Abstract
Neutralizing antibody protection against HIV-1 may require broad and potent antibodies targeting multiple epitopes. We tested 7 monoclonal antibodies (MAbs) against 45 viruses of diverse subtypes from early infection. The CD4 binding site MAb NIH45-46W was most broad and potent (91% coverage; geometric mean 50% inhibitory concentration [IC50], 0.09 μg/ml). Combining NIH45-46W and a V3-specific MAb, PGT128, neutralized 96% of viruses, while PGT121, another V3-specific MAb, neutralized the remainder. Thus, 2 or 3 antibody specificities may prevent infection by most HIV-1 variants.
TEXT
Studies in nonhuman primate models have demonstrated that passively infused neutralizing antibodies (NAbs) can protect against HIV-1 infection when present at the time of exposure (1, 3, 15, 18, 19, 21). However, an enormous challenge to preventing infection in naturally exposed populations is the requirement for NAb responses to recognize diverse circulating variants. Recently identified HIV-1 monoclonal antibodies (MAbs) capable of potently neutralizing diverse variants have spurred optimism for a NAb-based vaccine, as these MAbs may define key targets for protective NAb responses and may also be candidates for gene delivery (2, 16) and potentially for passive immunization to prevent or modify the course of infection (23). However, it is unclear how effective these MAbs are specifically against transmitted variants, which may comprise a unique subset of HIV variants (24) that have distinct characteristics compared to variants in chronic infection, such as shorter variable loop lengths and fewer potential N-linked glycosylation sites (PNGS) (7, 8, 25, 29) and, in some cases, different neutralization profiles compared to nontransmitted variants (8, 9, 29, 31).
We analyzed the neutralization profiles of 45 HIV-1 envelope variants of diverse subtypes (A, C, D), which were obtained soon after heterosexually acquired infection (median, 59 days postinfection) (4, 5, 17), against 7 recently identified broadly neutralizing MAbs targeting several distinct epitopes. These included the following: VRC01, which targets the CD4 binding site (CD4bs) (30); NIH45-46W (10), which also targets the CD4bs but is an engineered mutant that improves the neutralization breadth and potency of MAb NIH45-46, a clonal variant of VRC01 (26); PG9, PG16, and PGT145, which recognize a glycan-dependent quaternary epitope in V1/V2 and V3 (27, 28); and MAbs PGT121 and PGT128 (27), which form another class of antibodies targeted to glycan-dependent epitopes in V3. Serial dilutions of all MAbs were tested at a starting concentration of 1 μg/ml against envelope pseudoviruses in the TZM-bl assay as described previously (29). This starting MAb concentration was chosen due to the limited reagent availability and the reported breadth of the MAbs, even at low concentrations (10, 27, 28, 30).
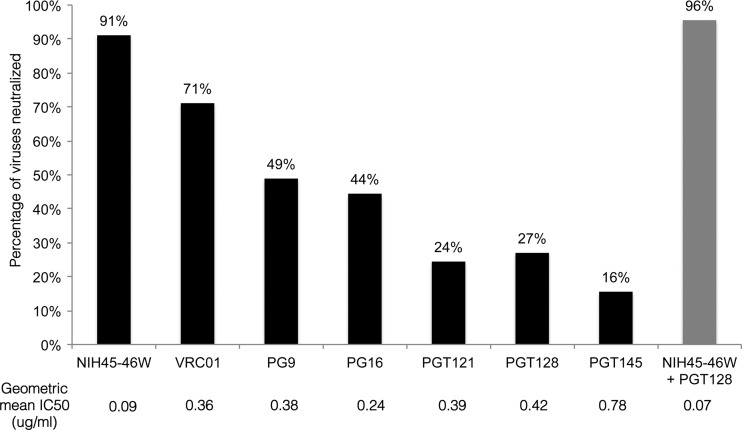
The MAbs had differing neutralizing activities against the panel viruses, with 50% inhibitory concentration (IC50) values ranging by more than 3 orders of magnitude from 0.0003 to >1 μg/ml (Fig. 1). The CD4bs MAb NIH45-46W neutralized 91% of variants with a geometric mean IC50 of 0.09 μg/ml, while VRC01, another CD4bs MAb, neutralized 71% of variants with a geometric mean IC50 of 0.36 μg/ml (Fig. 2). The glycan-dependent PG and PGT MAbs were less broad and potent than the CD4bs MAbs, neutralizing only 16% to 49% of variants with a geometric mean IC50 of 0.24 to 0.78 μg/ml.
