Abstract
Paramyxoviruses produce pleiomorphic particles containing variable amounts of genetic material that correlate with virion diameter by electron microscopy. However, the infectious nature of these particles is unknown, and functional genomes are indistinguishable from defective RNA. A quantitative approach to paramyxovirus packaging revealed a majority of infectious Newcastle disease viruses contain one functional genome. Virions encapsidating two or three genomes (approximately 25% of total) were also observed by utilizing three different recombinant viruses expressing unique fluorescent reporters.
TEXT
Newcastle disease virus (NDV) belongs to the family Paramyxoviridae of the order Mononegavirales. Other viruses in the family include a variety of human pathogens, such as measles, mumps, human parainfluenza and respiratory syncytial viruses, which are among the most common etiologic agents of pediatric infections. NDV infects domestic and wild avian species, and zoonotic infections cause mild conjunctivitis and influenza-like symptoms in patients (2). Virulent strains may cause epidemic outbreaks resulting in significant economic losses for the global poultry industry (1, 2). Paramyxoviruses encode their genomes on a single strand of negative-sense RNA. Evidence for multiploid paramyxoviruses, including NDV, emerged in the mid-20th Century (4). Notably, the paramyxoviral ribonucleoproteins (vRNPs) are characteristically disordered, in contrast to some other negative-sense RNA viruses, like influenza A virus with its distinctive “7 + 1” RNP arrangement, and more research is needed to refine our understanding of paramyxovirus genomic content (6, 8, 10, 14). Various electron microscopy (EM) studies of paramyxoviruses, including NDV and measles, parainfluenza, and Sendai viruses, have produced images of pleiomorphic particles ranging in size from approximately 100 nm to greater than 500 nm in diameter (5, 10, 11, 17). Cryo-electron tomography captured Sendai virions containing variable quantities of genomic material correlating with particle diameter, and it produced an estimate that particles package between one and six genomic copies (10). While EM studies have characterized particle morphology in detail and offer estimates of genomic contents, these studies do not differentiate between infectious and noninfectious particles, nor can they adequately distinguish functional, full-length genomes from defective interfering RNAs.
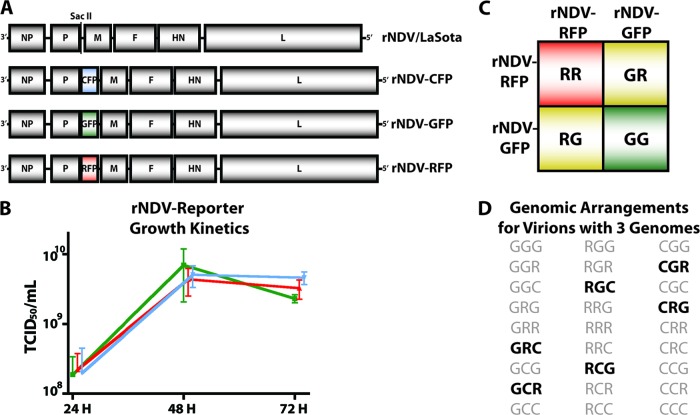
In order to better appreciate the genomic contents of infectious paramyxoviruses, several recombinant NDV (rNDV) strain LaSota reporter viruses expressing fluorescent proteins were utilized. Recombinant NDV strain LaSota cDNA was engineered to carry either cyan fluorescent protein (CFP) (Clontech) or red fluorescent protein (RFP) (courtesy of R. Y. Tsien) and rescued as previously reported for rNDV-GFP, which expresses green fluorescent protein (GFP) (3, 12, 13) (Fig. 1A). Each rNDV strain is replication competent and reaches a similar endpoint titer with similar kinetics when grown in embryonated eggs (Fig. 1B). Considering a simplified model of a diploid virus packaging rNDV-GFP and rNDV-RFP genomes with equal efficiency, 50% of virions carrying two genomes (GR or RG) would be detectable by the presence of both reporters (Fig. 1C). Particles carrying two copies of the GFP or RFP reporter (GG or RR) would be indistinguishable from those packaging a single genome (not depicted in the model). Notably, these reporters differ from genomes used in complementation studies of paramyxovirus packaging in which two genomes are essential to the formation of viable particles (7, 16). Therefore, artificial selective pressure should not favor particles with multiple genomes under the assay conditions of the present study.
Fig 1.
