Abstract
Shiga toxin 2 (Stx2)-producing Escherichia coli (STEC) O104:H4 caused one of the world's largest outbreaks of hemorrhagic colitis and hemolytic uremic syndrome in Germany in 2011. These strains have evolved from enteroaggregative E. coli (EAEC) by the acquisition of the Stx2 genes and have been designated enteroaggregative hemorrhagic E. coli. Nucleotide sequencing has shown that the Stx2 gene is carried by prophages integrated into the chromosome of STEC O104:H4. We studied the properties of Stx2-encoding bacteriophages which are responsible for the emergence of this new type of E. coli pathogen. For this, we analyzed Stx bacteriophages from STEC O104:H4 strains from Germany (in 2001 and 2011), Norway (2006), and the Republic of Georgia (2009). Viable Stx2-encoding bacteriophages could be isolated from all STEC strains except for the Norwegian strain. The Stx2 phages formed lysogens on E. coli K-12 by integration into the wrbA locus, resulting in Stx2 production. The nucleotide sequence of the Stx2 phage P13374 of a German STEC O104:H4 outbreak was determined. From the bioinformatic analyses of the prophage sequence of 60,894 bp, 79 open reading frames were inferred. Interestingly, the Stx2 phages from the German 2001 and 2011 outbreak strains were found to be identical and closely related to the Stx2 phages from the Georgian 2009 isolates. Major proteins of the virion particles were analyzed by mass spectrometry. Stx2 production in STEC O104:H4 strains was inducible by mitomycin C and was compared to Stx2 production of E. coli K-12 lysogens.
INTRODUCTION
In Germany between May and July 2011, Shiga toxin-producing E. coli (STEC) O104:H4 was identified as the cause of one of the world's largest outbreaks of STEC (19). The outbreak spread to other European and non-European countries and finally accounted for 3,842 cases of human infections, with a high percentage of patients developing the life-threatening hemolytic-uremic syndrome (HUS) (17). Until 2011, O104:H4 was considered a rare STEC serotype (8, 47), with only a few sporadic cases of human STEC O104:H4 infection reported in some European and Asian countries (51). The first STEC O104:H4 strains were isolated in 2001 from the feces of siblings with hemorrhagic colitis (HC) and HUS in Cologne, Germany (7, 17).
The causative STEC O104:H4 strain recovered in the 2011 outbreak differs from E. coli O157:H7 and other enterohemorrhagic E. coli (EHEC) strains, because it is negative for the EHEC virulence plasmid and for the genes encoding the attaching/effacing (A/E) mechanism (7, 51). On the genome level, STEC O104:H4 strains were shown to be most similar to the previously characterized enteroaggregative E. coli (EAEC) strain 55989 and were found to carry typical virulence genes of EAEC strains (7, 46). Accordingly, the term enteroaggregative hemorrhagic E. coli (EAHEC) was proposed for this new group of human-pathogenic E. coli (10). Interestingly, EAEC-specific virulence genes were not found in all STEC O104:H4 isolates collected before 2011, and significant differences in the EAEC-specific virulence genes and EAEC plasmids were found by comparing the German STEC O104:H4 strains from 2001 to those from 2011 (7, 26).
Similar to many classical EHEC strains, the STEC O104:H4 outbreak isolate produces the Shiga toxin 2a (Stx2a). This toxin type is associated with classical EHEC (16, 38, 46), in particular with EHEC O157:H7 strains. The stx2a genes are located on the genome of lambdoid bacteriophages integrated in the chromosome of their bacterial hosts (31, 38, 39). Augmented colonization of the humans by STEC O104:H4 through aggregative adherence fimbriae might result in large quantities of Stx2a being delivered into the host intestine. This was suggested as the causative mechanism leading to the high numbers of patients with severe clinical illness in STEC O104:H4 infection (7).
Stx2a was previously shown to be associated with severe clinical illness in humans (20, 21), and the amount of Stx2a produced by EHEC O157:H7 strains was found to correlate with the development of severe clinical illness in a gnotobiotic piglet model (3). The production of Stx by STEC can be significantly enhanced by certain antimicrobial and DNA-damaging agents, such as ciprofloxacin and mitomycin C, respectively (6, 13, 48). An increase in Stx production caused by exposure to agents such as ciprofloxacin likely results from induction of the prophage. Stx production was also found to be stimulated by neutrophils and norepinephrine in the gastrointestinal tract, indicating that the inducibility of Stx plays a role in in vivo pathogenesis (14, 54). The genetic composition of Stx-encoding bacteriophages could have an influence on bacterial host specificity and on Stx production (13, 48, 55). A number of Stx2-encoding bacteriophages have been analyzed for the nucleotide sequence, and significant differences in global nucleotide composition and in the regulatory genes affecting Stx production were found (31, 39, 45, 49). Genomic sequencing of the STEC O104:H4 strains revealed that they carry Stx2a-encoding prophages (40, 43).
In this work, we characterized the Stx2a-encoding short-tailed DNA bacteriophage (podovirus) P13374 from the STEC O104:H4 strain CB13374, which was isolated in the course of outbreak investigations from an opened package of retail sprouted seeds from the household of an infected patient (53). We analyzed P13374 for its genetic composition, biological properties, and bacterial host specificity. Genomic sequencing and analysis of restriction endonuclease digestion patterns indicate that bacteriophage P13374 is highly similar to the Stx2a phages carried by STEC O104:H4 strains isolated from human patients in Germany in 2001 and 2011. Additionally, a comparison to phages which are present in Stx2a-producing STEC O104:H4 strains from Norway and Georgia revealed close similarities.
MATERIALS AND METHODS
Bacteria and culture conditions.
Bacterial strains used in this work were from the collection of the National Reference Laboratory for Escherichia coli (NRL-E. coli; Federal Institute for Risk Assessment [BfR] Berlin, Germany). STEC O104:H4 strains were serotyped for the O and H antigens and confirmed by real-time PCR for the presence of stx2, aggR, terB, and wzxO104 genes as previously described (53). To discriminate between different virulotypes of STEC O104:H4, the strains were additionally investigated for the EAEC heat-stable enterotoxin (astA) and for sequence encoding aggregative adherence fimbriae I (aggA) and III (agg3A) as described previously (7). The strains used for induction and isolation of Stx2-encoding bacteriophages are listed in Table 1. The STEC O104:H4 strain RKI 11-022027 (CB13344) was isolated from a female patient with HUS at the onset of the STEC O104:H4 outbreak in May 2011 in northern Germany (19). Strain CB13374 was isolated from an opened package of sprouted seeds from the household of a patient infected with STEC O104:H4. The sprouts originated from the farm incriminated as the food source of the STEC O104:H4 2011 outbreak strain (4, 53). STEC strain CB8983 was isolated in 2001 from a 5-year-old girl with hemorrhagic colitis in Cologne, Germany. Subsequently, the girl developed HUS and was hospitalized. The clinical case report reveals that the strain HUSEC 41 (STEC O104:H4) was isolated from the same patient after she was hospitalized with HUS (7 and L. Beutin, unpublished data). STEC O104:H4 strain 1106-3060-1 (CB13437) originated from a patient in Norway who became infected in 2006. Two STEC O104:H4 strains, 2009EL-2050 (CB13771) and 2009EL-2071 (CB13772), were isolated in 2009 from patients with bloody diarrhea in the Republic of Georgia (12). A collection of 31 EAEC strains was used for susceptibility tests with the STEC O104:H4 Stx2a phage (see Table S1 in the supplemental material) and was characterized for serotypes and for the presence of the aggR gene as an indicator of the presence of the aggregative adherence mechanism (24). Escherichia coli K-12 strain C600 was used as the recipient for propagation and lysogenization of Stx bacteriophages (50). C600 derivative strain lysogenizated with different Stx phages and phage lambda was described previously, and the strains are listed in Table 3. Media for cultivation of bacteria, preparation of phage lysates, and phage susceptibility were used as previously described (50).
Table 1.
STEC O104:H4 strains used for isolation of lysogenic Stx-encoding bacteriophages
| Strain no. | Original designation | Source, origin, yr of isolation, and disease | Presence of virulence gene: |
|||||
|---|---|---|---|---|---|---|---|---|
| stx2 | aggR | wzxO104 | aggA | agg3A | astA | |||
| CB13344 | RKI 11-02027a | Human, Germany, 2011, HUS | + | + | + | + | − | − |
| CB13374 | Sprouted seeds, Germany, 2011 | + | + | + | + | − | − | |
| CB8983 | Human, Germany, 2001, HC | + | + | + | − | + | + | |
| CB13437 | 1106-3060-1b | Human, Norway, 2006, diarrhea | + | + | + | − | + | + |
| CB13771 | 2009EL-2050c | Human, Georgia, 2009, HC | + | + | + | + | − | − |
| CB13372 | 2009EL-2071c | Human, Georgia, 2009, HC | + | + | + | + | − | − |
Provided by the Robert Koch Institute, Berlin, Germany.
From the collection of the Norwegian Institute of Public Health.
From the collection of the Centers for Disease Control and Prevention, Atlanta, GA.
Table 3.