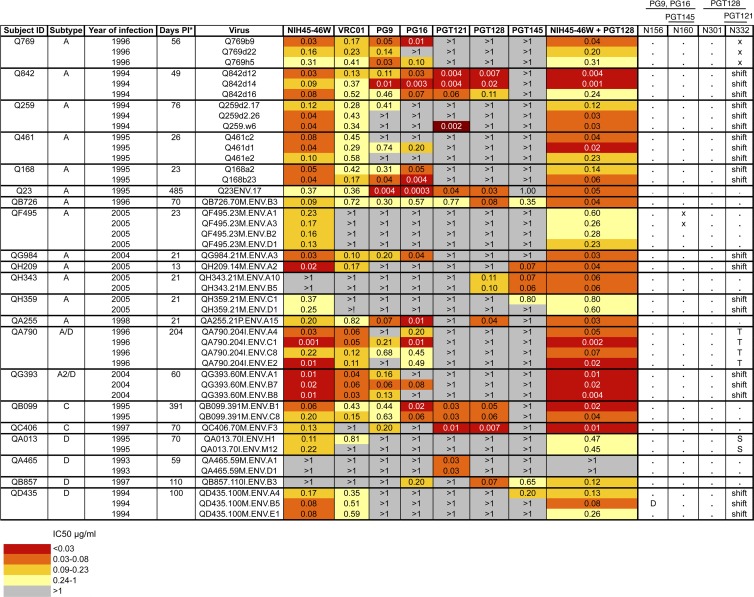
Fig 1.
Summary of neutralization profiles of panel viruses against MAbs. Subject ID, virus subtype based on V1-V5 envelope sequence, and calendar year of infection are shown in the first 3 columns. Each row shows the virus name, IC50 for MAbs tested, and known core residues based on HXB2 numbering required for neutralization by MAbs shown. Viruses were obtained as described previously (4, 5, 17). For some patients, multiple viruses were obtained from the same time point to represent the diversity of the virus population at that time point as determined by phylogenetic analysis (5). Symbols: asterisk, estimated days postinfection at which envelope clone was obtained; dot, amino acid is present; x, amino acid is present but not in a glycosylation sequon; shift, amino acid is present but in position 334; D, T, S, amino acid substitutions. Darker shading indicates increasing MAb potency, as indicated by the key at the bottom, grouped by quartiles of IC50 values for all virus-MAb combinations. Gray shading indicates that 50% neutralization was not achieved at the highest concentration of MAb tested (1 μg/ml). The combination of NIH45-46W and PGT128 was tested at a starting concentration of 1 μg/ml of each MAb. IC50 values shown are averages from at least 2 independent experiments performed in duplicate.
Fig 2.
Summary of neutralization breadth and potency of MAbs against 45 viruses. Percentages of viruses neutralized for each MAb are indicated at the bottom of the graph. Geometric mean IC50 value for each MAb is indicated below the MAb name. IC50 values greater than the highest MAb concentration tested (1 μg/ml) were assigned a value of 1 in the geometric mean IC50 calculations.
Because the PG and PGT MAbs failed to neutralize a majority of variants, we investigated whether these variants lacked the PNGS required for neutralization by these MAbs (Fig. 1). In some cases, resistance to these MAbs could be explained by the absence of a key PNGS. For example, variants isolated from a number of patients, including Q769, QG984, QH209, and QH359, which were resistant to PGT121 and PGT128, lacked the N332 residue required for neutralization (22, 27). Two of the four PG9/16-resistant variants, isolated from subject QF495, did not have the full glycosylation sequon that is a target for these MAbs, despite having the N160 residue (20, 28). Similarly, one of the QD435 variants resistant to these MAbs did not have the N156 residue required for recognition by PG9 (20). However, for all other variants resistant to PG9/16, the absence of known PNGS targets could not account for resistance, as these variants possessed key residues required for neutralization (N156, N160) (18, 26). Moreover, the presence of positively charged residues at positions 168, 169, and 171, which have been reported to be important for recognition by PG9/16 (11), did not always predict sensitivity to these MAbs (data not shown).