Recombinant NDV viruses and genomic packaging models. (A) rNDV-CFP and rNDV-RFP were generated in the LaSota strain background to study the ability of an NDV virion to carry multiple genomic copies. The fluorescent reporters were cloned into the SacII restriction site between the P and M proteins, and the viruses were rescued as previously described for rNDV-GFP (12, 13). (B) All reporter viruses grow to similar endpoint titers with similar kinetics when 100 PFU of rNDV is inoculated into 9-day-old embryonated chicken eggs. The growth curves are shaded to correspond to the fluorescent reporter carried by each virus. Error bars represent standard deviations. TCID50, 50% tissue culture infective dose. (C) In a simplified model, assuming that NDV packages two genomic copies with an equal preference for the GFP and RFP reporter genomes, 50% of virions carrying two genomes (GR and RG) are detectable by the presence of both GFP and RFP, while 50% (GG and RR) are indistinguishable from virions with one genomic copy (G or R, respectively). (D) In a second model, assuming that NDV packages three genomes per virion with no preference for any reporter, 6 out of 27 possible genomic arrangements in the virions are detectable by the presence of all three reporters (boldface type).
In order to test the model presented in Fig. 1C, DF1 cells (UMNSAH/DF1, an embryonic chicken fibroblast cell line obtained from ATCC [CRL-12203]) were coinfected with rNDV-GFP and rNDV-RFP at a multiplicity of infection (MOI) of 5 each and incubated at 37°C for 24 h in Dulbecco's modified Eagle's medium (DMEM)–10% allantoic fluid, which contains trypsin to facilitate efficient viral budding by cleaving the F protein. Trypsin is essential for maturation of infectious virions for avirulent NDV strains such as LaSota (15). An MOI of 5 for each virus was chosen to maximize the number of cells infected by both reporters without applying so much virus as to kill the DF1 cells before significant viral replication occurs. Control DF1 cells were infected individually with either rNDV-GFP or rNDV-RFP alone at an MOI of 10. All viral supernatants were harvested after 24 h and filtered (0.45-μm polyvinylidene difluoride [PVDF] filter) to remove fluorescent DF1 cells. The supernatants from the individually infected control cells were mixed 1:1 (mixed control group), and all samples were allowed to stand overnight at 4°C in order to control for virions randomly adhering to one another in the growth medium. Supernatants were serially diluted in phosphate-buffered saline (PBS) (containing 1% bovine albumin, 0.835 mM CaCl2, 1.05 mM MgCl2) and subsequently used to infect human embryonic kidney 293T cells (HEK293T; ATCC catalog no. CRL-11268), which were incubated overnight in DMEM–10% fetal calf serum (FCS) (to inhibit viral egress and multicycle replication) at 37°C to allow for reporter expression. HEK293T cells were chosen as they are readily infected by rNDV and express the fluorescent reporters. FCS is included in the HEK293T medium rather than allantoic fluid to prevent F protein cleavage for assurance that multicycle replication does not occur.
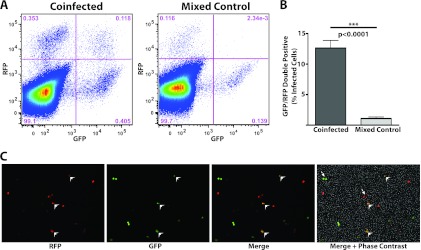
HEK293T cells treated with coinfected viral supernatant were detected expressing either GFP or RFP alone at a near 1:1 ratio, and cells expressing both reporters were observed by flow cytometry and live-cell fluorescence microscopy (Fig. 2A and C). The percentage of infected cells coexpressing GFP and RFP was approximately 13% by flow cytometry in the coinfected samples, significantly above the background signal of approximately 1% detected in the mixed controls (Fig. 2A and B). Using the model presented in Fig. 1C and assuming that each reporter genome is packaged with equal efficiency, as suggested by the approximately 1:1 ratio of GFP- to RFP-expressing cells (Fig. 2A), one might estimate that approximately 23% of virions contain two functional genomes after correcting for background. This number was calculated by subtracting the background from the average of three replicate experiments (12.65% − 1.075% = 11.575%) and multiplying the result by 2 (23.15%) to account for the particles packaging two genomic copies that are not detectable by the assay (schematized in Fig. 1C as GG and RR). Notably, our quantification indicates that a majority of infectious rNDV particles, approximately three-quarters, package one functional genome. In all samples, several dilutions of the DF1 supernatants were made such that an effective MOI of between 0.01 and 0.001 was analyzed for each replicate. This ensures that HEK293T cell expression of colocalized GFP and RFP is likely representative of a single infection event by a virion carrying two genomes rather than coinfection by two virions. The low background levels of colocalized reporter expression suggest that the particle/PFU ratio for NDV is relatively low and that genomic complementation between noninfectious particles occurs infrequently. It is feasible that a viable vRNP may be copackaged with an incomplete reporter genome such that complementation permits the latter's expression. We believe this occurs infrequently as previously published Northern blot data demonstrate that NDV efficiently produces full-length genomes (7).