E. coli K-12 (C600) Stx phage lysogenic derivative strains
| Strain | Phage susceptible to lysogeny | Phage source strain (reference or source) | Shiga toxin | Sensitivity to: |
|
|---|---|---|---|---|---|
| P8983, P13344, P13374 | P13371, P13372 | ||||
| C600 | 49 | + | + | ||
| TPE2385 | P13344 | CB13344 (53) | Stx2a | − | − |
| TPE2364 | P13374 | CB13374 (53) | Stx2a | − | − |
| TPE2369 | P8983 | CB8983 (this work) | Stx2a | − | − |
| TPE2381 | P13771 | CB13771 (this work) | Stx2a | + | − |
| TPE2383 | P13772 | CB13772 (this work) | Stx2a | + | − |
| TPE1923 | P933w | EDL933 (49) | Stx2a | + | + |
| TPE1914 | PH19-B | H19 (50) | Stx1 | + | + |
| TPE1916 | Lambda | + | + | ||
| TPE1841 | P6220 | CB6220 (27) | Stx1c | + | + |
| TPE1874 | CB2851 | CB2851 (49) | Stx2c | + | + |
Isolation, propagation, and purification of Stx2 phages from STEC O104:H4 strains.
Single colonies of the respective E. coli strains were grown in Luria broth to exponential growth. Induction of lysogenic bacteriophages was performed by adding 0.5 μg/ml mitomycin C to the growing bacterial cultures, which were further incubated overnight. Bacteriophages were isolated from single plaques grown on the E. coli K-12 strain C600 as described previously (27). High-titer phage lysates (109 to 1011 PFU/ml) were obtained from E. coli C600 as described previously (50) and were used for isolation of phage DNA, characterization of phage envelope proteins, and phage infection studies.
Lysogenization of the E. coli K-12 strain C600 with Stx2a-encoding bacteriophages from STEC O104:H4 strains.
Serial dilutions of phages released in mitomycin C-induced cultures of O104:H4 strains were spotted on a lawn of the stx-negative E. coli K-12 strain C600. Isolation of lysogenic C600 (Stx phage) strains was performed as previously described (50). The presence of the Stx phage in C600 was confirmed by stx2-specific PCR as described previously (53) and by mitomycin C induction of viable Stx2 phages from C600 lysogens.
Susceptibility of EAEC to the Stx2 phage from CB13374.
Cultures of 31 EAEC strains belonging to 22 different serotypes from the collection of the NRL-E. coli (see Table S1 in the supplemental material) were grown overnight at 37°C in LB. One hundred μl of the culture was added to 3 ml of molten LC-Top agar and immediately poured onto LB agar as described previously (50). Twenty-μl portions of P13374 lysates (1011 PFU/ml) and of freshly mitomycin C-induced cultures of the STEC O104:H4 strain CB13374 were spotted on EAEC strains, and the plates were incubated for 20 to 22 h at 37°C. Lysis of bacteria and plaque formation was monitored after overnight incubation. Strains C600 and CB13374 were used as positive and negative controls for plaque formation, respectively. To investigate the possible transfer of the stx2 gene to EAEC strains by transduction, bacteria grown in zones spotted with P13374 were subcultured on LB agar and grown overnight at 37°C. DNA extracted from subcultured bacteria was investigated for the presence of the stx2 gene by real-time PCR as described previously (53).
Determination of Stx2 production.
Stx production by bacteria was measured semiquantitatively by the Ridascreen verotoxin enzyme immunoassay (Ridascreen-EIA; R-biopharm, Darmstadt, Germany). Bacterial samples for the Stx EIA were grown at 37°C in tryptic soy broth (TSB) supplemented with 0.5 μg/ml mitomycin C or left unsupplemented under aeration as described previously (5). Undiluted and serially diluted bacterial culture fluids were tested from overnight cultures of bacteria, showing titers of about 109 CFU/ml. The Ridascreen EIA results were recorded photometrically with a Dynatech MRX microplate reader at 450 nm with a 630-nm reference wavelength.
Statistical analysis.
Independent t tests were used to test for a difference (mean value of the optical density at 450 nm [OD450]) between two independent groups (STEC wild-type strains grown in the presence or absence of mitomycin C as well as C600 Stx phage lysogens grown in the presence or absence of mitomycin C, respectively). All analyses were performed with PASW Statistics (v18.0.2). The statistical analyses were 2-tailed, and P < 0.05 was considered significant.
Morphology of isolated bacteriophages.
To determine the morphological characteristics of isolated phages by transmission electron microscopy (TEM), the negative staining procedure was used. Briefly, Pioloform-carbon-coated, 400-mesh copper grids (Plano GmbH, Wetzlar, Germany) were placed in drops from supernatants of samples collected and primed for TEM for 10 min, fixed with a 2.5% aqueous glutaraldehyde (Taap Laboratories, Aldermaston, United Kingdom) solution for 1 min, and stained with 2% aqueous uranyl acetate (Merck, Darmstadt, Germany) solution for 1 min.
In addition, the ultrathin sectioning method was performed for electron microscopic investigations of the E. coli K-12 strain TPE2364 (C600) infected with phages lysates of the sprout strain CB13374. For these investigations, infected and mitomycin C-induced bacterial cell suspensions were concentrated by centrifugation. The culture supernatant was removed, and the bacterial pellet was suspended in a 2.5% aqueous glutaraldehyde (Taap Laboratories) solution for 24 h and centrifuged at 8,000 rpm for 5 min. The supernatant was removed and the pellet was mixed with a 3% aqueous low-melting-point agarose (MP Biomedicals, Eschwege, Germany) solution, centrifuged shortly at 14,000 rpm for 5 s, and immediately refrigerated for 10 min. This agarose/bacterial cell pellet was cut into little cubes and soaked in 1% aqueous osmium tetroxide (Electron Microscopic Science, Hatfield, United Kingdom) solution before embedding in Epon resin (Agar Scientific, Stansted, United Kingdom) according to standard procedures. Thereafter, ultrathin sections were cut and then postcontrasted with 2% aqueous uranyl acetate (Merck) solution for 20 min and 2.7% alkaline lead citrate (Serva, Heidelberg, Germany) work solution for 15 min. The negatively stained specimens and the ultrathin sections were examined with a JEM-1010 electron microscope (Jeol, Tokyo, Japan) at an 80-kV acceleration voltage. Photographs and measurements were performed using a MegaView II digital camera and a computer-assisted analysis program (SIS Analysis; Olympus, Münster, Germany).
SDS-PAGE and in-gel digestion of P13374 proteins.
Phage P13374 proteins were extracted from concentrated phage particles and reduced with 10 mM dithiothreitol (DTT) for 5 min at 95°C, and thiol (SH) groups were subsequently alkylated with 50 mM iodoacetamide for 45 min in the dark at room temperature. Samples were heated for 5 min in sample buffer at 95°C and subjected to SDS-PAGE according to the procedure of Laemmli (29). Protein samples were electrophoresed on precast gradient gels (4 to 20% Criterion Tris-HCl; Bio-Rad, Munich, Germany) in a Bio-Rad Protean II electrophoresis system. Gels were washed with water and stained using BioSafe Coomassie stain (Bio-Rad). Bands of interest were excised and cut into smaller pieces using a clean scalpel. Gel pieces were washed one after another with water, 50% acetonitrile (ACN), and 20 mM NH4HCO3. Gel pieces were washed twice with 20 mM NH4HCO3-ACN (50:50, vol/vol) to destain the proteins, dehydrated with 100% ACN, and dried. Tryptic in-gel digestion was carried out overnight at 37°C using a 12.5 ng/ml solution of proteomics-grade recombinant trypsin (Roche, Mannheim, Germany) in 25 mM NH4HCO3–10% ACN–5 mM CaCl2. Tryptic peptides were extracted with 50% ACN in 5% trifluoroacetic acid (TFA) for 30 min. The extracted peptides were dried, reconstituted with 0.3% TFA in 60% ACN, and directly subjected to tandem matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF-TOF MS/MS) analysis. Alternatively, dried peptides were reconstituted in 0.1% TFA and purified using a C18 Zip-Tip according to the manufacturer's instructions (Millipore, Schwalbach, Germany) and subjected to MALDI-TOF-TOF MS/MS analysis.
MALDI sample preparation and MALDI-TOF-TOF MS/MS analysis.
One μl of extracted P13374 peptide was spotted on a MALDI stainless steel target plate and mixed with 1 μl of matrix solution (5 mg/ml α-cyano-4-hydroxycinnamic acid in 0.3% TFA–60% ACN). Zip-Tip-purified samples were eluted with 5 μl 50% ACN in 0.1% TFA, and 1 μl was spotted onto a 800-μm AnchorChip Target plate and dried. Subsequently, 1 μl of matrix solution was added (0.7 mg/ml α-cyano-4-hydroxycinnamic acid in 0.1% TFA–85% ACN–1 mM NH4H2PO4). For external calibration, Peptide Calibration Standard II was used (Bruker Daltonics). MALDI-TOF-TOF MS/MS analysis was performed on an Ultraflex II TOF/TOF instrument (Bruker Daltonics) operated in the reflector mode for MALDI-TOF peptide mass fingerprint (PMF) or LIFT mode for MALDI-TOF-TOF analysis. PMF and LIFT spectra were interpreted with the Mascot software (Matrix Science Ltd., London, United Kingdom). Database (NCBInr) searches using Mascot were performed via BioTools 3.0 software (Bruker Daltonics) using combined PMF and MS/MS (peptide fragment fingerprinting [PFF]) data sets. The peptide sequences obtained were localized in the annotated P13374 genome sequence using WU-blast 2.0 (http://www.proweb.org/).