Some viruses, such as those from QF495, QH343, and QA465, had key PNGS for PGT121 and PGT128 recognition (N301 and N332) (20) yet were resistant to one or both of these MAbs. For other viruses, such as those isolated from Q259, Q168, and QD435, the shift of PNGS at position 332 to position 334 may account for resistance to PGT121 and PGT128. However, this shift in PNGS did not always predict resistance to PGT121 and PGT128, as exemplified by Q842 variants, which had a shift of PNGS to position 334 yet were sensitive to these MAbs. Thus, the presence of known residues important for neutralization by the PG and PGT MAbs did not fully explain differing neutralization profiles among these early variants, suggesting that there may be other determinants of sensitivity to these MAbs. Indeed, the fact that some viruses, such as those from QF495 and QA465, had the expected epitope targets yet were resistant to most MAbs suggests that these viruses may have altered conformations that result in global neutralization resistance, as was observed for another early subtype A virus from heterosexual transmission (6) and for subtypes A and A/D vertically transmitted variants (14). Of note, variants that possessed the canonical epitopes for PG and/or PGT MAbs (QA013, QB857, QF495 QH343, and QH359 variants) were still not neutralized even when these MAbs were tested at a higher starting concentration of 10 μg/ml (data not shown). An alignment of V1-V3 sequences of all variants did not readily reveal signature sequences that would predict sensitivity to these MAbs (see Fig. S2 in the supplemental material), although QF495 variants had a large insertion in V2, including the addition of multiple PNGS, that could explain their resistance to most MAbs tested here.
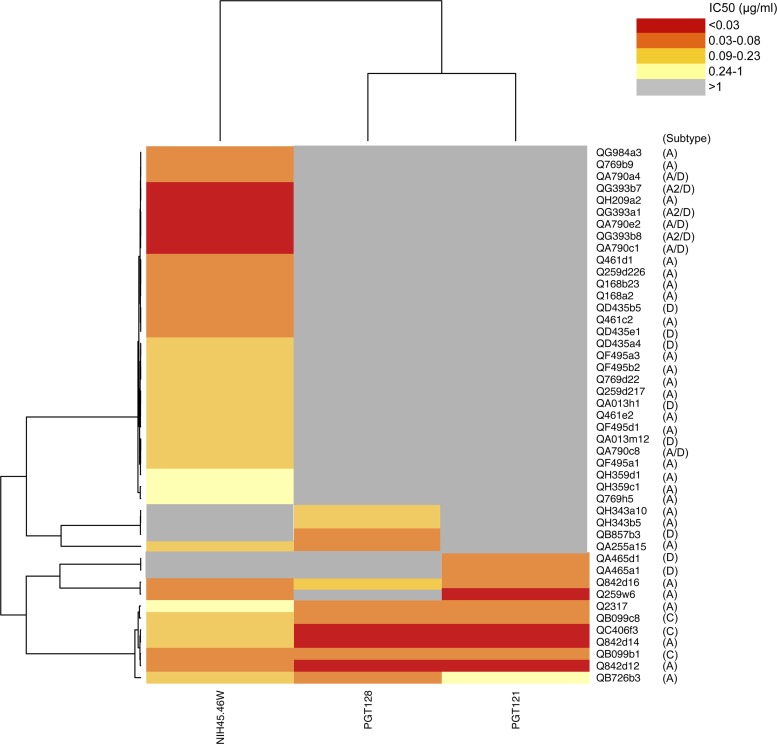
NIH45-46W neutralized all but 5 viruses in the panel, including some viruses that were not neutralized by any other MAb (QF495, QH359, and QA013) (Fig. 1). Interestingly, although MAbs PGT121, PGT128, and PGT145 displayed limited breadth at the highest concentration tested in these experiments (1 μg/ml), they potently neutralized variants that were resistant to all other MAbs, including NIH45-46W. (Fig. 1; see also Fig. S1 in the supplemental material). Specifically, QA465 and QH343 variants were only neutralized by PGT121, PGT128, and/or PGT145 (IC50 ≤ 0.11 μg/ml). Hierarchical clustering analyses suggested that combining NIH45-46W and PGT128 would neutralize all but 2 variants, which were recognized by PGT121 (Fig. 3).
Fig 3.
Hierarchical clustering of MAbs NIH45-46W, PGT128, and PGT121 (bottom) and panel viruses (right). A heatmap of IC50 values for each virus-MAb combination is shown, with darker shading indicating increasing potency, as indicated by the key. Gray shading indicates that 50% neutralization was not achieved at the highest concentration of MAb tested (1 μg/ml).