Fig 2.
rNDV-GFP and rNDV-RFP genomes are packaged into a single virion. (A) Flow cytometry of HEK239T cells infected with virus derived from DF1 cells coinfected with rNDV-GFP and rNDV-RFP (left panel) as well as HEK293T cells infected with a mixture of two supernatants from DF1 cells infected individually with rNDV-GFP or RFP (mixed control). Dot plots are representative of three independent infections of DF1 cells. (B) Quantification of flow cytometry analysis of three independent infections of HEK293T cells with either rNDV derived from coinfected DF1 cell supernatant or the mixed control. GFP-RFP double-positive cells are graphed as the percentage of total infected cells. This was calculated by dividing the percentage of cells expressing both reporters by the sum of the percentages of cells expressing GFP, RFP, or GFP-RFP: e.g., [0.118/(0.118 + 0.353 + 0.405)] × 100% = 13.47%. A t test shows a statistically significant difference between the coinfected samples and the mixed control with P < 0.0001. Error bars indicate the standard deviations. (C) Live-cell microscopy of coinfected HEK293T cells demonstrates a population of infected cells expressing both reporters when infected at an MOI of <0.01. White arrowheads in all four panels indicate cells infected by a single virion that incorporated functional copies of both reporters. Immediately adjacent cells expressing the same reporter likely result from a single infection event followed by cellular division, and examples are marked by white arrows in the far-right panel, which is a composite of previous images with the addition of phase contrast.
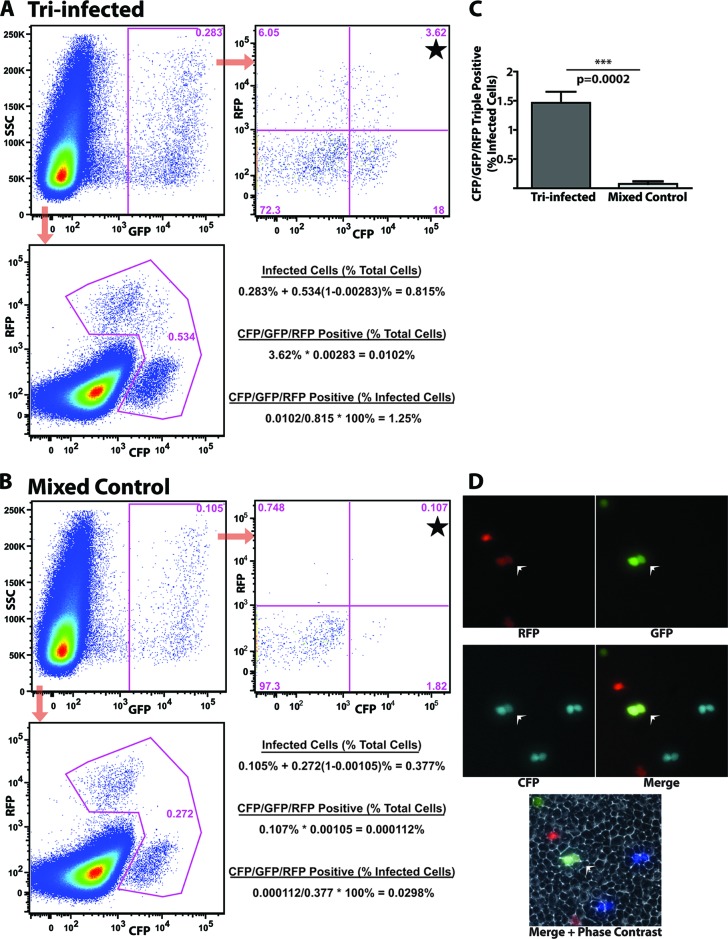
Clearly, the model presented in Fig. 1C is oversimplified and does not account for virions with more than two functional genomes. For example, a virion carrying two GFP reporters and an RFP reporter (GFP-GFP-RFP) is indistinguishable from another packaging one copy of each (GFP-RFP) by this methodology. In order to determine whether infectious virions package three functional genomes, rNDV-CFP was utilized in addition to the other reporter viruses (Fig. 1A and B). The experiments were repeated with DF1 cells infected either by all three rNDV reporter viruses at an MOI of 5 each (tri-infected) or individually at an MOI of 15. Supernatants were harvested and filtered, the individual viral supernatants were mixed 1:1:1 (mixed control), and samples were left to stand overnight at 4°C to control for adhering virions. The supernatants were serially diluted to infect HEK293T cells for reporter expression and analysis by flow cytometry and live-cell fluorescence microscopy (Fig. 3A, B, and D). Detection of HEK293T cells expressing all three fluorophores in the tri-infected samples after infection at a low MOI indicates that a population of NDV particles packages a minimum of three functional genomes. Assuming, again, that each reporter is packaged at random and with equal efficiency, particles encapsidating all three reporters represent 6 out of 27 possible genomic arrangements (Fig. 1D). In the tri-infected samples, 1.46% of infected cells expressed all three reporters compared to a background of 0.08% in the mixed controls in an average of three independent experiments (Fig. 3C). After correcting for background (1.46% − 0.08% = 1.38%) and multiplying by 27/6, an estimated 6.1% of rNDV particles package three genomes. A particle packaging all three reporters (CGR) is indistinguishable by this assay from a particle packaging four genomes with the arrangement CCGR, for example. For reasons unknown, some preference is detected for copackaging of certain pairs of reporter genomes. For example, GFP and CFP were colocalized at 3 times the rate of GFP and RFP in HEK293T cells infected with rNDV derived from tri-infected DF1 cells (Fig. 3A). Therefore, this assay deviates from the idealized models discussed, and exact quantification of virions packaging one, two, three, or more genomes is not straightforward, but a clear trend has certainly emerged.