Isolation, sequencing, and bioinformatic analysis of the CB13374 Stx prophage.
The Stx prophage sequence of P13374 was deduced from one contig of the whole-genome sequencing (WGS) project (unpublished data) of E. coli O104:H4 strain CB13374. Genomic DNA of the bacteria was extracted and purified by the use of the RTP Bacteria DNA minikit (Stratec, Berlin, Germany). Determination of the CB13374 genomic sequence was performed with the Roche 454 genome sequencer FLX system. Libraries for 454 sequencing were prepared and sequenced according to Roche's 454 protocols and as previously described (44). Assembling of the reads resulted in several contigs with an average sequence coverage of more than 30-fold per consensus base. The genome sequence of the prophage was confirmed by partial nucleotide sequencing of DNA from isolated phage particles. From these data, the final P13374 genome sequence of 60,882 bp was identified and used for further analyses. Protein-encoding open reading frames (ORFs) were predicted using the algorithms of Accelrys Gene version 2.5 (Accelrys Inc.) and ORF-Finder (http://www.ncbi.nlm.nih.gov/) with manual curation. For the ORF analysis, the criteria were the presence of ATG, GTG, or TTG as a potential start codon and a length of at least 25 encoded amino acids. To compare the gene products of the P13374 ORFs to the annotated proteins of the nonredundant GenBank database, BLASTp searches (http://www.ncbi.nlm.nih.gov/blast/) were performed at the NCBI homepage. For gene products with low e values (>0.001) or no sequence similarity, the PSI-BLAST algorithm was used (1). Identification of putative tRNA genes was performed using tRNAscan-SE (30). Putative Rho-independent transcription terminators were identified using TRANSTERM (9, 15).
Analysis of integration sites of Stx2 phages.
STEC O104:H4 strains and C600 Stx2 phage lysogens were investigated for possible prophage insertion sites with primers developed previously (11) (Table 2). PCR amplification products obtained with STEC O104:H4 strains and E. coli K-12 C600 phage lysogens were investigated by nucleotide sequencing.
Table 2.
PCR primers for detection of the wrbA gene, the Stx2 phage integration site into wrbA, and circular phage DNA
| Gene and/or integration site | GenBank accession no. | Position in sequence | Primer | Nucleotide sequence (5′–3′) | Melting temp (°C) | PCR product (bp) |
|---|---|---|---|---|---|---|
| wrbA; integrase prophage P13374 (ORF01) | AE005174 | 1,330,538–1,330,556 | ML1 | GTGGAACTAAAGACGCTCG | 55.0 | 500 |
| 1,331,037–1,331,016 | ML2 | ATTTATTGCATCACAGATGGGG | ||||
| wrbA; prophage P13374 (ORF79) | HE664024 | 60,619–60,638 | orf79 | ACGGTAAATACCCTACCCAG | 50.8 | 314 |
| AP009048 | 1,068,130–1,068,108 | wrbA-R | ATGGCTAAAGTTCTGGTGCTTTA | |||
| wrbA | AP009048 | 1,067,534–1,067,550 | wrbA-F | TTAGCCGTTAAGTTTAACTGCC | 57 | 596 |
| 1,068,130–1,068,108 | wrbA-R | ATGGCTAAAGTTCTGGTGCTTTA | ||||
| Prophage P13374 (ORF79); integrase prophage P13374 (ORF01) | HE664024 | 60,619–60,638 | orf79 | ACGGTAAATACCCTACCCAG | 50.8 | 474 |
| 190–210 | ML2 | ATTTATTGCATCACAGATGGGG |
Accession numbers.
The complete nucleotide sequence of phage P13374 was submitted to EMBL under accession number HE664024. The sequences of the left and right integration sites of the investigated Stx2a phages in wild-type EAEHEC and in E. coli K-12 strains were submitted (left site, accession numbers HE775132 to HE775141; right site, accession numbers HE775266 to HE775275). Additionally, the phage attachment sites in the viral genomes were deposited under accession numbers HE775127 to HE775131 (see Table S2 in the supplemental material).
RESULTS
Isolation of Stx2-encoding bacteriophages from STEC O104:H4 strains.
Six STEC O104:H4 strains from Germany (isolated in 2001 and 2011), Norway (2006), and the Republic of Georgia (2009) were used as sources for isolation of temperate Stx2-encoding bacteriophages (Table 1). All strains were found to be positive by PCR for stx2, agr, and wzxO104 genes, but the presence of astA, aggA, and agg3A genes varied by strain (Table 1).
Induction of temperate bacteriophages using mitomycin C was successful for all O104:H4 strains. Stx2a-encoding bacteriophages were isolated from the German (CB8983, CB13344, and CB13374) and the Georgian (CB13771 and CB13772) STEC O104:H4 strains. The Norwegian strain CB13437 released only bacteriophages which did not carry stx2 genes (see below). All phages formed pinhead-sized plaques on the lawn of strain C600. The presence of the stx2a gene in the purified bacteriophage DNA was confirmed by PCR as described previously (53). C600 lysogenic for Stx2 phages was isolated as described in Materials and Methods. The bacteriophage released from CB13437 did not form lysogens on C600.
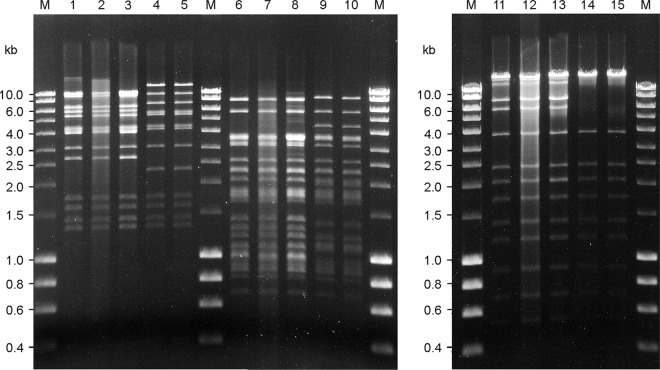
To study the relatedness of the phages, purified phage DNAs were subjected to restriction enzyme digestion with PvuI, EcoNI, and BsrBI. The Stx2a phages isolated from strains CB8983 (Germany 2001) as well as CB13344 and CB13374 (both Germany 2011) had identical restriction profiles for all enzymes tested (Fig. 1). Similar findings were obtained with Stx2a phages from other STEC O104:H4 strains collected from human patients in the 2011 outbreak in Germany (data not shown). The restriction profiles of the Stx2a phages isolated from the Georgian strains CB13771 and CB13772 were highly similar to each other but showed differences in their restriction patterns with all three enzymes tested compared to the Stx2a phages isolated from the German 2001 and 2011 isolates (Fig. 1). The Norwegian O104:H4 strain CB13437 did not release Stx phages, although the strain was positive for Stx2a phage-associated sequences at the attachment site in the wrbA locus.
Fig 1.
Restriction endonuclease digestion of Stx bacteriophage DNA. Restriction nuclease-digested bacteriophage DNA separated on 0.7% agarose digested with EcoNI (lanes 1 to 5), BsrBI (lanes 6 to 10), and PvuI (lanes 11 to 15). Lanes 1, 6, and 11, phage P8983; 2, 7, and 12, phage P13344; 3, 8, and 13, phage P13374; 4, 9, and 14, phage P13771; and 5, 10, and 15, phage P13772. M, molecular size standard (range, 0.2 to 10 kb).
Lysogenization of E. coli with Stx2 phages isolated from STEC O104:H4.
Stx2a-encoding bacteriophages derived from the German and Georgian O104:H4 strains were found to lysogenize C600, resulting in derivative strains TPE2364 (derived from phage P13374, which is released by strain CB13374), TPE2369 (phage P8983), TPE2381 (phage P13771), TPE2383 (phage P13772), and TPE2385 (phage P13344) (Table 3). The C600 lysogens were positive for the stx2a gene and converted to Stx2 production, as tested by Stx EIA (Fig. 2). Induction of lysogenic strains with mitomycin C resulted in release of Stx2a phages which were morphologically similar to those of their wild-type host strains. The integration site of all Stx2a phages in C600 was identical to those found in the STEC wild-type strains (see below).
Fig 2.