To test the hypothesis that NIH45-46W and PGT128 would complement rather than interfere with each other's neutralizing ability, we investigated the neutralization profiles of a subset of viruses against NIH45-46W and PGT128 alone or in a 1:1 combination. We chose viruses that were either (i) neutralized by one but not the other MAb (Q168.A2 and QH343.21 M.ENV.A10) or (ii) neutralized by both MAbs (QB099.391 M.ENV.B1 and QC406.70 M.ENV.F3). The presence of one MAb did not interfere with the activity of the other MAb regardless of whether the virus tested was sensitive to one (see Fig. S3A in the supplemental material) or both (see Fig. S3B) MAbs, with no increase in IC50 values for MAbs in a 1:1 combination compared to MAbs tested alone. Because NIH45-46W is an engineered antibody, we also confirmed that a naturally occurring broad and potent CD4bs MAb, VRC01, would not interfere with PGT128 neutralization (see Fig. S3). These results demonstrate that broad and potent MAbs targeting the CD4bs and V3 do not compete for neutralization.
Overall, the combination of NIH45-46W and PGT128 neutralized 96% of variants with a geometric mean IC50 of 0.07 μg/ml (Fig. 1 and 2). The remaining 2 variants not neutralized by NIH45-46W and PGT128 alone or in combination were potently neutralized by PGT121 (Fig. 3). Because PGT121 and PGT128 have previously been shown to compete for binding (27), we investigated whether these MAbs would interfere with each other's neutralizing capacity against these 2 viruses. The combination of PGT121 and PGT128, with or without NIH45-46W present, neutralized both viruses, which were sensitive to PGT121 but resistant to PGT128, to a similar extent as PGT121 alone (see Fig. S4 in the supplemental material). These results demonstrate that the presence of PGT128 does not interfere with PGT121 neutralization against these variants. It is possible that these observations reflect an absence of binding of PGT128 to the viruses tested rather than a lack of competition between the MAbs.
We observed generally similar neutralization breadth for the individual MAbs NIH45-46W and VRC01 and only slightly lower breadth for PG9 and PG16 compared to previous reports (10, 28, 30). However, the neutralization breadth of PGT121, PGT128, and PGT145 (16 to 27%) was 2- to 3-fold lower than that observed previously (27). In the prior study by Walker et al. (27), these MAbs were tested against a panel of viruses weighted toward variants from chronic infection, potentially suggesting differences in efficacy of the MAbs against variants in early versus chronic infection. However, differences in assays used and the subtypes of viruses examined could also be relevant and studies that directly compare these variables will be needed to understand differences in efficacy against different virus panels.
Prior studies of a subset of MAbs tested here, including CD4bs and V1/V2 MAbs, suggested that the combination of PG9 and VRC01 provided almost universal coverage of the viruses tested, which included subtype B viruses (13) and viruses from diverse subtypes (12). When PG9 and VRC01 were tested at 1 μg/ml, 26% of viruses in our panel were resistant to both MAbs (Fig. 1), which is more than what was reported in the prior cross-clade study (∼10%) (12). This difference could again reflect differences in the virus panel, which included viruses from both acute and chronic infection from a study by Doria-Rose et al. (12). Additionally, differences in the calendar period from which viruses were isolated could also influence sensitivity to these MAbs (13), but our sample size and distribution over the sampling period (Fig. 1) were not adequate to rigorously address this issue. Finally, the study by Doria-Rose et al. used higher MAb concentrations (50 μg/ml) and a larger dilution range, and at this higher concentration, we have observed a tendency to obtain lower IC50 values than those obtained with the lower starting concentration (1 μg/ml) and tighter dilution range used here for viruses that were potently neutralized (IC50 < 0.1 μg/ml). However, this would not have altered our overall results, which focused on whether MAbs could neutralize variants at1 μg/ml, although it could lead to small differences in the geometric mean IC50. Notably, at the lower MAb concentration used here, only 4% of viruses in our panel were resistant to NIH45-46W and PGT128, two MAbs that were not included in previous studies, implying that these MAbs may be among the most effective against variants found early in infection. Although 7/45 viruses were obtained later in infection (Q23, QA790, and QB099), and thus may not be representative of recently transmitted variants, removing these viruses from the analysis did not significantly affect the results of our study. For example, even by excluding these viruses, NIH45-46W was still the most broad and potent MAb, while PGT121, PGT128, and PGT145 were the least broad and potent MAbs against our panel.