Fig 3.
Infectious rNDV particles can package at least three functional genomes. (A and B) Flow cytometry of HEK293T cells infected with either supernatant derived from DF1 cells infected at an MOI of 5 with each of the three reporter viruses (tri-infected) (A) or the mixed control (B). The upper left panel in both panels A and B depicts gating for GFP-positive cells, and this population is analyzed for CFP and RFP expression in the upper right panel of both panels A and B. The populations expressing all three reporters are marked with black stars. In the lower left panel of panels A and B, GFP-negative cells are analyzed for CFP and RFP expression, and the percentage of infected cells is the sum of the GFP-positive population plus the RFP- and CFP-positive populations. Dot plots are representative of three independent infections of DF1 cells. In the lower-right-hand panel of panels A and B, a sample calculation is made for the percentage of total cells that are infected, the percentage of total cells that are CFP-GFP-RFP triple positive, and the percentage of infected cells that are triple positive. (C) Quantification of flow cytometry analysis of three independent infections of HEK293T cells with rNDV derived from tri-infected DF1 viral supernatant or the mixed control according to the sample calculations provided in the lower right panels of panels A and B. In the tri-infected group, an average of 1.46% of infected cells were positive for all three reporters compared to a background of 0.08% for the mixed control. This is a statistically significant difference with P = 0.0002 by t test. Error bars indicate standard deviations. (D) Live-cell microscopy of HEK293T cells infected with viral supernatant derived from DF1 cells infected with all three reporter viruses. Two adjacent cells expressing all three reporters are indicated by the white arrowheads in all four panels. Immediately adjacent cells expressing the same reporter likely result from a single infection event followed by cellular division. The lower panel is a composite of the other four images with the addition of phase contrast.
The data presented here suggest that a majority of infectious NDV virions, estimated at nearly three-quarters, contain a single functional genome, with particles packaging two, three, or potentially more functional genomes accounting for smaller minorities of infectious particles. We propose a model in which infectious virions encapsidating a single functional vRNP are the most abundant species and increasingly larger particles containing additional genomic copies exist along a declining population gradient (Fig. 4). Based upon the volume of the largest measles virus particles, it has been postulated that >30 genomes could be packaged in a particle (16). While the data presented here cannot exclude the existence of paramyxoviruses packaging large amounts of genomic material, indications are that infectious NDV particles containing a high copy number of functional genomes are quite rare.