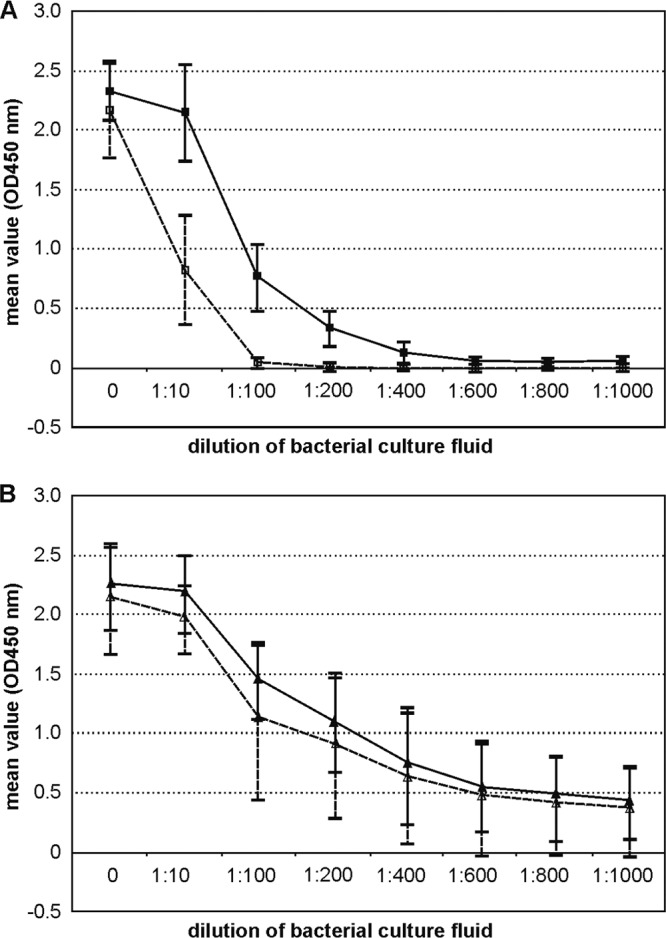
Stx production of STEC O104:H4 wild-type strains and C600 phage lysogens. Extinction values (OD450) obtained by measuring serial dilutions of the six STEC O104:H4 wild-type strains (Table 1) and five E. coli K-12 lysogens (Table 3) by the Stx EIA. (A) ■, STEC wild-type strains listed in Table 1 grown in the presence of mitomycin C; □, STEC wild-type strains grown without mitomycin C induction. (B) ▲, C600 Stx phage lysogens grown in the presence of mitomycin C; △, C600 Stx phage lysogens grown without mitomycin C.
Quantification of Shiga toxin production in cultures of STEC O104:H4 and C600 lysogens with and without mitomycin C treatment.
Stx production was measured with bacteria grown in TSB supplemented with mitomycin C or left unsupplemented. All six STEC O104:H4 strains listed in Table 1 produced significantly more (P = 0.033) Stx in the presence of mitomycin C than untreated cultures. For Stx production there were no significant differences found (P = 0.074) between the tested STEC O104:H4 strains, and the results obtained for the six strains are summarized in Fig. 2A. In the presence of mitomycin C, Stx was still detectable in culture fluid diluted to between 1:400 and 1:600. In cultures without mitomycin C treatment, Stx was not detectable in dilutions of 1:100 and higher.
C600 lysogens harboring Stx2 phages from STEC O104:H4 wild-type strains (TPE2364, TPE2369, TPE2381, TPE2383, and TPE2385; Table 3) produced significantly larger amounts of Stx than the wild-type STEC O104:H4 strains when grown in the presence (P = 0.023) or absence (P < 0.001) of mitomycin C (summarized in Fig. 2B).
In contrast to the STEC O104:H4 wild-type strains, there were no significant differences for Stx production between the C600 lysogens for growth with or without mitomycin C (P = 0.397). To determine if this was due to an increasing spontaneous induction of Stx phages in the P13374 K-12 lysogen, we explored the differences in the spontaneous phage release between TPE2364 (Table 3) and CB13374 during a 24-h bacterial growth period. This was investigated in three independent experiments and different time intervals (6, 12, and 24 h) by titration of cell-free supernatants of both strains on E. coli C600. Compared to the STEC strain CB13374, a higher level (one to two orders of magnitude) of released phages in cell-free supernatants of TPE2364 was observed (data not shown). These findings could explain the relatively higher Stx2 levels found in the C600 lysogens.
The amounts of Stx2a produced by STEC wild-type strains were found to be similar to that produced by an Stx2a EHEC O157:[H7] strain (CB6161; stx2a) in cultures with (P = 0.605) and without (P = 0.81) mitomycin C (data not shown).
Morphology of phage particles.
Negative staining shows that all investigated strains except CB13437 (Table 1) release phages with a short tail and a hexagonal head (indicative of the virus family Podoviridae). Phage tails were 12.7 nm (±1.4 nm; n = 20) in width, and their lengths were up to 40 nm (approximately 26 nm on average). The diameters of the phage heads were 68.5 nm (±2.3 nm) as measured between the parallel sides and 70.7 nm (±1.4 nm) as measured diagonal to the tail. The phages from all strains (CB8983, CB13344, CB13374, CB13771, and CB13772) as well as from their C600 lysogens (Table 3) were morphologically very similar to each other (Fig. 3), resembling the Stx2a phage 933W of EHEC O157:H7 strain EDL933, which is also a podovirus (34, 39, 41, 56). In ultrathin sections of C600 infected with P13374, several stages of phage development were observed (phage attachment, propagation, and liberation by bacteriolysis) (Fig. 4A and B). Upon mitomycin C induction, the Norwegian strain CB13437 yielded a morphologically different Stx-negative phage of the virus family Myoviridae (Fig. 5).
Fig 3.
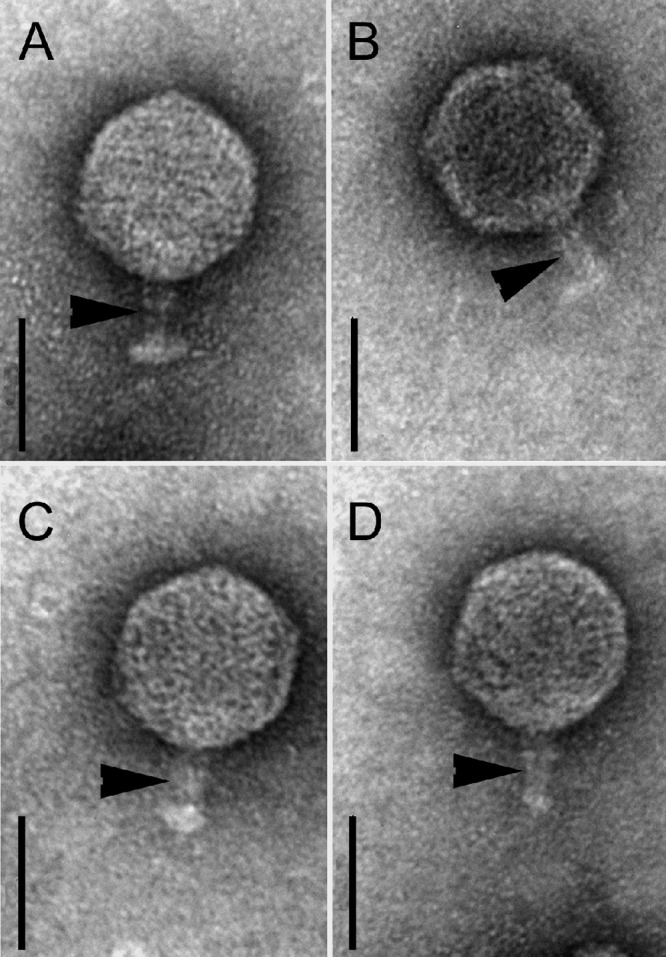
TEM of Stx phages from STEC O104:H4 strains. Shown are negatively stained phages with a short tail (arrowheads) and hexagonal head isolated from E. coli K-12 (C600) after infection with phage lysates of strains CB8983 (A), CB13344 (B), CB13771 (C), and CB13374 (D). Bars, 50 nm.
Fig 4.
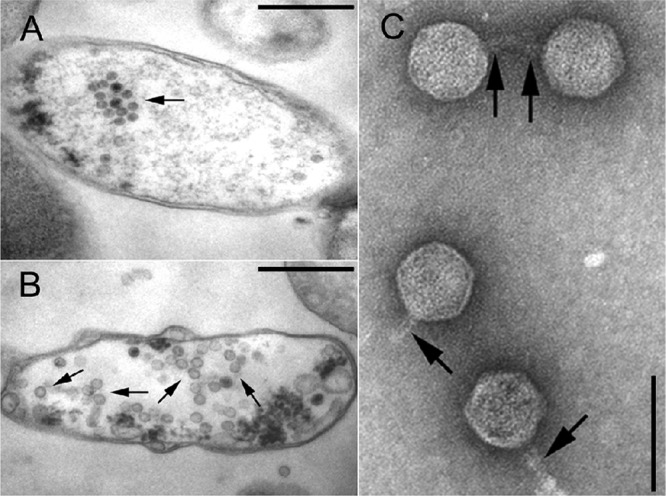
TEM of P13374 induction from lysogenic strain TPE2364. Ultrathin sections of two bacterial cells (TPE2364) with maturating virion particles within the cytoplasm indicated by arrows (bars, 500 nm). The cells show signs of bacteriolysis (loss of cytoplasm) (A) and damaged cell walls, as well as the formation of vesicles or opaque clumps (B). (C) TEM of CsCl-purified, negatively stained P13374 particles released by strain TPE2364 (bar, 100 nm). Four phages with a short tail (arrows) and a hexagonal head are shown. In the upper part, two phages with tail-to-tail interactions are shown.
Fig 5.
