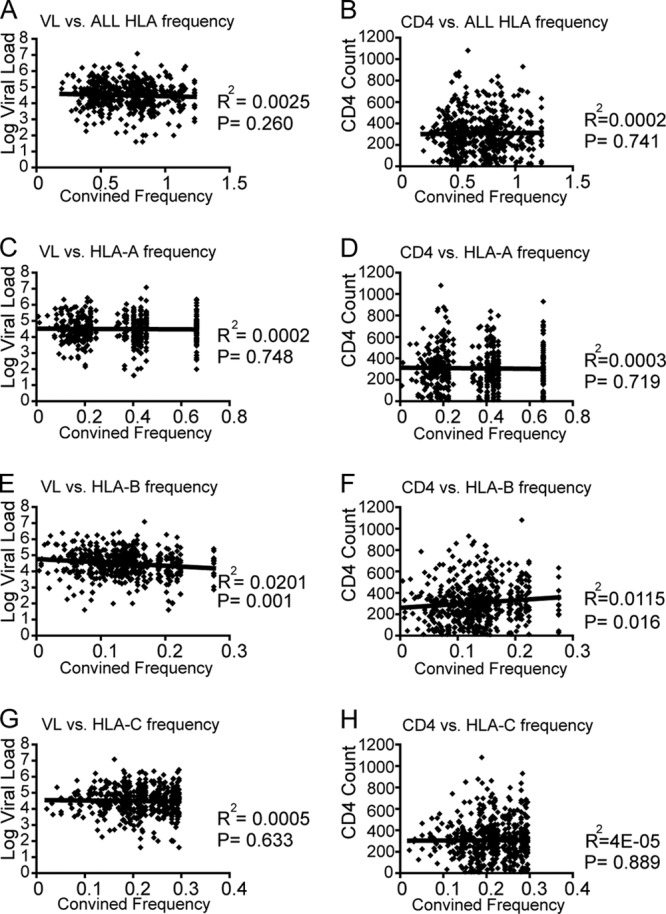
HLA frequency-dependent effects on viral load and CD4 counts. The correlations between VL or CD4 count with combined frequencies for all subjects (n = 504) are shown. HLA frequencies used to calculate the combined frequencies for each individual were derived from the overall allele frequencies in the entire cohort of 504 subjects. Linear regression lines are included in the plots. The distribution of HLA class I frequencies among 504 chronically HIV-1-infected Japanese individuals used in this analysis is shown in Table S4 in the supplemental material.
Fig 3.
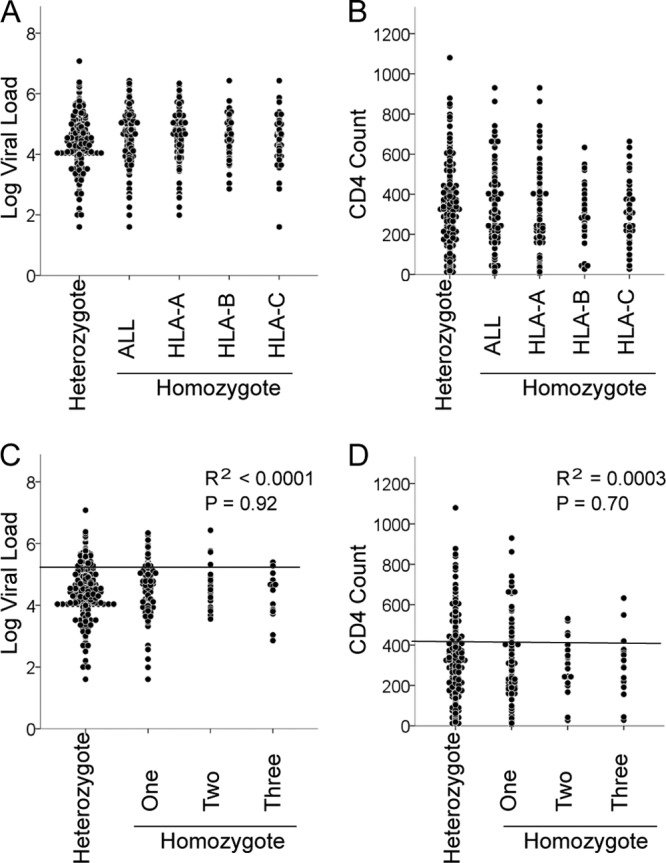
Effect of HLA class I homozygosity on VL and CD4. The comparison between heterozygotes at all three class I loci (n = 349) and homozygotes (n = 155) for at least one HLA allele is shown (A and B). (A and B) Homozygotes are grouped according to homozygosity for the HLA-A (n = 106), -B (n = 42), or -C (n = 64) locus. No significant difference was observed between these groups. Median VLs and CD4 counts, respectively, are 32,500 and 289 (fully heterozygous), 44,000 and 280 (HLA-A homozygous), 35,500 and 290 (HLA-B homozygous), 29,500 and 305 (HLA-C homozygous), and 42,000 and 288 (homozygous for at least one HLA locus). (C and D) Homozygotes were grouped according to homozygosity for one (n = 115), two (n = 23), or three (n = 17) HLA class I loci. Median VLs and CD4 counts, respectively, are 32,500 and 289 (fully heterozygous), 47,000 and 284 (homozygous at a single locus), 28,000 and 335 (homozygous at two loci), and 31,000 and 288 (homozygous at all three loci). No significant difference was observed between these groups. The lines in panels C and D are linear regression lines.
We also analyzed 147 ART-naïve Japanese individuals with clinical AIDS. There were no strong associations of HLA alleles with either VL or CD4 count (see Table S3 in the supplemental material). We excluded these individuals in the present study for the following reasons: (i) There were radical differences in VL and CD4 count between AIDS and non-AIDS groups, and (ii) since AIDS represents an effective breakdown of the immune response, a putative association observed in the non-AIDS group would not be comparable to that in the AIDS group even if HLA class I alleles had an effect on VL and CD4 count.