Fig 4.
Model for infectious NDV particle ploidy and size. In this model, NDV particles with a single functional genome are the most prominent species, and increasingly larger particles packaging two, three, or more functional genomes exist along a declining population gradient.
Paramyxoviruses may derive an evolutionary advantage by producing polyploid particles as a mechanism for generating genomic diversity and increasing the likelihood that a given particle delivers an infectious genome to its target cell (10). Increasing the number of infectious genomes packaged beyond two or three may hypothetically yield diminishing returns because larger particles are less stable, as well as being energetically more costly to produce because they require more viral protein and genomic material to form (9, 17). Therefore, paramyxovirus particle size may be governed by opposing selective pressures, whereby the advantage of polyploidy is in equilibrium with the necessity of efficiently generating high infectious viral titers. The methodology presented here may be readily applied to other paramyxoviruses in order to better understand the genomic composition of infectious particles.
ACKNOWLEDGMENTS
We are grateful to R. Y. Tsien for kindly providing the RFP construct used to generate rNDV-RFP.
This work was partially supported by Northeast Biodefense Center grant U54 AI057158-06 (principal investigator, W. I. Lipkin).
Footnotes
Published ahead of print 11 July 2012
REFERENCES
- 1. Ababneh MMK, Dalab AE, Alsaad SR, Al-Zghoul MB, Al-Natour MQ. Molecular characterization of a recent Newcastle disease virus outbreak in Jordan. Res. Vet. Sci., in press [DOI] [PubMed] [Google Scholar]
- 2. Alexander DJ. 2011. Newcastle disease in the European Union 2000 to 2009. Avian Pathol. 40:547–558 [DOI] [PubMed] [Google Scholar]
- 3. Campbell RE, et al. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 99:7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dahlberg JE, Simon EH. 1969. Physical and genetic studies of Newcastle disease virus: evidence for multiploid particles. Virology 38:666–678 [DOI] [PubMed] [Google Scholar]
- 5. DiNapoli JM, et al. 2007. Immunization of primates with a Newcastle disease virus-vectored vaccine via the respiratory tract induces a high titer of serum neutralizing antibodies against highly pathogenic avian influenza virus. J. Virol. 81:11560–11568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fournier E, et al. 2012. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 40:2197–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao Q, Park MS, Palese P. 2008. Expression of transgenes from Newcastle disease virus with a segmented genome. J. Virol. 82:2692–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harris A, et al. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 103:19123–19127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Homann HE, Hofschneider PH, Neubert WJ. 1990. Sendai virus gene expression in lytically and persistently infected cells. Virology 177:131–140 [DOI] [PubMed] [Google Scholar]
- 10. Loney C, Mottet-Osman G, Roux L, Bhella D. 2009. Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of Sendai virus. J. Virol. 83:8191–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lund GA, Tyrrell DL, Bradley RD, Scraba DG. 1984. The molecular length of measles virus RNA and the structural organization of measles nucleocapsids. J. Gen. Virol. 65:1535–1542 [DOI] [PubMed] [Google Scholar]
- 12. Maamary J, et al. 2011. Newcastle disease virus expressing a dendritic cell-targeted HIV Gag protein induces a potent Gag-specific immune response in mice. J. Virol. 85:2235–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakaya T, et al. 2001. Recombinant Newcastle disease virus as a vaccine vector. J. Virol. 75:11868–11873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noda T, et al. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439:490–492 [DOI] [PubMed] [Google Scholar]
- 15. Park MS, Steel J, GarcíA-Sastre A, Swayne D, Palese P. 2006. Engineered viral vaccine constructs with dual specificity: avian influenza and Newcastle disease. Proc. Natl. Acad. Sci. U. S. A. 103:8203–8208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rager M, Vongpunsawad S, Duprex WP, Cattaneo R. 2002. Polyploid measles virus with hexameric genome length. EMBO J. 21:2364–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Terrier O, et al. 2009. Parainfluenza virus type 5 (PIV-5) morphology revealed by cryo-electron microscopy. Virus Res. 142:200–203 [DOI] [PubMed] [Google Scholar]