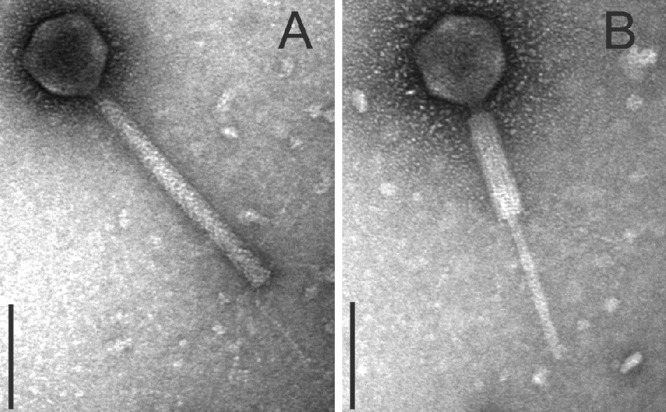
TEM of myovirus phages isolated from the Norwegian STEC O104:H4 strain CB13437. Negatively stained phages comprise a hexagonal head, contractile tail, and kinked tail fibers (Myoviridae). The phages were propagated on E. coli K-12 strain C600 using phage lysates of the Norwegian strain CB13437. (A) Myovirus with noncontracted tail. (B) Myovirus with contracted tail (bars, 100 nm).
Complete nucleotide sequencing and annotation of Stx2 prophage P13374.
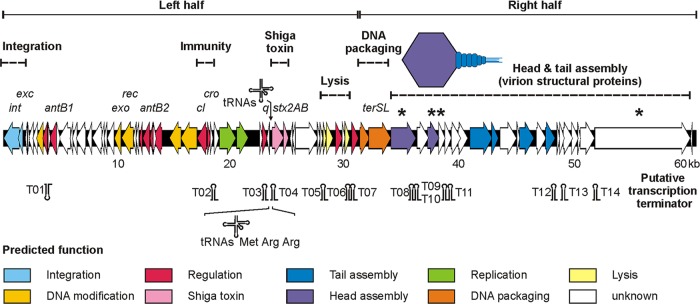
The complete nucleotide sequence of the Stx2 prophage P13374 was deduced from the whole-genome sequence of the German E. coli O104:H4 outbreak strain CB13374 and confirmed by partial sequencing of relevant regions of the phage DNA of virion particles. Based on this analysis, a genomic sequence of 60,894 bp with a mean G+C content of 50.3% for the temperate phage P13374 was inferred. The annotated prophage genome also includes the left and right core sequences of the attachment sites (13 nucleotides). From detailed bioinformatic analyses (for details, see Materials and Methods), 79 open reading frames (ORFs) with good coding potential were predicted. Furthermore, the investigated prophage genome comprises 14 putative Rho-independent transcription terminators and three transfer RNAs (tRNAs) (see Table S3 in the supplemental material). The modular organization of the prophage and the specific features of the bioinformatic analyses are summarized in Fig. 6. Further detailed information is given in Table S3 in the supplemental material. Out of 79 gene products that have been predicted for phage P13374, 62 gene products revealed values of identity to annotated proteins of previously characterized Stx-encoding phages ranging from 38 to 100% (see Table S3). According to the predicted function of the suggested gene products on P13374, the genome can be divided into two parts (Fig. 6), left and right halves. Moreover, the whole-genome organization of P13374 is strongly reminiscent of the enterobacterial phage sequences TL-2011c (JQ011318.1) and VT2phi_272 (HQ424691.1), which originate from E. coli pathotypes O103:H25 and O157:H7, respectively (28). The annotation of the P13374 genome was arranged according to the organization of the prophage sequence starting with the putative genes int (ORF01) and exc (ORF02), coding for a phage integrase and excisionase, respectively. Both genes are closely related to their functional counterparts on a cryptic prophage sequence of the E. coli O26:H11 strain 11368 (NC_013361.1) and previously described Stx-encoding phages (933W, VT2-Sakai, Stx1-converting phage, and Stx2-converting phages I and II). Further, genes on the left half of the genome code for proteins involved in regulatory circuits, antitermination, replication, nucleotide metabolism, Shiga toxin production, and host cell lysis. The core sequences of the genetic switch of P13374 show an organization similar to that of lambda-like phages but consist of only the two antagonistic repressor genes cI and cro, which code for the prophage repressor CI and the lytic repressor Cro. Whereas the two repressor proteins could be identified due to their close relationships to lambda-like phage repressors, it was not possible to find operator sites in the intergenic region between these repressor genes. Furthermore, genes encoding proteins like CII and CIII, which are involved in the regulation of the genetic switch of lambda-like phages, could not be identified on the genome of P13374. This finding suggests that P13374 carries a regulation mechanism for the lysogenic/lytic cycle which is only distantly related to lambda-like phages. Further analyses have to be performed to determine the specificity of the repressors (Cro and CI) to its operator sequences. Downstream of the repressor genes cI and cro, functional counterparts of genes for a bacteriophage-related helicase and primase were identified. Both gene products might be of key importance for the replication of the phage genome. While helicases act on DNA to separate the double-stranded DNA into single strands, primases catalyze the synthesis of short RNA or single-strand DNA fragments to initiate complementary strand synthesis. Downstream of the genes encoding Shiga toxin subunits A and B, the functional module for cell wall degradation and the lysis of the host cell was identified. This module consists of the two essential genes which are typically transcribed from adjacent ORFs and serve as functional counterparts to holins (Gp50) and endolysins (Gp51).
Fig 6.
Genome organization of prophage P13374. Putative genes are colored according to the predicted functions of their products. The positions of putative Rho-independent transcription terminators and transfer RNAs (tRNAs) are indicated. Further detailed information on the structural components of P13374 are given in Table S3 in the supplemental material. P13374 virion proteins which were identified by SDS-PAGE and mass spectrometry are marked by asterisks.
The right half of the P13374 genome begins with the small and large subunits of the terminase genes (terS and terL, respectively) (Fig. 6). Both putative proteins are closely related to their functional counterparts on phage genomes 933W, VT2-Sakai, and other Shiga toxin-converting phages (86, Stx1, Stx2-I, and Stx2-II). Further, genes of the right half of the P13374 genome might represent a cluster of structural proteins involved in virion morphology and synthesis (Fig. 6). Almost all annotated P13374 gene products encoding proteins for phage morphology have functional counterparts to proteins of the virion structure module of Stx-converting phages (Fig. 6). An unusual feature of the tail gene cluster (gp65, gp66, and gp69) is the 2,043-bp open reading frame ORF65. The putative gene product of this ORF has functional counterparts in characterized phages which are of key importance for the adsorption specificity of a phage to its host cell. From the bioinformatic analyses it can be suggested that the specificity of phage P13374 follows the presence of short (gp65) and distal (gp66 and gp69) tail fibers. Self-comparison analyses of the phage tail fiber protein (gp65) shows a large insertion sequence which consists of several imperfect direct repeats (data not shown). Some putative gene products of P13374 were confirmed by SDS-PAGE and mass-spectrometric analyses as the main structural proteins of the virion particle (see below).
Genetic similarity of phage P13374 to other E. coli phages.
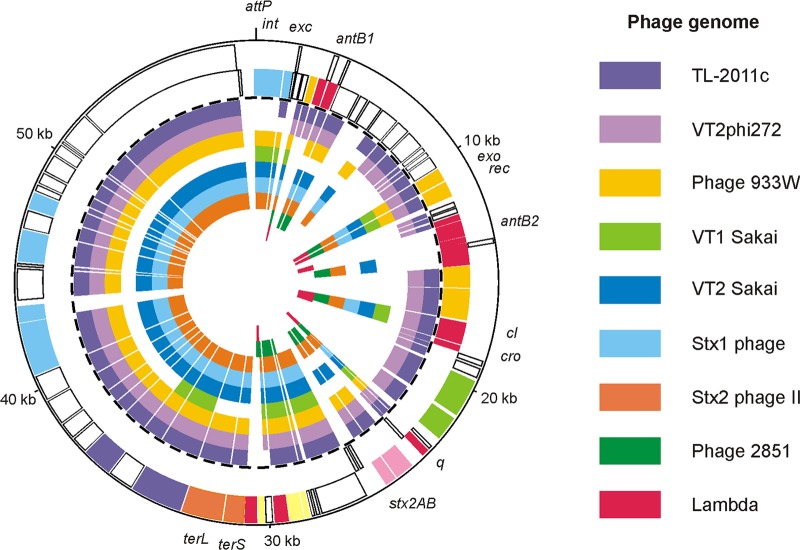
Overall, the gene content and structural organization of several functional modules are similar to most of the previously described Stx-converting phages. As summarized in Fig. 7, gene products of TL-2011c and VT2phi272 show the highest degree of similarity to the predicted P13374 proteins. Fifty-eight and 57 putative gene products of phage P13374 (79 gene products in total) are related (51 to 100%) to TL-2011c and VT2phi272, respectively. The highest degree of similarity to previously characterized Stx-converting phages was to 933W (AF125520), Min27 (NC_010237), VT2-Sakai (AP000422), and Stx1- and Stx2-converting bacteriophages (AP005153 and AP0044202). The enterobacterial phages HK97 (NC_002167) and lambda (J02459) and the Stx2c phage 2851 (FM180578) share only lower numbers of related proteins with P13374 (see Table S3 in the supplemental material).
Fig 7.