In summary, the HLA-B locus appears to have the strongest allelic effects on VL and CD4 counts in this Japanese cohort, with the HLA-B*52:01-C*12:02 haplotype showing the greatest significance. These findings in a Japanese cohort highlight the differences of the effects of HLA class I alleles on HIV-1 control between Japanese and Africans/Caucasians.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Global COE program “Global Education and Research Center Aiming at the Control of AIDS,” launched as a project commissioned by the Ministry of Education, Science, Sports, and Culture, Japan, and by a grant-in-aid for scientific research from the Ministry of Health (no. H21-AIDS-010 and H24-AIDS-007), Japan. This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
We have no conflicting financial interests.
We thank Sachiko Sakai for her secretarial assistance.
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Altfeld M, et al. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581–2591 [DOI] [PubMed] [Google Scholar]
- 2. Carrington M, et al. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752 [DOI] [PubMed] [Google Scholar]
- 3. Carrington M, O'Brien SJ. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535–551 [DOI] [PubMed] [Google Scholar]
- 4. Costello C, et al. 1999. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS 13:1990–1991 [DOI] [PubMed] [Google Scholar]
- 5. Fellay J, et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao X, et al. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668–1675 [DOI] [PubMed] [Google Scholar]
- 7. Kiepiela P, et al. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775 [DOI] [PubMed] [Google Scholar]
- 8. Lazaryan A, et al. 2006. Human leukocyte antigen B58 supertype and human immunodeficiency virus type 1 infection in native Africans. J. Virol. 80:6056–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leslie A, et al. 2010. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J. Virol. 84:9879–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Migueles SA, et al. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Brien SJ, Gao X, Carrington M. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7:379–381 [DOI] [PubMed] [Google Scholar]
- 12. Saito S, Ota S, Yamada E, Inoko H, Ota M. 2000. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens 56:522–529 [DOI] [PubMed] [Google Scholar]
- 13. Trachtenberg E, et al. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928–935 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.