In summary, we demonstrated that recently identified broadly neutralizing HIV-1 MAbs have variable activity against variants found early in infection. NIH45-46W, an engineered mutant of NIH45-46 that targets the hydrophobic CD4 binding cavity in gp120 (10), displayed remarkable breadth and potency against these viruses. However, this MAb was unable to neutralize ∼10% of the viruses in the panel, which were potently neutralized by glycan-dependent MAbs PGT121, PGT128, and/or PGT145. PGT128 and NIH45-46W displayed no competition for neutralization, and a combination of these MAbs neutralized 96% of variants, with PGT121 neutralizing the only 2 viruses not neutralized by this combination. Our results suggest that optimal neutralization coverage of transmitted variants may be achieved by combining a potent CD4bs NAb with one or more NAbs directed to glycan-dependent epitopes in V3. It is currently unclear whether this particular combination of broad and potent NAbs can develop within a patient during natural infection, and it is likely that eliciting such responses will be challenging. However, the results presented here provide motivation to focus on these epitopes, given that the antibody combination against them can neutralize viruses representing recently transmitted variants.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michelle Long, Catherine Blish, Ozge Dogan, Minh-An Nguyen, and Stephanie Rainwater for generating envelope plasmids used in our panel, the IAVI Neutralizing Antibody Consortium for providing MAbs PG9, PG16, PGT121, PGT128, and PGT145, Xueling Wu and John Mascola for providing VRC01, and Ron Diskin, Paola Marcovecchio, and Pamela Bjorkman for providing NIH45-46W.
This work was supported by NIH grant HD058304/AI103981. L.G. was supported in part by a Fred Hutchinson Cancer Research Center Interdisciplinary Research Fellowship.
Footnotes
Published ahead of print 25 July 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Baba TW, et al. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200–206 [DOI] [PubMed] [Google Scholar]
- 2. Balazs AB, et al. 2012. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481:81–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barouch DH, et al. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blish C, Nedellec R, Mandaliya K, Mosier D, Overbaugh J. 2007. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS 21:693–702 [DOI] [PubMed] [Google Scholar]
- 5. Blish CA, et al. 2009. Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D envelope variants of human immunodeficiency virus type 1. J. Virol. 83:7783–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blish CA, Nguyen MA, Overbaugh J. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5:e9 doi:10.1371/journal.pmed.0050009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chohan B, et al. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 79:6528–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Derdeyn CA, et al. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 9. Dickover R, et al. 2006. Role of maternal autologous neutralizing antibody in selective perinatal transmission of human immunodeficiency virus type 1 escape variants. J. Virol. 80:6525–6533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diskin R, et al. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doria-Rose NA, et al. 23 May 2012. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J. Virol. doi:10.1128/JVI.00808–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doria-Rose NA, et al. 2012. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J. Virol. 86:3393–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Euler Z, et al. 2011. Activity of broadly neutralizing antibodies, including PG9, PG16, and VRC01, against recently transmitted subtype B HIV-1 variants from early and late in the epidemic. J. Virol. 85:7236–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goo L, Milligan C, Abel C, Nduati R, Overbaugh J. 27 June 2012. Neutralizing antibody escape during HIV-1 mother-to-child transmission involves conformational masking of distal epitopes in envelope. J. Virol. doi:10.1128/JVI.00953–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hessell AJ, et al. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15:951–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson PR, et al. 2009. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 15:901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Long EM, Rainwater SM, Lavreys L, Mandaliya K, Overbaugh J. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retroviruses 18:567–576 [DOI] [PubMed] [Google Scholar]
- 18. Mascola JR, et al. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mascola JR, et al. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207–210 [DOI] [PubMed] [Google Scholar]
- 20. McLellan JS, et al. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parren PW, et al. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pejchal R, et al. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Safrit JT, et al. 2004. Immunoprophylaxis to prevent mother-to-child transmission of HIV-1. J. Acquir. Immune Defic. Syndr. 35:169–177 [DOI] [PubMed] [Google Scholar]
- 24. Sagar M. 2010. HIV-1 transmission biology: selection and characteristics of infecting viruses. J. Infect. Dis. 202(Suppl 2):S289–S296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sagar M, et al. 2009. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J. Infect. Dis. 199:580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scheid JF, et al. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walker LM, et al. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker LM, et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu X, et al. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80:835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang H, et al. 2010. Functional properties of the HIV-1 subtype C envelope glycoprotein associated with mother-to-child transmission. Virology 400:164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.