Relationship of P13374 to Stx phages and phage lambda. The circular synthetic plot shows the P13374 genome organization and the similarity of predicted gene products to proteins of related Stx phages. The two outermost circles represent the plus and minus strands of the P13374 genome, respectively (coloring according to Fig. 6). The inner circles show phage TL-2011c (accession no. JQ011318), phage VT2phi272 (HQ424691), phage 933W (AF125520), phage VT1-Sakai (AP000400), phage VT2-Sakai (AP000422), Stx1-converting phage (AP005153), Stx2-converting phage (AP0044202), phage 2851 (FM180578), and phage lambda (J02459).
Except for three singular stretches (genome regions 5,250 to 6,750, 29,100 to 31,300, and 40,900 to 43,800) of the P13374 genome that exhibit no significant homology to known Stx-encoding phages, the remaining P13374 DNA is 99 to 100% identical to E. coli phage TL-2011c (Fig. 7). On average, P13374 and TL-2011c DNAs are 90% identical. Interestingly, the one major difference between the two phage genomes concerns the putative outer membrane lipoprotein (Bor protein precursor) homolog, the coding sequence of which is present on the TL-2011c genome and which is widespread among a number of other lambdoid coliphages. A functional counterpart of the TL-2011c Bor-like protein which might promote bacterial resistance to serum complement killing could not be identified on P13374. Further differences were identified in the tail gene clusters of P13374 and TL-2011c. In contrast to P13374, which contains the information for a long tail fiber adhesin (gp66) downstream of the tail fiber protein encoding ORF65, a related gene product on TL-2011c could not be identified. Therefore, it can be speculated that TL-2011c and P13374 form different types of tail fibers. Further differences between the two phages are restricted to sequences for which no function can be predicted.
Analysis of P13374 proteins.
To verify the bioinformatic analysis of the phage genome, major protein bands from phage preparations were excised from one-dimensional SDS-PAGE gels and subjected to tryptic in-gel digestion followed by tandem MALDI-TOF-TOF MS/MS identification. In this way, four abundant proteins could be assigned to putative coding regions (ORF58, ORF60, ORF61, and ORF78) of the genome sequence of phage P13374 (Fig. 6 and Table 4). These ORFs most likely represent structural phage proteins.
Table 4.
Peptide identification by PMF and PFF using MALDI-TOF-TOF MS/MS and MASCOT database searching on tryptic digests of P13374 proteins
| P13374 ORF | Nucleotide position | Molecular mass (kDa) |
No. of peptides found (method used) | GenBank accession no. (GI) | Function, name | |
|---|---|---|---|---|---|---|
| Calculated | Estimateda | |||||
| 58 | 34,002–36,146 | 81.030 | 70–90 | 11 (PMF) | EGR59966 (340730675) | Putative portal protein (Escherichia coli O104:H4 strain 01-09591) |
| 60 | 37,335–38,549 | 44.877 | 46–50 | 28 (PMF); 5 (PFF) | NP_049514 (9632520) | Hypothetical protein 933Wp54 (Enterobacteria phage 933W) |
| 61 | 38,605–38,994 | 13.496 | <14 | 7 (PMF); 2 (PFF) | NP_049515 (9632521) | Hypothetical protein 933Wp55 (Enterobacteria phage 933W) |
| 78 | 51,952–60,372 | 309.315 | >250 | 3 (PFF) | NP_049532 (9632538) | Hypothetical protein 933Wp72 (Enterobacteria phage 933W) |
Data are from SDS-PAGE analysis.
The E. coli wrbA locus is the integration site of Stx2a phages from STEC O104:H4 strains.
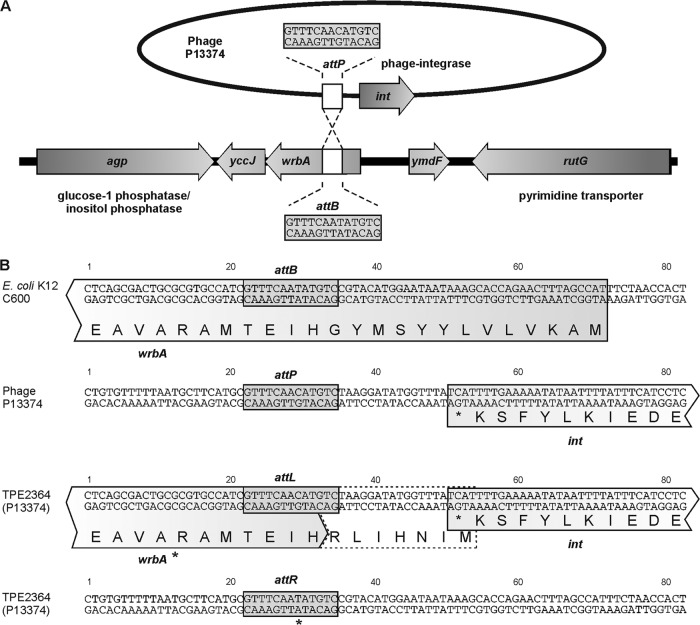
STEC O104:H4 strains and C600 Stx2 phage lysogens were investigated by PCR for different known Stx phage insertion sites as previously described (11) (Fig. 8. A 500-bp product was obtained for all O104:H4 strains and C600 Stx2 phage lysogens (Table 3) with primers ML1 and ML2 (Table 2) spanning the region between the E. coli wrbA gene and the phage integrase gene, thus defining the left integration site of the prophage. Nucleotide sequence analysis of the PCR fragments obtained with ML1 and ML2 primers indicated that the Stx2 prophages are integrated in the 5′ end of the coding region of the E. coli wrbA gene (AP009048 in E. coli K-12 W3110). The core region for attachment in the bacterial genome attB is a 13-bp-long sequence, GTTTCAATATGTC. Due to the insertion of the Stx2 phage, the coding region of the wrbA gene is altered.
Fig 8.
Integration of Stx phage P13374 into E. coli K-12 C600. (A) Integration of phage P13374 into the wrbA gene of E. coli C600. Selected genes and the attachment site of the bacteria (attB) and the phage (attP) are indicated. (B) Sequences flanking the attachment sites in E. coli strain C600, phage P13374, and the left (attL) and right (attR) core sequences in lysogenic C600 strains. wrbA* indicates a putative modified wrbA gene.
Primer orf79 binding to the 3′ region of the P13374 prophage sequence and primer wrbA-R8 (Table 2) were used to identify the right integration site of the prophage in the interrupted wrbA gene. A 256-bp PCR fragment was investigated for its nucleotide sequence. PCR amplification of the phage DNA from P13374 virion particles using primers orf79 and ML2 (int gene) yielded a PCR product of 473 bp and revealed the phage attachment site attP (GTTTCAACATGTC). The attachment sites attB and attP were found to be different in one nucleotide position (underlined nucleotides) (Fig. 8).
From lysates of phages P8983, P13344, P13374, P13771, and P13772, the same PCR product of 473 bp was amplified, whereas no PCR product was obtained with the stx-negative phage from strain CB13437 as the template. Nucleotide sequencing showed that all phage genomes in virion particles are closed between ORF79 (hypothetical protein) and the integrase (ORF01) gene. Thus, the sequence analyses of phage DNAs revealed that the prophage genomes between ORF79 and the integrase gene are joined, which leads to circular permuted genomes of the phage.
Immunity of C600 Stx phage lysogens to superinfection with Stx2a phages from STEC O104:H4 strains.
To investigate the immunity between the Stx2 phages from STEC O104:H4 strains and other Stx-encoding bacteriophages, plaque assays were performed. For these examinations, phage lysates originating from the German and Georgian STEC O104:H4 strains were used on different C600 Stx1- and Stx2-encoding phage lysogens (Table 3). C600 was used as a phage-negative control. The Stx2 phages from the German STEC O104:H4 strains did not form plaques on C600 lysogens carrying the phages P8983, P13344, and P13374. On all other C600 derivatives, including strains TPE2381 and TPE2383, which carry the Stx2a phages from the Georgian STEC O104:H4 strains, lytic phage activity was determined. In contrast, the Stx2 phages from the Georgian O104:H4 strains (P13771 and P13772) did not lyse C600 lysogen carrying the Stx2 phages from the German outbreak strains (TPE2364, TPE2369, and TPE2385) or from the Georgian O104:H4 strains (TPE2381 and TPE2383).
Investigation of EAEC for susceptibility to the Stx2a-encoding bacteriophage P13374.
To obtain information about the host range of the Stx2a phage P13374, phage preparations with approximately 2 × 109 PFU were plated on enteroaggregative E. coli strains. However, none of 31 EAEC strains belonging to 22 different O:H serotypes (see Table S1 in the supplemental material) showed plaque formation or lysis. C600 was used as a phage-sensitive control. Subcultures of bacteria grown in zones spotted with P13374 preparations were negative for the stx2 gene by PCR, indicating that lysogenization with P13374 had not occurred. The 31 STEC strains were further investigated by PCR for the presence of an intact wrbA gene, which is the E. coli host integration site for P13374. All but one of the EAEC strains (CB7373; O111:H10) yielded a 596-bp PCR product with primers wrbA-F and wrbA-R, which amplify the complete wrbA gene encompassing the bacterial attachment site for P13374 integration. These results indicate that despite the presence of the potential phage integration site, phage P13374 is restricted for its host range among E. coli strains belonging to the EAEC group.
DISCUSSION
STEC O104:H4 is a new type of emerging pathogen which has been identified as the causative agent in one of the world's largest outbreaks of food-borne illness in humans, resulting in 855 HUS and 53 fatal cases (7, 19). There are several models for the evolution of the German STEC O104:H4 strains. STEC O104:H4 could have evolved from an enteroaggregative E. coli (EAEC) ancestor by the uptake of Stx2 phages into the chromosome of the bacterium (10, 40). It is also possible that an Stx2 O104:H4 strain was the progenitor of both HUSEC41 (Germany 2001) and the 2011 outbreak strains (LB226692), as described by Mellmann et al. (32). The latter model could explain our findings that the Stx2 phages found in CB8983 (Germany 2001), CB13344, and CB13374 (Germany 2011) were highly similar to each other (this work). The increased human virulence potential of the STEC O104:H4 strain compared to any EAEC O104:H4 progenitor probably results from production of phage-encoded Stx2a combined with the efficient enteroaggregative colonization of the human intestine. These properties in combination are believed to be the cause of maximum delivery of Stx into the human host, resulting in augmented numbers of cases with severe clinical illness (7).
The presence of Stx phages in EAEC strains is unusual, and apart from EAEC O104:H4, only a few other EAEC serotype strains were reported to carry stx2 genes (25, 33). In this work, we were interested to explore the characteristics of the Stx phages present in STEC O104:H4 strains isolated in the 2011 German outbreak and from earlier, epidemiologically unrelated episodes of human infections with STEC O104:H4.
Contaminated, sprouted seeds produced at a horticultural farm in northern Germany were identified as the food source of the STEC O104:H4 2011 outbreak strain (4, 18). We determined the nucleotide sequence of the Stx2 phage present in the STEC O104:H4 strain CB13374, which was isolated from sprouted seeds produced at this farm. As this strain is believed to be the origin of the pandemic spread of STEC O104:H4 in May/June 2011, we compared its Stx2 phage to those present in STEC O104:H4 strains from human patients in Germany (2001 and 2011), Norway (2006), and Georgia (2009). Although all of the STEC O104:H4 strains carried stx2 genes, Stx2 phages could be isolated only from the German and the Georgian O104:H4 strains. All of these phages were identified as podoviruses and were morphologically similar to the Stx2 phage 933W that was carried by EHEC O157:H7 strain EDL933 (34). Similar to phage 933W, the Stx2a phages harbored by the German and the Georgian O104:H4 strains were found to be integrated in the wrbA locus in the wild-type STEC O104:H4 and in E. coli K-12 phage-lysogenic strains (37). Although we found evidence for the presence of a similar Stx2a prophage in the Norwegian STEC O104:H4 strain CB13437, viable Stx phages could not be isolated. Instead, a morphologically different bacteriophage of the Myoviridae family which does not encode Stx was released from CB13437. Interestingly, myoviruses were not found in mitomycin C-induced cultures of the German and the Georgian O104:H4 strains.
Surprisingly, the Stx2 phages isolated from the 2011 German outbreak STEC O104:H4 strains (sprouts and patients) were indistinguishable for their restriction endonuclease profiles from the Stx2 phage harbored by the STEC O104:H4 strain CB8983, which was isolated in 2001 from the feces of a child with HC in Germany. Phage genome sequencing of P8983 revealed >99.9% identity to the genome of P13374 (R. Reinhardt, unpublished results). In contrast, Stx2 phages from STEC O104:H4 strains from two patients in Georgia (2009) showed clearly altered endonuclease restriction profiles compared to the German STEC O104:H4 strains. Differences between the German and the Georgian O104:H4 Stx2 phages were also found in their bacterial host range and superinfection susceptibility. The phage-encoded prophage repressor protein binds efficiently to specific sites in the immunity region on the phage genome. This mechanism is highly specific, as most of the temperate phages only bind to their own DNA and not that from other closely related phages.
All Stx2 phages from STEC O104:H4 strains showed lysis and plaque formation on E. coli K-12 strains harboring phage lambda or different lambdoid Stx1 and Stx2 phages. These observations suggest that the Stx phages from the STEC O104:H4 strains and previously characterized Stx1- and Stx2-encoding phages from other STEC serotypes exhibit different prophage repressor specificities. This is in strong concordance with the findings obtained from the DNA sequence analysis of the primary immunity region of phage P13374. No related lambda-like or other Stx phage-like prophage repressor or Cro repressor binding sites (operators) could be identified in the predicted immunity region (cI to cro) of P13374.
Differences between the German and the Georgian E. coli O104:H4-derived Stx2 phages were also found in their bacterial host range and superinfection susceptibility. The Stx2 phages from the German O104:H4 strains possessed a more extended lysis spectrum than the Stx2 phages from the Georgian O104:H4 strains, pointing to possible differences in their immunity regions. This needs to be confirmed by nucleotide analysis of the Stx2 phages harbored by the Georgian STEC O104:H4 strains.
We therefore investigated EAEC strains of different serotypes for their susceptibility to P13374 as a prototype of the highly virulent Stx2 phage from the German STEC O104:H4 strains, and despite the presence of an undisrupted wrbA gene in most of the EAEC recipient strains, these strains were found to be resistant to a high infective dose of P13374. We do not know the mechanisms which confer resistance to the Stx2 phage, but this observation indicates that the generation of STEC by uptake of Stx phages in nature is a rare event. Nevertheless, under unknown conditions Stx phages are able to infect EAEC strains, thus pointing to an extended host range. Previous findings about the presence of Stx phages and stx genes in Enterobacteriaceae other than E. coli also show this possibility (35, 36, 52).
It is remarkable that despite the genomic differences found between the German STEC O104:H4 strains isolated in 2001 and 2011, both harbor the same Stx2 phage. Strain CB8983 from 2001 differs from the outbreak 2011 strains in the composition of its EAEC plasmid, the presence of a plasmid encoding heat-stable enterotoxins, and the presence of genes conferring antibiotic resistance (7).
The original source of human infections and outbreaks with STEC O104:H4, which has been reported from different countries, has not been identified in any case. As EAEC and STEC strains were found to be associated only with humans and not animals, a human source is supposed as most likely (4). Based on similarities between STEC O104:H4 strains that caused the 2011 outbreak in Germany and a satellite outbreak in France in June of the same year, a batch of fenugreek seeds imported from Egypt which served for sprout production in Germany and France was suspected as the origin of the outbreaks in both countries (18). Analysis of SNPs (single-nucleotide polymorphisms) of STEC O104:H4 strains isolated in Germany and France in 2011 revealed a higher diversity in the French strains than in the German isolates. This is surprising, as the French outbreak was confined to only 15 persons. This finding raises questions about the analysis of the source of the STEC O104:H4 outbreak in Europe in 2011 (22).
The high similarity observed between the Stx2 phages present in STEC O104:H4 strains isolated in Germany in 2001 and 2011 is surprising in light of the Egyptian origin of the suspect seeds, the amount of time between their isolations, the considerable genetic heterogeneity generally seen among Stx phages in nature (23), and the finding that the phages harbored by the Georgian isolates showed distinct differences from the German STEC O104:H4 phages. The sequence of the Norwegian Stx2 prophage in strain CB13437 has yet to be determined.
It has previously been shown that STEC O104:H4 can survive in a viable but nonculturable (VBNC) state for long periods of time (2). During and after the outbreak, it has also been shown that asymptomatic human carriers may serve as a natural reservoir for STEC O104:H4 strains (4, 42). Further research is needed on Stx2 phages present in other STEC O104:H4 strains to determine their potential for long-term survival outside the bacterial host.
As with many STEC and EHEC strains, Stx production in STEC O104:H4 strains could be increased with mitomycin C, resulting in a significant enhancement of Stx2 production and delivery (5, 48). Similar results were reported with ciprofloxacin as an inducer for stx2 transcription, Stx2 production, and release of Stx phages from STEC O104:H4 strains (6). However, a basal level of Stx2 production was still observed in the absence of mitomycin C in STEC O104:H4 wild-type strains, confirming previous findings made for other STEC serotype strains harboring Stx2-encoding bacteriophages (13). In our study, the amount of Stx2 produced by the STEC O104:H4 strains was not significantly different from that of an Stx2a-producing EHEC O157:H7 strain. Similar results were reported when ciprofloxacin was used as an inducer to compare Stx2 production of STEC O104:H4 and O157:H7 strains (6). These findings suggest that the augmented virulence of STEC O104:H4 is not related to enhanced production of Stx2 but rather to the efficient colonization of the STEC strains allowing sustained delivery of Stx2a into the host intestine. Genetic studies of additional Stx prophages from STEC O104:H4 strains will allow us to determine if P13374 is a prototype phage for the conversion of EAEC strains to Shiga toxin production.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Sabine Haby, Antje Konietzny, Karin Pries, Katja Steege, and Maria-Margarida Vargas for technical assistance.
J. A. Hammerl was supported by a grant of the BMBF-funded project SiLeBAT.
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aurass P, Prager R, Flieger A. 2011. EHEC/EAEC O104:H4 strain linked with the 2011 German outbreak of haemolytic uremic syndrome enters into the viable but non-culturable state in response to various stresses and resuscitates upon stress relief. Environ. Microbiol. 13:3139–3148 [DOI] [PubMed] [Google Scholar]
- 3. Baker DR, Moxley RA, Francis DH. 1997. Variation in virulence in the gnotobiotic pig model of O157:H7 Escherichia coli strains of bovine and human origin. Adv. Exp. Med. Biol. 412:53–58 [DOI] [PubMed] [Google Scholar]
- 4. Beutin L, Martin A. 2012. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J. Food Prot. 75:408–418 [DOI] [PubMed] [Google Scholar]
- 5. Beutin L, et al. 2007. Comparative evaluation of the Ridascreen verotoxin enzyme immunoassay for detection of Shiga-toxin producing strains of Escherichia coli (STEC) from food and other sources. J. Appl. Microbiol. 102:630–639 [DOI] [PubMed] [Google Scholar]
- 6. Bielaszewska M, et al. 2012. Effects of antibiotics on Shiga toxin 2 production and bacteriophage induction by epidemic Escherichia coli O104:H4 strain. Antimicrob. Agents Chemother. 56:3277–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bielaszewska M, et al. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11:671–676 [DOI] [PubMed] [Google Scholar]
- 8. Blanco J, et al. 2001. Epidemiology of verocytotoxigenic Escherichia coli (VTEC) in ruminants, p 113–148 In Duffy G, Garvey P, McDowell DA. (ed), Verocytotoxigenic E. coli. Food & Nutrition Press, Inc., Trumbull CT [Google Scholar]
- 9. Brown CM, Dalphin ME, Stockwell PA, Tate WP. 1993. The translational termination signal database. Nucleic Acids Res. 21:3119–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brzuszkiewicz E, et al. 2011. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Arch. Microbiol. 193:883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bugarel M, Beutin L, Scheutz F, Loukiadis E, Fach P. 2011. Identification of genetic markers for differentiation of Shiga toxin-producing, enteropathogenic and avirulent strains of Escherichia coli O26. Appl. Environ. Microbiol. 77:2275–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CDC 2011. Investigation update: outbreak of Shiga toxin-producing E. coli O104 (STEC O104:H4) infections associated with travel to Germany. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ecoli/2011/ecolio104/ [DOI] [PubMed] [Google Scholar]
- 13. de Sablet T, et al. 2008. Differential expression of stx2 variants in Shiga toxin-producing Escherichia coli belonging to seropathotypes A and C. Microbiology 154:176–186 [DOI] [PubMed] [Google Scholar]
- 14. Dowd SE. 2007. Escherichia coli O157:H7 gene expression in the presence of catecholamine norepinephrine. FEMS Microbiol. Lett. 273:214–223 [DOI] [PubMed] [Google Scholar]
- 15. Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27–33 [DOI] [PubMed] [Google Scholar]
- 16. Ethelberg S, et al. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 10:842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. European Food Safety Authority 2011. Shiga toxin-producing E. coli (STEC) O104:H4 2011 outbreaks in Europe: taking stock. EFSA J. 9:2390 [Google Scholar]
- 18. European Food Safety Authority 2011. Tracing seeds, in particular fenugreek (Trigonella foenum-graecum) seeds, in relation to the Shiga-toxin-producing E. coli (STEC) O104:H4 outbreaks in Germany and France. European Food Safety Authority, Parma, Italy [Google Scholar]
- 19. Frank C, et al. 2011. Large and ongoing outbreak of haemolytic uraemic syndrome, Germany, May 2011. Euro Surveill. 16:19878. [PubMed] [Google Scholar]
- 20. Friedrich AW, et al. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 21. Friedrich AW, et al. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41:2448–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grad YH, et al. 2012. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc. Natl. Acad. Sci. U. S. A. 109:3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herold S, Karch H, Schmidt H. 2004. Shiga toxin-encoding bacteriophages-genomes in motion. Int. J. Med. Microbiol. 294:115–121 [DOI] [PubMed] [Google Scholar]
- 24. Huang DB, Mohanty A, Dupont HL, Okhuysen PC, Chiang T. 2006. A review of an emerging enteric pathogen: enteroaggregative Escherichia coli. J. Med. Microbiol. 55:1303–1311 [DOI] [PubMed] [Google Scholar]
- 25. Iyoda S, et al. 2000. Inducible stx2 phages are lysogenized in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patients. FEMS Microbiol. Lett. 191:7–10 [DOI] [PubMed] [Google Scholar]
- 26. Kim J, et al. 2011. Escherichia coli O104:H4 from 2011 European outbreak and strain from South Korea. Emerg. Infect. Dis. 17:1755–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koch C, Hertwig S, Lurz R, Appel B, Beutin L. 2001. Isolation of a lysogenic bacteriophage carrying the stx(1(OX3)) gene, which is closely associated with Shiga toxin-producing Escherichia coli strains from sheep and humans. J. Clin. Microbiol. 39:3992–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. L'Abee-Lund TM, et al. 2012. The highly virulent 2006 Norwegian EHEC O103:H25 outbreak strain is related to the 2011 German O104:H4 outbreak strain. PLoS One 7:e31413 doi:10.1371/journal.pone.0031413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 30. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Makino K, et al. 1999. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 74:227–239 [DOI] [PubMed] [Google Scholar]
- 32. Mellmann A, et al. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751 doi:10.1371/journal.pone.0022751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morabito S, et al. 1998. Enteroaggregative, Shiga toxin-producing Escherichia coli O111:H2 associated with an outbreak of hemolytic-uremic syndrome. J. Clin. Microbiol. 36:840–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Brien AD, Marques LR, Kerry CF, Newland JW, Holmes RK. 1989. Shiga-like toxin converting phage of enterohemorrhagic Escherichia coli strain 933. Microb. Pathog. 6:381–390 [DOI] [PubMed] [Google Scholar]
- 35. Ooka T, et al. 2012. Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 18:488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paton AW, Paton JC. 1996. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic syndrome. J. Clin. Microbiol. 34:463–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perna NT, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 38. Persson S, Olsen KE, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Plunkett G, III, Rose GDJ, Durfee TJ, Blattner FR. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rasko DA, et al. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rietra PJ, et al. 1989. Comparison of vero-cytotoxin-encoding phages from Escherichia coli of human and bovine origin. J. Gen. Microbiol. 135:2307–2318 [DOI] [PubMed] [Google Scholar]
- 42. Robert Koch Institut 2011. Abschließende Darstellung und Bewertung der epidemiologischen Erkenntnisse im EHEC O104:H4 Ausbruch, Deutschland, p 1–43 Robert Koch Institut, Berlin, Germany [Google Scholar]
- 43. Rohde H, et al. 2011. Open-source genomic analysis of Shiga toxin-producing E. coli O104:H4. N. Engl. J. Med. 365:718–724 [DOI] [PubMed] [Google Scholar]
- 44. Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E. 2011. Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100. J. Bacteriol. 193:1745–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sato T, et al. 2003. Distinctiveness of the genomic sequence of Shiga toxin 2-converting phage isolated from Escherichia coli O157:H7 Okayama strain as compared to other Shiga toxin 2-converting phages. Gene 309:35–48 [DOI] [PubMed] [Google Scholar]
- 46. Scheutz F, et al. 2011. Characteristics of the enteroaggregative Shiga toxin/verotoxin-producing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Euro Surveill. 16:19889. [DOI] [PubMed] [Google Scholar]
- 47. Scheutz F, Strockbine NA. 2005. Genus I. Escherichia, p 607–624 In Garrity GM, Brenner DJ, Krieg NR, Staley JT. (ed), Bergey's manual of systematic bacteriology, 2nd ed Springer-Verlag, New York, NY [Google Scholar]
- 48. Shimizu T, Ohta Y, Noda M. 2009. Shiga toxin 2 is specifically released from bacterial cells by two different mechanisms. Infect. Immun. 77:2813–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strauch E, et al. 2008. Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infect. Immun. 76:5466–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Strauch E, Lurz R, Beutin L. 2001. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect. Immun. 69:7588–7595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takkinen J, et al. 2011. Shiga toxin/verotoxin-producing Escherichia coli in humans, food and animals in the EU/EEA, with special reference to the German outbreak strain STEC O104. ECDC & EFSA, Stockholm, Sweden [Google Scholar]
- 52. Tschape H, et al. 1995. Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: green butter as the infection source. Epidemiol. Infect. 114:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tzschoppe M, Martin A, Beutin L. 2012. A rapid procedure for the detection and isolation of enterohaemorrhagic Escherichia coli (EHEC) serogroup O26, O103, O111, O118, O121, O145 and O157 strains and the aggregative EHEC O104:H4 strain from ready-to-eat vegetables. Int. J. Food Microbiol. 152:19–30 [DOI] [PubMed] [Google Scholar]
- 54. Wagner PL, Acheson DW, Waldor MK. 2001. Human neutrophils and their products induce Shiga toxin production by enterohemorrhagic Escherichia coli. Infect. Immun. 69:1934–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wagner PL, et al. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957–970 [DOI] [PubMed] [Google Scholar]
- 56. Willshaw GA, Smith HR, Scotland SM, Field AM, Rowe B. 1987. Heterogeneity of Escherichia coli phages encoding Vero cytotoxins: comparison of cloned sequences determining VT1 and VT2 and development of specific gene probes. J. Gen. Microbiol. 133:1309–1317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.